Abstract
Uterine carcinosarcoma (UCS) is a form of endometrial cancer simultaneously exhibiting carcinomatous and sarcomatous elements, but the underlying molecular and epigenetic basis of this disease is poorly understood. We generated complete DNA methylomes for both the carcinomatous and the sarcomatous components of three UCS samples separated by laser capture microdissection and compared DNA methylomes of UCS with those of normal endometrium as well as methylomes derived from endometrioid carcinoma, serous endometrial carcinoma, and endometrial stromal sarcoma. We identified epigenetic lesions specific to carcinosarcoma and specific to its two components. Hallmarks of DNA methylation abnormalities in UCS included global hypomethylation, especially in repetitive elements, and hypermethylation of tumor suppressor gene promoters. Among these, aberrant DNA methylation of MIR200 genes is a key feature of UCS. The carcinoma component of UCS was characterized by hypermethylation of promoters of EMILIN1, NEFM, and CLEC14A, genes that are associated with tumor vascularization. In contrast, DNA methylation changes of PKP3, FAM83F, and TCP11 were more characteristic of the sarcoma components. Our findings highlight the epigenetic signatures that distinguish the two components of UCS, providing a valuable resource for investigation of this disease.
Introduction
Cancer is a disease of epigenetic lesions as well as genetic lesions. Human cancers display abnormal DNA methylation patterns including genome-wide hypomethylation and site-specific hypermethylation [1]. Locus-specific DNA methylation alterations of promoters or CpG islands have demonstrated effects on expression of nearby genes (e.g., tumor suppressor genes), which have important clinical significance [2], [3]. Globally, hypomethylation of most genomic transposable elements (TEs) leads to chromosome instability [4], whereas alterations in methylation levels of other TEs contribute to tumor initiation or progression [5]. The scope of aberrant methylation of distal enhancers continues to receive attention in many cancers [6], [7].
Uterine carcinosarcoma (UCS) is an aggressive variant of endometrial cancer with potential for local recurrence and metastasis, accounting for approximately 15% of uterine cancer–associated deaths in the United States [8]. Women with UCS survive for less than 2 years on average, which is worse than either endometrioid adenocarcinoma or high-grade serous carcinoma [9].
Histologically, UCS is composed of an admixture of malignant epithelial and sarcomatous elements. The sarcoma component exhibits differentiation along the lines of mesenchymal cell types normally present in the uterus, with histologic features of leiomyosarcoma, endometrial stromal sarcoma (ESS), or fibrosarcoma (“homologous” sarcomas), or may resemble rhabdomyosarcoma, chondrosarcoma, osteosarcoma, or other heterologous sarcomas [10]. The ability of UCS to adopt sarcomatoid morphologies may be linked to its clinical aggressiveness, as epithelial–mesenchymal transition by carcinoma cells is known to contribute to their metastatic potential [11], and UCS displays a mesenchymal phenotype even prior to metastasis. Thus, understanding the mechanism by which UCS adopts sarcomatoid morphology could lead to targeted therapy.
Genetic and epigenetic profiling of UCS has previously focused on bulk tumor, presumably due to the challenge of separating the intimately admixed carcinomatous and sarcomatous components [12]. Characteristic mutations in TP53, KRAS, and PIK3CA have been reported in UCS [13], [14]. When separate carcinomatous and sarcomatous components have been studied, the components have shown concordant “root” mutations [15] with additional private “stem” mutations present in one component or the other [16]. Given the role of epigenetic marks in enforcing gene expression patterns and cellular phenotypes, we hypothesized that the components might differ at an epigenetic, as well as genetic, level. We therefore undertook an epigenetic analysis aimed at elucidating genome-wide DNA methylation patterns in UCS, analyzing the results as they relate to cancer initiation and progression.
