Abstract
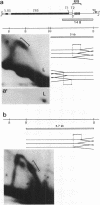
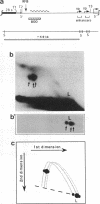
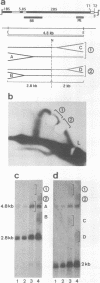
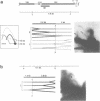
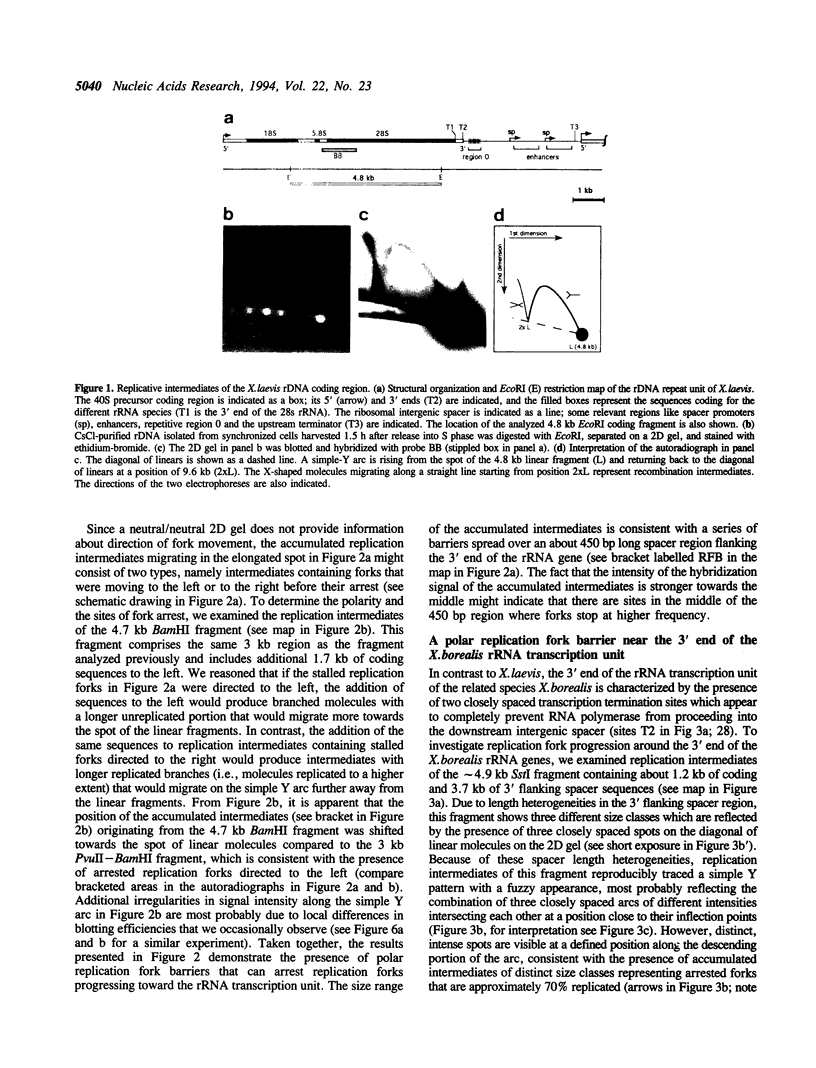
To investigate replication fork progression along the tandemly repeated rRNA genes of Xenopus laevis and Xenopus borealis, rDNA replication intermediates from dividing tissue culture cells were analyzed by two-dimensional gel electrophoresis. Analysis of the direction of replication in the rRNA coding regions revealed replication forks moving in both directions. However, in both frog species, polar replication fork barriers (RFB) arresting forks approaching the rRNA transcription units from downstream were identified. Whereas in X. borealis the RFB maps to a defined site close to the transcription terminator, in X. laevis the arrest of fork movement can occur at multiple positions throughout a 3' flanking repetitive spacer region. A short DNA element located near the respective RFB sites is shared between these two related frog species, suggesting its possible involvement in the arrest of replication fork movement. In a subset of rDNA repeats, these barriers cause an absolute block to replication fork progression, defining the sites where replicon fusion occurs, whereas in the remainder repeats, most probably in the non-transcribed gene copies, the replication machinery can pass the RFB sequences and replicate the rRNA transcription unit in a 3'-to-5' direction.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brewer B. J., Fangman W. L. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988 Nov 18;55(4):637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Lockshon D., Fangman W. L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992 Oct 16;71(2):267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Huberman J. A. DNA replication origins in animal cells: a question of context? Science. 1994 Feb 4;263(5147):639–640. doi: 10.1126/science.8303270. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Brehm S. L. Replication of the extrachromosomal ribosomal RNA genes of Tetrahymena thermophilia. Nucleic Acids Res. 1981 Jul 24;9(14):3531–3543. doi: 10.1093/nar/9.14.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Sogo J. M., Ryan C. A. Ribosomal gene clusters are uniquely proportioned between open and closed chromatin structures in both tomato leaf cells and exponentially growing suspension cultures. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5256–5260. doi: 10.1073/pnas.89.12.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Widmer R. M., Koller T., Sogo J. M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989 Jun 2;57(5):753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J. M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993 May 25;21(10):2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. The ribosomal spacer in Xenopus laevis is transcribed as part of the primary ribosomal RNA. Nucleic Acids Res. 1986 Aug 11;14(15):6041–6051. doi: 10.1093/nar/14.15.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L. Origins of DNA replication in metazoan chromosomes. J Biol Chem. 1993 Jan 5;268(1):1–4. [PubMed] [Google Scholar]
- Dhar V., Schildkraut C. L. Role of EBNA-1 in arresting replication forks at the Epstein-Barr virus oriP family of tandem repeats. Mol Cell Biol. 1991 Dec;11(12):6268–6278. doi: 10.1128/mcb.11.12.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Vaughn J. P., Hamlin J. L. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991 Aug;11(8):3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Brewer B. J. Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol. 1991;7:375–402. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992 Nov 20;258(5086):1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- Gahn T. A., Schildkraut C. L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989 Aug 11;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L. Mammalian origins of replication. Bioessays. 1992 Oct;14(10):651–659. doi: 10.1002/bies.950141002. [DOI] [PubMed] [Google Scholar]
