Abstract
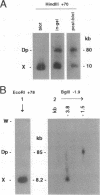
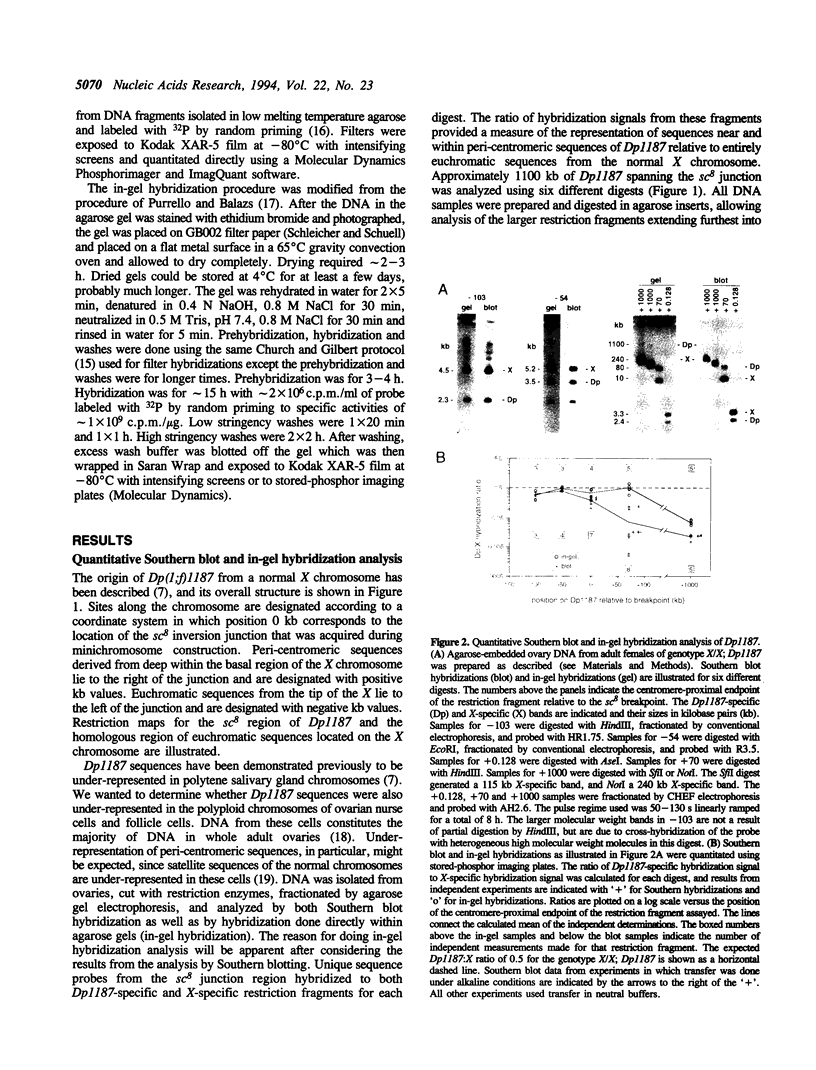
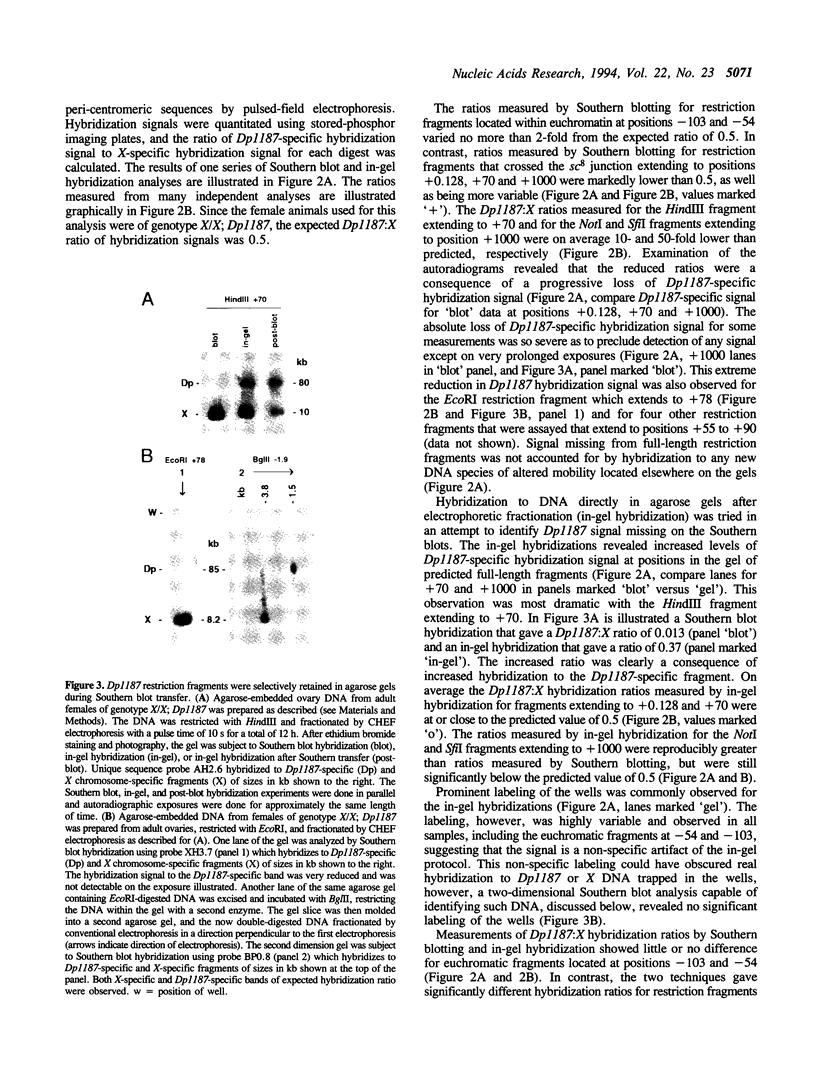
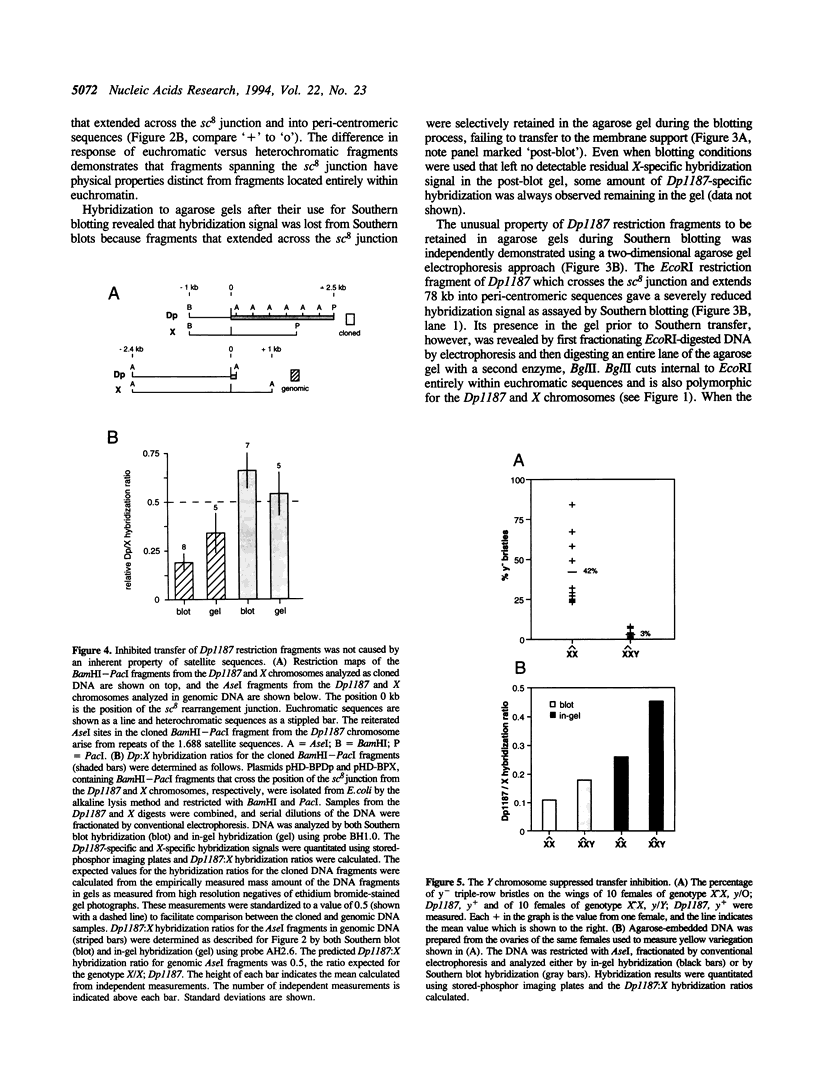
While investigating the copy number of minichromosome Dp(1;f)1187 sequences in the polyploid chromosomes of ovarian nurse and follicle cells of Drosophila melanogaster we discovered that restriction fragments spanning the euchromatic-heterochromatic junction of the chromosome and extending into peri-centromeric sequences had the unusual property of being selectively resistant to transfer out of agarose gels during Southern blotting, leading to systematic reductions in Dp1187-specific hybridization signals. This property originated from the peri-centromeric sequences contained on the junction fragments and was persistently associated with Dp1187 DNA, despite attempts to ameliorate the effect by altering experimental protocols. Transfer inhibition was unlikely to be caused by an inherent physical property of repetitive DNA sequences since, in contrast to genomic DNA, cloned restriction fragments spanning the euchromatic-heterochromatic junction and containing repetitive sequences transferred normally. Finally, the degree of inhibition could be suppressed by the addition of a Y chromosome to the genotype. On the basis of these observations and the fact that peri-centromeric regions of most eukaryotic chromosomes are associated with cytologically and genetically defined heterochromatin, we propose that peri-centromeric sequences of Dp1187 that are incorporated into heterochromatin in vivo retain some component of heterochromatic structure during DNA isolation, perhaps a tightly bound protein or DNA modification, which subsequently causes the unorthodox properties observed in vitro.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avramova Z., Tsanev R. Stable DNA-protein complexes in eukaryotic chromatin. J Mol Biol. 1987 Jul 20;196(2):437–440. doi: 10.1016/0022-2836(87)90704-2. [DOI] [PubMed] [Google Scholar]