In the present study, we used laser capture microdissection (LCM) to separate the two components of UCS samples from three patients. We used two complementary next-generation sequencing–based methods, methylated DNA immunoprecipitation sequencing (MeDIP-seq) and methylation-sensitive restriction enzyme digestion sequencing (MRE-seq) [17], to construct genome-wide DNA methylation maps at single-CpG resolution in these components. Compared to array-based methods, which query a preselected probe set, our method profiles a much larger, unbiased, and complete set of CpG sites. By comparing to the DNA methylome maps of normal endometrium, we identified differentially methylated regions (DMRs) associated with the two distinct components of UCS. Many DMRs were found in CpG islands and promoters, which were associated with aberrant expression of nearby genes. Globally, UCS exhibited hypomethylation of TEs, especially in L1 elements. By comparing our findings to other types of uterine cancer—endometrial serous and endometrioid carcinoma (UPSC and EAC) and ESS—we defined two sets of cancer-type specific DMRs: carcinoma-associated DMRs (CADs) and sarcoma-associated DMRs (SADs). We found that both CADs and SADs were enriched in regulatory elements and both hypermethylated CADs and SADs were associated with developmental genes. However, CADs and SADs were associated with different groups of developmental genes, suggesting that they might have different developmental history. The tumor suppressor MIR200 family exhibited differential DNA methylation in carcinoma and sarcoma, including in the UCS components. Methylation-associated silencing of the MIR200 family may explain the ectopic activation of mesenchymal pathways in UCS.
Materials and Methods
Sample Collection
The study was approved by the Human Research Protection Office, Washington University School of Medicine (protocol 201201013). Three prospectively banked fresh-frozen UCS samples were retrieved from the institutional biosample bank. Top slides were reviewed by a pathologist (I.S.H.) to confirm neoplastic cellularity >80% and necrosis <10%. All three carcinoma components resembled high-grade serous carcinoma; all three sarcoma components were homologous. Carcinoma and sarcoma components were separated by LCM of hematoxylin and eosin–stained frozen section slides, performed by a skilled technician under direct supervision by I. S. H., followed by DNA extraction using a QIAamp spin column method to yield a mean of 0.68 μg DNA/specimen. Data previously generated from 3 EACs, 3 UPSCs, 3 UCSs, 3 ESSs, and 10 pooled normal endometrium specimens by the same MeDIP-seq/MRE-seq approach were used for comparison [7]. Data for 34 normal endometria, 80 EACs, 33 UPSCs, 57 UCSs, and 94 sarcomas from The Cancer Genome Atlas (TCGA) project were retrieved as validation cohorts.
Global Methylation Analysis
Genomic DNA from tumor tissues and normal endometrium was extracted using the DNeasy Tissue kit (Qiagen, Valencia, CA). MeDIP and MRE sequencing libraries were constructed as previously described [18]. Sequencing reads were aligned to hg19 with BWA [19]. MRE reads were normalized to account for differences in enzyme efficiency, and scoring consisted of tabulating reads with CpGs at each fragment end. All data methylation values were predicted by methylCRF based on MeDIP and MRE signals according to published methods [20].
DMR Identification
The methylMnM package (http://epigenome.wustl.edu/MnM/) was used to identify DMRs in the R 2.0.15 environment. Default parameters were used, and a statistical cutoff of q value <10−9 was applied to select UCS shared DMRs from each pairwise comparison at a resolution of 500 bp. UCS carcinoma component DMRs and sarcoma component DMRs were defined such that the same genomic region must have been called a DMR and have the same direction of DNA methylation change in at least two out of the three cancers versus normal pairwise comparisons. Similarly, DMRs which were recurrent in 6 out of 9 carcinoma samples were called carcinoma-associated DMRs (q value <10−5). DMRs which were recurrent in 4 out of 6 sarcomas were called sarcoma-associated DMRs (q value <10−5). RepeatMasker annotations, CpG islands, and RefSeq Gene coding locus features were all downloaded from the UCSC Genome Browser [21]. Annotations of enhancers by all available roadmap data encompassing 127 reference epigenomes across multiple tissue and cell types (111 from Roadmap and 16 from ENCODE) were downloaded from the Roadmap Epigenomics project website (http://www.roadmapepigenomics.org) (Supplementary Material). One-kilobase core promoters were defined as 1 kb around the most 5′ transcription start site (500 bp upstream and 500 bp downstream of TSS) of any RefSeq gene annotation. miRNA loci were downloaded from mirBASE. The microRNA gene cluster TSSs were download from mirStart (http://mirstart.mbc.nctu.edu.tw/). lincRNA loci were download from the Human lincRNA Catalog (http://www.broadinstitute.org/genome_bio/human_lincrnas/) [22].
