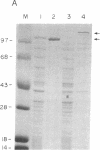
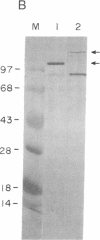
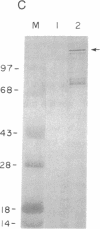
Abstract
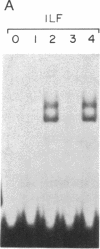
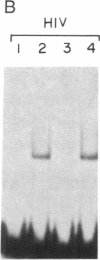
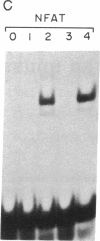
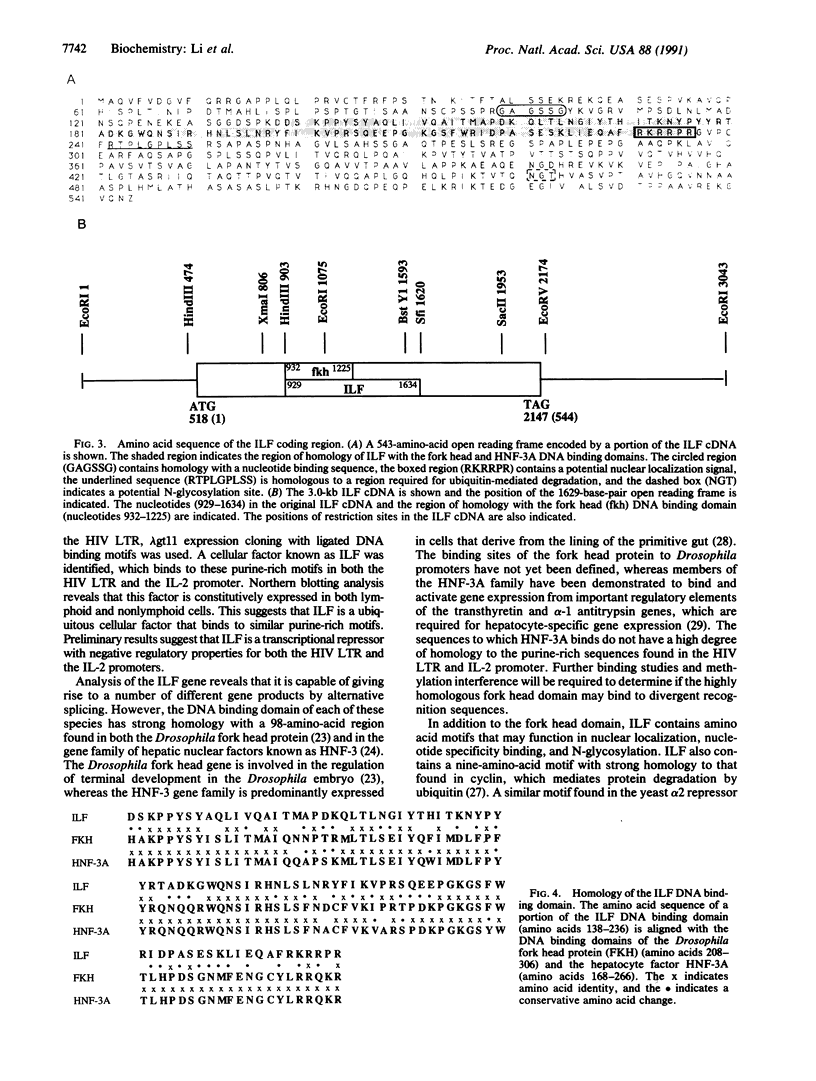
Human immunodeficiency virus (HIV) gene expression is regulated by both general transcription factors and factors induced by activation of T lymphocytes such as NF-kappa B and the nuclear factor of activated T cells (NFAT). Within the HIV long terminal repeat (LTR), two purine-rich domains between nucleotides -283 and -195 have homology to a regulatory region found in the interleukin 2 promoter, which binds NFAT and other cellular factors. In the HIV LTR, this region has been demonstrated to have both positive and negative regulatory effects on HIV gene expression. In an attempt to clone genes encoding cellular factors that bind to these NFAT-like elements in the HIV LTR, we used lambda gt11 expression cloning with oligonucleotides corresponding to these binding motifs. A ubiquitously expressed cDNA encoding a 60-kDa protein, which we termed interleukin binding factor (ILF), binds specifically to these purine-rich motifs in the HIV LTR. This factor also binds to similar purine-rich motifs in the interleukin 2 promoter, through with lower affinity than to HIV LTR sequences. Sequence analysis reveals that the DNA binding domain of ILF has strong homology to the recently described fork head DNA binding domain found in the Drosophila homeotic protein fork head and a family of hepatocyte nuclear factors, HNF-3. Other domains found in ILF include a nucleotide binding site, an N-glycosylation motif, a signal for ubiquitin-mediated degradation, and a potential nuclear localization signal. These results describe a DNA binding protein that may be involved in both positive and negative regulation of important viral and cellular promoter elements.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bürglin T. R., De Robertis E. M. The nuclear migration signal of Xenopus laevis nucleoplasmin. EMBO J. 1987 Sep;6(9):2617–2625. doi: 10.1002/j.1460-2075.1987.tb02552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Darnell J. E., Jr Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989 Apr;9(4):1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree G. R. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989 Jan 20;243(4889):355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- Emmel E. A., Verweij C. L., Durand D. B., Higgins K. M., Lacy E., Crabtree G. R. Cyclosporin A specifically inhibits function of nuclear proteins involved in T cell activation. Science. 1989 Dec 22;246(4937):1617–1620. doi: 10.1126/science.2595372. [DOI] [PubMed] [Google Scholar]
- Fujita T., Shibuya H., Ohashi T., Yamanishi K., Taniguchi T. Regulation of human interleukin-2 gene: functional DNA sequences in the 5' flanking region for the gene expression in activated T lymphocytes. Cell. 1986 Aug 1;46(3):401–405. doi: 10.1016/0092-8674(86)90660-4. [DOI] [PubMed] [Google Scholar]
- Garcia J. A., Harrich D., Soultanakis E., Wu F., Mitsuyasu R., Gaynor R. B. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 1989 Mar;8(3):765–778. doi: 10.1002/j.1460-2075.1989.tb03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor R. B., Kuwabara M. D., Wu F. K., Garcia J. A., Harrich D., Briskin M., Wall R., Sigman D. S. Repeated B motifs in the human immunodeficiency virus type I long terminal repeat enhancer region do not exhibit cooperative factor binding. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9406–9410. doi: 10.1073/pnas.85.24.