- Hernández P., Martín-Parras L., Martínez-Robles M. L., Schvartzman J. B. Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J. 1993 Apr;12(4):1475–1485. doi: 10.1002/j.1460-2075.1993.tb05791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Méchali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993 Dec;12(12):4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Méchali M. Plasmid replication in Xenopus eggs and egg extracts: a 2D gel electrophoretic analysis. Nucleic Acids Res. 1992 Apr 11;20(7):1463–1469. doi: 10.1093/nar/20.7.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Hidaka M., Nishizawa M., Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992 Jun;233(3):355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Functional difference between the sites of ribosomal 40S precursor 3' end formation in Xenopus laevis and Xenopus borealis. Nucleic Acids Res. 1990 Sep 11;18(17):5271–5277. doi: 10.1093/nar/18.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang W. H., Reeder R. H. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R. D., Platt T. H., Schildkraut C. L. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993 Oct;13(10):6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Lucchini R., Sogo J. M. Chromatin structure and transcriptional activity around the replication forks arrested at the 3' end of the yeast rRNA genes. Mol Cell Biol. 1994 Jan;14(1):318–326. doi: 10.1128/mcb.14.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini R., Sogo J. M. Different chromatin structures along the spacers flanking active and inactive Xenopus rRNA genes. Mol Cell Biol. 1992 Oct;12(10):4288–4296. doi: 10.1128/mcb.12.10.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani H. M., Paull T., Elder J. K., Blow J. J. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 1992 Apr 11;20(7):1457–1462. doi: 10.1093/nar/20.7.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Parras L., Hernández P., Martínez-Robles M. L., Schvartzman J. B. Unidirectional replication as visualized by two-dimensional agarose gel electrophoresis. J Mol Biol. 1991 Aug 20;220(4):843–853. doi: 10.1016/0022-2836(91)90357-c. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Bustin M., Miller O. L., Jr Electron microscopic analysis of chromosome metabolism in the Drosophila melanogaster embryo. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):741–754. doi: 10.1101/sqb.1978.042.01.075. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr, Beatty B. R. Visualization of nucleolar genes. Science. 1969 May 23;164(3882):955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- Moss T., Boseley P. G., Birnstiel M. L. More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res. 1980 Feb 11;8(3):467–485. doi: 10.1093/nar/8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H. Regulatory elements of the generic ribosomal gene. Curr Opin Cell Biol. 1989 Jun;1(3):466–474. doi: 10.1016/0955-0674(89)90007-0. [DOI] [PubMed] [Google Scholar]
- Saffer L. D., Miller O. L., Jr Electron microscopic study of Saccharomyces cerevisiae rDNA chromatin replication. Mol Cell Biol. 1986 Apr;6(4):1148–1157. doi: 10.1128/mcb.6.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Vogt V. M., Braun R. The replication of ribosomal DNA in Physarum polycephalum. Eur J Biochem. 1977 Nov 1;80(2):557–566. doi: 10.1111/j.1432-1033.1977.tb11912.x. [DOI] [PubMed] [Google Scholar]