TCGA DNA Methylation Data
Processed DNA methylation data (Infinium HumanMethylation450 BeadChip platform), and mRNA-seq and miRNA-seq data of 34 normal endometria, 80 EACs, 33 UPSCs, 57 UCSs, and 94 sarcomas were downloaded from TCGA (http://cancergenome.nih.gov/). The methylation beta value of each probe within DMRs was isolated for further analysis in the R 2.15 environment. Reads per kilobase of transcript per million reads mapped values were computed for each gene using TCGA's mRNA sequencing data. The lengths of transcripts were obtained from NCBI Genbank. Reads per million values for each microRNA gene were computed using TCGA's microRNA sequencing data. Sample histology information was obtained from the supplementary materials of Kandoth et al. [23]. The microarray data of GSE32507 [24] and GSE28866 [25] were downloaded from the NCBI Gene Expression Omnibus database. A total of 46 chips derived from endometrial cancers were available, including 14 UCSs, 24 ECs, and 8 ESSs. The probe-level data were converted into the corresponding genetic symbols based on the relationship of the genes and the matching probes on platform GPL6480. By taking the average expression value, the expression values of all probes for each gene were normalized to signal intensity. Normalized 3SEQ expression data for the 66 cancer libraries and the 27 normal libraries included coding, lncRNA, and other known transcripts. Expression data were normalized, and the square root of each value was taken to reduce the effect of outliers.
Enrichment Calculation
The binding site enrichment score (ES) for each genomic feature, DHS, and transcription factor with respect to DMRs was calculated as:
where n_hit is the number of DMRs that contain a specific genomic feature, experimentally annotated DHS, or TFBS; n_DMR is the total number of DMRs; N_hit is the number of genomic windows with a specific genomic feature; and N_all is the number of 500-bp windows in the human genome (hg19).
Gene Ontology (GO) Enrichment Analysis
GO analyses for biological processes were performed using the GREAT Tool [26] with default gene regulatory regions spanning 5 kb upstream to 1 kb downstream of the TSS (regardless of other nearby genes). Gene regulatory domains were extended in both directions to the nearest gene's basal domain but no more than a maximum extension in one direction.
Availability of Supporting Data
Sequencing data have been deposited in the NCBI's Gene Expression Omnibus repository under accession number GSE86505. The methylation data of normal endometrium, EAC, and UPSC were obtained from the GSE51565 data set in the Gene Expression Omnibus database. All the processed data and DMRs were uploaded to the WashU Epigenome Browser (http://epigenomegateway.wustl.edu/browser/?genome=hg19&publichub=UCS).
Results
Global DNA Methylome Patterns in Normal and Tumor Tissues
UCSs are characterized by an admixture of at least two histologically distinct components, one resembling carcinoma and another resembling sarcoma. With the aid of LCM, we separated the two different components from three UCS cases, thus enabling the detection of their specific epigenetic lesions. We combined DNA methylation enrichment (MeDIP-seq) and methylation-sensitive restriction enzyme digestion sequencing methods (MRE-seq) to obtain the whole DNA methylomes of the two components. We used methylCRF, a conditional random fields-based algorithm, to estimate DNA methylation levels at single-CpG resolution [20], [27] (Figure 1). Genome-wide CpG methylation in somatic cells typically follows a bimodal distribution, as observed for normal endometrial cells (Figure 2A). However, UCS exhibited a clear deviation from this pattern, reflecting a dramatic loss of DNA methylation (Figure 2A). In normal endometrium, 80% of the 28M CpGs were methylated (i.e., methylation level ≥ 0.8), whereas in UCS samples, this ratio fell to 60% to 70% (Supplemental Table 1). Global hypomethylation is known to be a hallmark of many cancer genomes, and here we confirmed this pattern for UCS (Supplemental Table 1).
