The study of human microbiota emerged as one of the most active fields of microbiology over the past 10 years [1]. This research has been fueled by the extensive use of culture, an approach that we named “culturomics” [2]. As an example, starting culturomics in 2009 [2] Lagier et al. isolated 337 gut bacterial species, including 31 new species. They were published, but the names of only 17 of them have officially been validated to date. This rate of discovery will accelerate with the rebirth of culture [3] as it has been estimated that only 30% of this microbiota has been characterized so far so that many new species are expected to be described in the coming years.
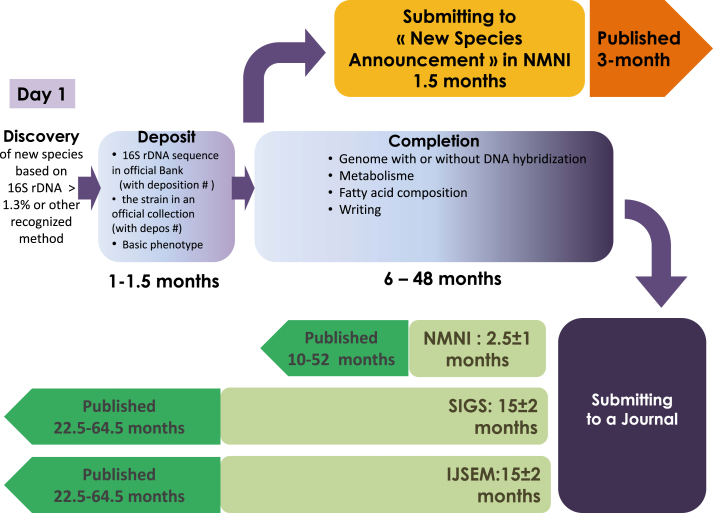
We now are facing the problem of rapidly communicating the scientific community this tremendous quantity of new species. The current process of description and validation of new species can take from 2 to 5 years before scientists are aware that a new species has been discovered! And that the type strain has been made available for further researches (Fig. 1). This situation is not scientifically fair as most of the knowledge about microbes and their roles are deriving from isolate-based observations and experiments. Moreover, this situation is at the exact antipodes of the current movement of open data in sciences.
Fig. 1.
Current process for publishing a new species. Delays here reported are based on the review of papers listed in “Supplementary reference list”. Green, current process; orange, new process by “New Species Announcement” in NMNI. Abbreviations list: IJSEM, International Journal if Systematic Evolutionary Microbiology; NMNI, New Microbes and New Infections; SIGS, Standards in Genomic Science.
In order to cope with the delay between the scientific discovery and the officialization of the proposed bacterial names, New Microbes New Infections now proposes an intermediate format between open data, in which all raw information would be made available, and the complete description of the new species. In our experience it takes in average 2.5 months to be released in New Microbes and New Infections, and it takes 15 months to be released in IJSEM and SIGS (see supplementary reference list). Such an intermediate article format already exists for genome sequences under the form of the Genome Announcement Journal. The new format that we propose aims at providing the scientific community with essential data on a putative new species, including main phenotypic characteristics and degree of 16S rRNA gene sequence identity with their closest phylogenetic relatives. It will make it possible to release this information within the 3 months following the first identification. The 16S rRNA gene sequence GenBank accession number is provided, and the type strain is made available by deposit in an official culture collection. This new format will not prevent microbiologists from further publishing a complete description of any new species.
We believe that this new format will enable researchers in the process of describing new bacterial species to rapidly inform the scientific community of their finding. It may help avoiding bacterial name redundancy for a given species.
We name this new format “New Species Announcement” and New Microbes and New Infections, as well as Human Microbiome, a new online journal to come, are proud of launching this initiative. Further information can be found at http://www.newmicrobesnewinfections.com/content/authorinfo.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.nmni.2016.04.006.
Contributor Information
Didier Raoult, Email: didier.raoult@gmail.com.
Michel Drancourt, Email: michel.drancourt@univ-amu.fr.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Sankar S.A., Lagier J.C., Pontarotti P., Raoult D., Fournier P.E. The human gut microbiome, a taxonomic conundrum. Syst Appl Microbiol. 2015;38:276–286. doi: 10.1016/j.syapm.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.