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991 Jan 10;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990 May 18;61(4):697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- Karim F. D., Urness L. D., Thummel C. S., Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A., Gunther C. V., Nye J. A. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990 Sep;4(9):1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990 Apr 6;61(1):113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E., Prezioso V. R., Smith E., Litvin O., Costa R. H., Darnell J. E., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990 Aug;4(8):1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- Lai E., Prezioso V. R., Tao W. F., Chen W. S., Darnell J. E., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991 Mar;5(3):416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- Lu Y. C., Touzjian N., Stenzel M., Dorfman T., Sodroski J. G., Haseltine W. A. Identification of cis-acting repressive sequences within the negative regulatory element of human immunodeficiency virus type 1. J Virol. 1990 Oct;64(10):5226–5229. doi: 10.1128/jvi.64.10.5226-5229.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Li C., Kornuc M., Gaynor R. CREB regulation of cellular cyclic AMP-responsive and adenovirus early promoters. J Virol. 1990 Sep;64(9):4296–4305. doi: 10.1128/jvi.64.9.4296-4305.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G., Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987 Apr 16;326(6114):711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Novak T. J., Chen D., Rothenberg E. V. Interleukin-1 synergy with phosphoinositide pathway agonists for induction of interleukin-2 gene expression: molecular basis of costimulation. Mol Cell Biol. 1990 Dec;10(12):6325–6334. doi: 10.1128/mcb.10.12.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterson M., Schaffner W. A purine-rich DNA sequence motif present in SV40 and lymphotropic papovavirus binds a lymphoid-specific factor and contributes to enhancer activity in lymphoid cells. Genes Dev. 1987 Nov;1(9):962–972. doi: 10.1101/gad.1.9.962. [DOI] [PubMed] [Google Scholar]
- Randak C., Brabletz T., Hergenröther M., Sobotta I., Serfling E. Cyclosporin A suppresses the expression of the interleukin 2 gene by inhibiting the binding of lymphocyte-specific factors to the IL-2 enhancer. EMBO J. 1990 Aug;9(8):2529–2536. doi: 10.1002/j.1460-2075.1990.tb07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. P., Utz P. J., Durand D. B., Toole J. J., Emmel E. A., Crabtree G. R. Identification of a putative regulator of early T cell activation genes. Science. 1988 Jul 8;241(4862):202–205. doi: 10.1126/science.3260404. [DOI] [PubMed] [Google Scholar]
- Siekevitz M., Josephs S. F., Dukovich M., Peffer N., Wong-Staal F., Greene W. C. Activation of the HIV-1 LTR by T cell mitogens and the trans-activator protein of HTLV-I. Science. 1987 Dec 11;238(4833):1575–1578. doi: 10.1126/science.2825351. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Tong-Starksen S. E., Luciw P. A., Peterlin B. M. Human immunodeficiency virus long terminal repeat responds to T-cell activation signals. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6845–6849. doi: 10.1073/pnas.84.19.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Jäckle H. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell. 1990 Nov 2;63(3):455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jürgens G., Küttner F., Seifert E., Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989 May 19;57(4):645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Hol W. G. Predicted nucleotide-binding properties of p21 protein and its cancer-associated variant. Nature. 1983 Apr 28;302(5911):842–844. doi: 10.1038/302842a0. [DOI] [PubMed] [Google Scholar]