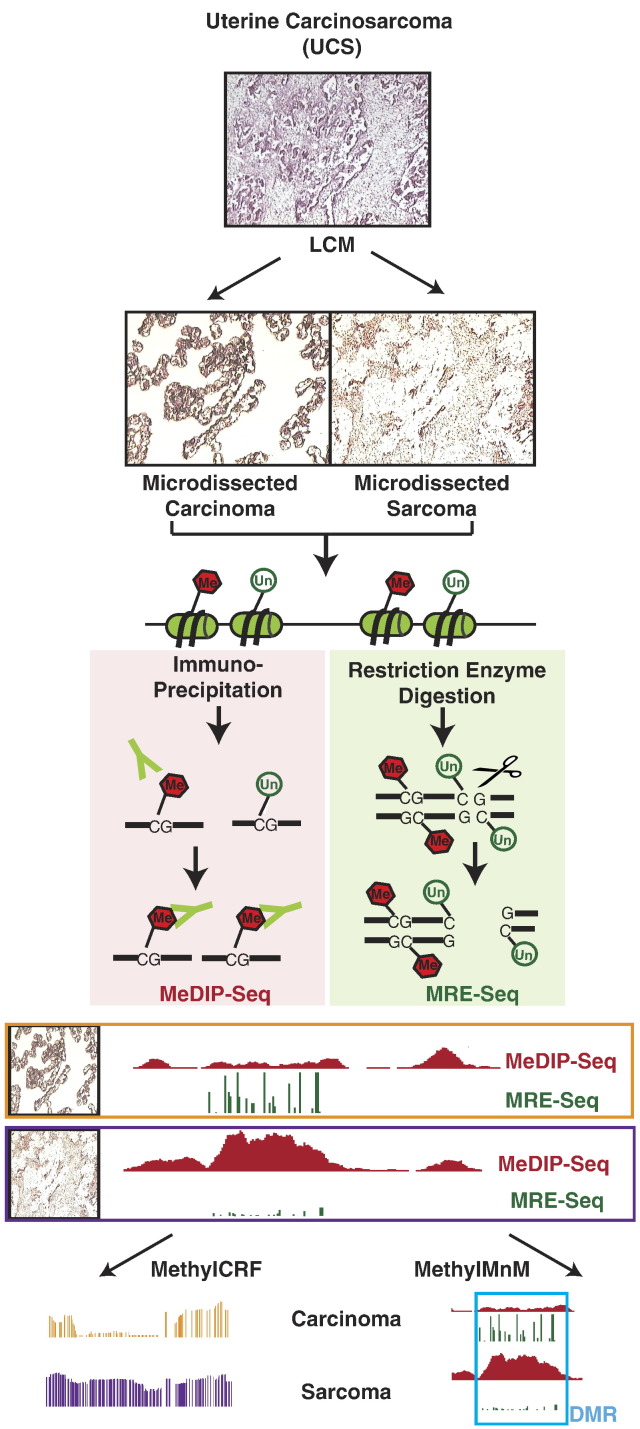
Fig. 1.
Basic workflow for DNA methylome sequencing of LCM samples. After LCM, distinct carcinoma components and sarcoma components dissected from UCS were sequenced using MeDIP-seq and MRE-seq. MeDIP-seq: After sonication, the methylated genomic DNA fragments were captured by a monoclonal anti–5′-methylcytosine antibody. Immunoprecipitated DNA fragments were analyzed by high-throughput DNA sequencing. MRE-seq: Instead of sonication, the genomic DNA was digested by multiple methylation-sensitive restriction enzymes. Specific size fragments were selected and then sequenced. The bioinformatics pipeline combined two algorithms (methylCRF and M&M) to estimate absolute methylation levels at single-CpG resolution and to detect DMRs between the two components, respectively.