- Bodnar J. W., Jones C. J., Coombs D. H., Pearson G. D., Ward D. C. Proteins tightly bound to HeLa cell DNA at nuclear matrix attachment sites. Mol Cell Biol. 1983 Sep;3(9):1567–1579. doi: 10.1128/mcb.3.9.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Carramolino L., Cabrera C. V., Ruíz-Gómez M., Villares R., Boronat A., Modolell J. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985 Feb;40(2):327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Glaser R. L., Karpen G. H., Spradling A. C. Replication forks are not found in a Drosophila minichromosome demonstrating a gradient of polytenization. Chromosoma. 1992 Dec;102(1):15–19. doi: 10.1007/BF00352285. [DOI] [PubMed] [Google Scholar]
- Gommers-Ampt J. H., Van Leeuwen F., de Beer A. L., Vliegenthart J. F., Dizdaroglu M., Kowalak J. A., Crain P. F., Borst P. beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993 Dec 17;75(6):1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- Hammond M. P., Laird C. D. Chromosome structure and DNA replication in nurse and follicle cells of Drosophila melanogaster. Chromosoma. 1985;91(3-4):267–278. doi: 10.1007/BF00328222. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Position-effect variegation after 60 years. Trends Genet. 1990 Dec;6(12):422–426. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Appels R. Pleiotropic effects associated with the deletion of heterochromatin surrounding rDNA on the X chromosome of Drosophila. Chromosoma. 1982;86(4):469–490. doi: 10.1007/BF00330122. [DOI] [PubMed] [Google Scholar]
- Hoheisel J. D. Extension of a pUC18-like polylinker by the octanucleotide recognition sites of NotI, FseI, SfiI and PacI. Trends Genet. 1990 Nov;6(11):346–346. doi: 10.1016/0168-9525(90)90261-4. [DOI] [PubMed] [Google Scholar]
- Karpen G. H., Spradling A. C. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992 Nov;132(3):737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G. H., Spradling A. C. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell. 1990 Oct 5;63(1):97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs E., Laemmli U. K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992 Feb;11(2):705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M. L., Hennig W. Heterochromatin: junk or collectors item? Chromosoma. 1990 Dec;100(1):3–7. doi: 10.1007/BF00337597. [DOI] [PubMed] [Google Scholar]