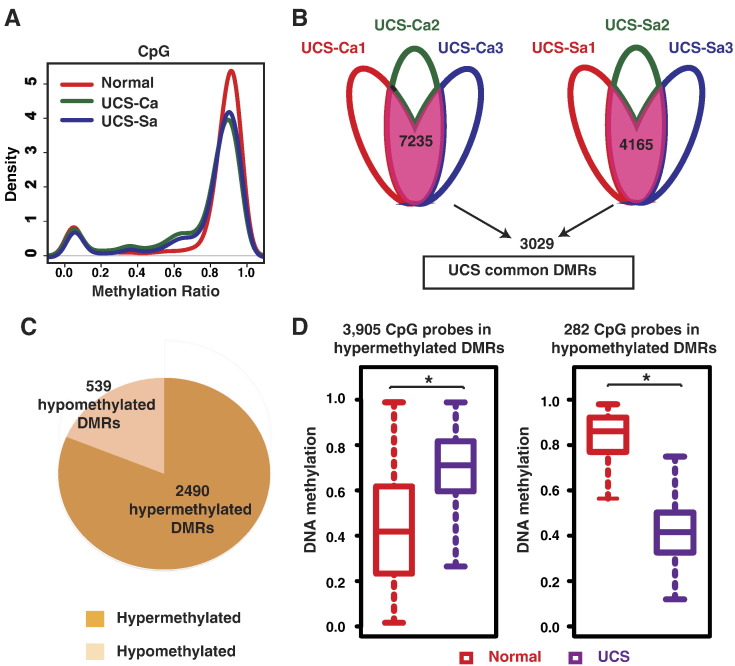
Fig. 2.
Global and local methylation changes in UCS samples. (A) Density plot of global DNA methylation level distribution estimated by methylCRF: normal endometrium (red line), UCS carcinoma components (green line), and UCS sarcoma components (blue line). (B) A total of 7235 DMRs were identified in at least 2 UCS carcinoma components; 4165 DMRs were identified in at least 2 UCS sarcoma components. A total of 3029 UCS common DMRs were recurrent in at least 2 UCS carcinoma components and 2 UCS sarcoma components. (C) Proportion of UCS common DMRs that were hypermethylated (dark orange) and hypomethylated (light orange). (D) Validation of UCS common DMRs using all probes located in DMRs in TCGA Infinium 450K data.
Discovery and Validation of DMRs in UCS
As endometrial cancers' tumorigenesis has been reported to reflect altered local epigenetic regulation [7], we next investigated the DMRs in UCS. Previously we described a computational algorithm (M&M) specifically designed to detect DMRs [28]. Here we applied M&M at a stringent statistical cutoff (false discovery rate <10−9) to detect DMRs in all 500-bp windows across the genome. This approach allowed us to discover a total of 7235 recurrent DMRs in at least 2 of the 3 carcinoma components, and 4165 DMRs in the sarcoma components, when compared to normal endometrium. Of these, 3029 DMRs were shared by both the carcinomatous and the sarcomatous components, which we defined as “UCS common DMRs” (Figure 2B). About 82% of UCS common DMRs were hypermethylated (2490 DMRs), and 18% were hypomethylated (539) (Figure 2C).
The TCGA Consortium has previously profiled the DNA methylation pattern of 57 bulk UCS samples and 34 normal controls using the Infinium HumanMethylation450 BeadChip platform, which interrogates roughly 450,000 CpGs in regions classically defined as “important” (i.e., promoters, CpG islands, etc.). These data provide an independent cohort of UCS samples which can serve as a validation panel for some of our discoveries (http://cancergenome.nih.gov/). A total of 4187 Infinium probes were located within the UCS common DMRs, including 3905 in the hypermethylated DMRs and 282 in the hypomethylated DMRs. Indeed, beta values of these CpGs across TCGA samples faithfully recapitulated the DNA methylation abnormalities we discovered with our UCS samples (Figure 2D). It is noteworthy that in addition to the high validation rate based on the TCGA data, our whole genome comparison allowed us to detect many more DMRs that the Infinium 450k platform is blind to (Supplementary Figure 1A). For example, 44.5% of the DMRs we detected do not even have a single probe on the Infinium 450k platform. This fraction is even more dramatic for the hypomethylated DMRs, where 66% of the DMRs do not contain probes.
We next examined expression levels of DNA methyltransferases using data provided by the TCGA Consortium. Interestingly, all three DNA methyltransferases, DNMT1, DNMT3a, and DNMT3b, exhibited significantly higher expression in UCS samples when compared to normal endometrium (P < .01, Student's t test; Supplemental Figure 1B). This is consistent with the detection of roughly three times more hypermethylated DMRs than hypomethylated DMRs in UCS.
Distinct DNA Methylation Patterns at Gene Promoters
Having validated our DNA methylation data on UCS with TCGA data, we set out to examine genomic distribution of DMRs. We found that the UCS-common DMRs are significantly enriched for gene promoters and CpG islands. This pattern is consistent with the hallmarks of cancer, which we confirmed for UCS [7], [29], [30] (Figure 3A). Specifically, we found 743 genes with hypermethylated DMRs within 2.5 kb of their transcription start site. In cancer cells, epigenetic inactivation of tumor-suppressor genes (TSGs) is a key tumorigenic event. The set of genes with hypermethylated promoters included 24 TSGs (KLF4, NDN, WT1, PROX1, PHOX2A, TBX5, CDX2, PCDH8, NKX2–8, CPNE7, BCL2L11, TFAP2A, MZB1, DND1, CLDN23, NNAT, CTNNA2, TNFRSF10B, CDO1, SLC39A4, PEG3, H19, MST1R, PLAGL1) (Supplementary Table 2) [31], of which 15 of 24 also had recurrent promoter hypermethylation in TCGA data (Figure 3B and Supplemental Figure 1C, Student's t test , P value < .01).
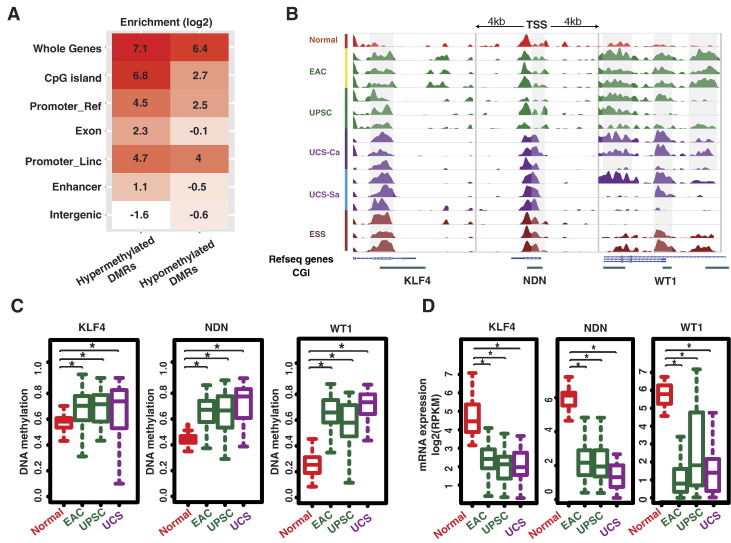
Fig. 3.
Genomic annotation of UCS common DMRs and examples of methylation-silenced TSGs. (A) Heatmap of DMR enrichment for different genomic regions. Promoter_Ref and Promoter_Linc mean 1-kb core promoter of RefSeq genes and lincRNA, respectively. (B) Epigenome Browser views of three TSG promoters with hypermethylated changes across uterine cancers: MeDIP tracks of normal endometrium (10 pooled healthy donors), 3 EACs, 3 UPSCs, 3 UCS carcinoma components, 3 UCS sarcoma components, and 3 ESSs are displayed. (C and D) Methylation levels and gene expression analysis of the same three TSG with hypermethylated promoters in normal endometrium, EAC, UPSC, and UCS samples using TCGA Infinium 450K methylation data and RNA sequencing data.
More specifically, the Wilms' tumor 1 (WT1) gene was one of the TSGs identified as harboring a hypermethylated DMR in its promoter (Figure 3B), and this was validated by TCGA methylation data (Figure 3C). TCGA mRNA expression data showed lower WT1 expression in UCS as well as other carcinomas (Figure 3D), supporting a functional effect for this aberrant state of methylation. Necdin (NDN) has been reported to be silenced by methylation in urothelial carcinoma [32]. NDN is regarded as a potential tumor suppressor gene due to its interactions with p53 and E2F-1. Like WT1, NDN showed promoter hypermethylation in MeDIP/MRE and TCGA data (Figure 3, B and C) and was downregulated at the mRNA level in UCS and other carcinomas (Figure 3D). Kruppel-like factor 4 (KLF4) serves as a prodifferentiation and antiproliferative protein and interacts with the tumor suppressor p53. By disrupting β-catenin–mediated recruitment of the coactivators p300/CBP, KLF4 is also involved in oncogenic Wnt signaling. KLF4, together with three other transcription factors, is capable of reprogramming fibroblasts into induced pluripotent stem cells that are similar to ES cells [33], hinting that KLF4 could promote cancer cells' adoption of a stemlike state. However, KLF9 and KLF4 protein levels have been reported to be decreased in endometrial tumors [34]. We found that KLF4 showed promoter hypermethylation in UCS, other carcinomas, and ESS (Figure 3, B and C) and was downregulated in UCS and other carcinomas (Figure 3D).