- Pfütz M., Gileadi O., Werner D. Identification of human satellite DNA sequences associated with chemically resistant nonhistone polypeptide adducts. Chromosoma. 1992 Oct;101(10):609–617. doi: 10.1007/BF00360538. [DOI] [PubMed] [Google Scholar]
- Purrello M., Balazs I. Direct hybridization of labeled DNA to DNA in agarose gels. Anal Biochem. 1983 Feb 1;128(2):393–397. doi: 10.1016/0003-2697(83)90391-3. [DOI] [PubMed] [Google Scholar]
- Rae P. M., Barnett T. R., Babbitt D. G. Factors influencing the yield of satellite DNA in extractions from Drosophila virilis and Drosophila melanogaster adults and embryos. Biochim Biophys Acta. 1976 May 3;432(2):154–160. doi: 10.1016/0005-2787(76)90157-x. [DOI] [PubMed] [Google Scholar]
- Reuter G., Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992 Sep;14(9):605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Skinner D. M., Triplett L. L. The selective loss of DNA satellites on deproteinization with phenol. Biochem Biophys Res Commun. 1967 Sep 27;28(6):892–897. doi: 10.1016/0006-291x(67)90062-9. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Karpen G. H. Sixty years of mystery. Genetics. 1990 Dec;126(4):779–784. doi: 10.1093/genetics/126.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A., Orr-Weaver T. Regulation of DNA replication during Drosophila development. Annu Rev Genet. 1987;21:373–403. doi: 10.1146/annurev.ge.21.120187.002105. [DOI] [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum Y., Sedat J., Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982 Sep 6;146(1):148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- Zhang P., Spradling A. C. Efficient and dispersed local P element transposition from Drosophila females. Genetics. 1993 Feb;133(2):361–373. doi: 10.1093/genetics/133.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]