To better understand the potential function of altered UCS methylation regions, we performed GO analysis on hypermethylated DMRs. This identified several biologic functional pathways that are potentially targeted by DNA methylation abnormalities in UCS (Supplementary Figure 1D). The highest-scoring functional annotation (P < 10−53) was composed of 128 genes known to be involved in embryonic morphogenesis, followed by gene clusters associated with pattern specification and cell adhesion.
Global Hypomethylation of TEs and Enrichment of Specific Subfamilies
TE-derived sequences are present in large quantities in almost all genomes, including those of humans. Global hypomethylation of TEs has long been regarded as a hallmark of cancers, with direct implication in cancer genome instability [35]. More recently, several groups reported abnormal expression of TE-derived sequences in cancers, which could also be a direct consequence of abnormal DNA methylation [36], [37], [38]. We thus characterized the DNA methylation pattern of TE-derived sequences in UCS. We found that all major classes of TEs exhibited global DNA hypomethylation in UCS, with LINEs exhibiting the largest effect size (Figure 4A). This effect was greater in UCS than in pure endometrial carcinomas and ESS (Figure 4A). Overall, 15.6% of hypermethylated and 40% of hypomethylated UCS-common DMRs were contributed by TEs (Figure 4B).
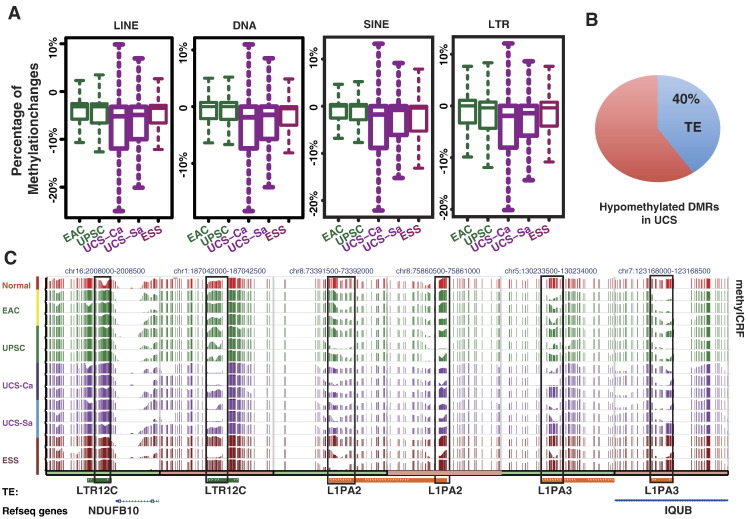
Fig. 4.
Abnormal DNA methylation of TEs. (A) Percentage of methylation shifts in different classes of TEs across uterine cancers compared to normal endometrium. (B and C) Percentage of hypomethylated DMRs overlapping TEs in UCS. (D) Epigenome Browser views of three subfamilies of TEs (LTR12C, L1PA2, and L1PA3). MethylCRF scores across different uterine cancers are displayed.
Interestingly, some TE subfamilies exhibited a mixture of hypermethylation and hypomethylation. For example, LTR12C is an endogenous retroviral element (ERV/LTR) recently reported to produce abnormal transcripts in liver cancer [38]. LTR12C contributed both hypermethylated and hypomethylated DMRs (Figure 4C). In contrast, most of the L1-derived DMRs were hypomethylated DMRs (Figure 4C). The unique DNA methylation abnormality of LTR12C highlights the complexity of TE DNA methylation in cancer and suggests detailed follow-up studies to determine their functional consequences.
Large-Scale DNA Hypermethylation of PCDH Gene Clusters in Endometrial Cancers
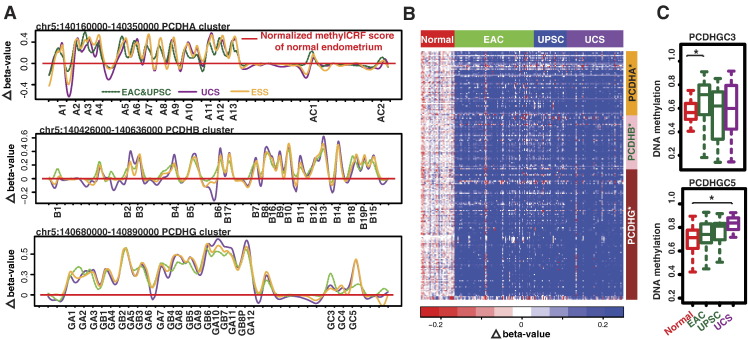
The protocadherin (PCDH) gene clusters are a superfamily of homophilic cadherin genes that encode cell-adhesion regulators with roles in neuron development and in the differentiation of many cancers [39]. Recurrent hypermethylated protocadherin genes in endometrial cancers occurred within 5q31, spanning three PCDH clusters (PCDHA, PCDHB, and PCDHG; Figure 5A). We detected an overall 20% increase in DNA methylation across PCDH clusters, and this result was supported by TCGA Infinium 450K data (Figure 5B). Interestingly, DNA methylation levels of PCDH clusters were similar across all endometrial cancers except for PCDHGC3 and PCDHGC5. Our data suggested that PCDHGC3 exhibited greater methylation changes in both EAC and UPSC than in UCS and ESS; in contrast, PCDHGC5 was more highly methylated in UCS and in ESS than in EAC and UPSC. TCGA methylation data were consistent with this pattern, albeit without reaching statistical significance (Figure 5C).
Fig. 5.
Long-range methylation changes of PCDH clusters in uterine cancers. (A) Normalized methylCRF scores of four different uterine cancers. (B) Heatmap of methylation scores of PCDH clusters based on TCGA Infinium 450K data. (C) Methylation level of PCDHGC3 and PCDHGC5 in normal endometrium, EAC, UPSC, and UCS samples using TCGA Infinium 450K methylation data.
Differentially Methylated Genes Between Carcinoma and Sarcoma Components
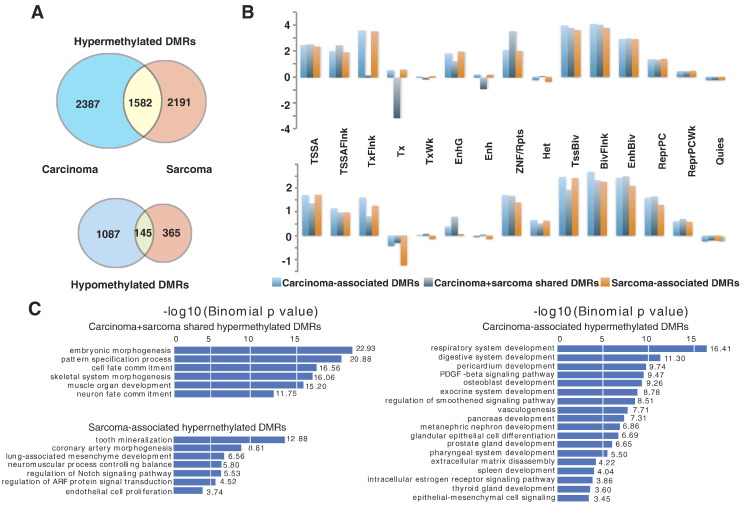
We hypothesized that the presence of DMRs within certain genes might be characteristic of carcinomatous or sarcomatous components of UCS. To identify these genes, we overlapped DMRs of UCS-Ca and EC to obtain a group of 3474 DMRs that we termed CADs (Methods). Of these, 2387 were hypermethylated and 1087 were hypomethylated (Figure 6A). To identify characteristic DMRs of sarcomatous components, we overlapped DMRs of UCS-Sa and ESS to obtain 2556 SADs. Of these, 2191 were hypermethylated and 365 were hypomethylated (Figure 6A). A total of 1727 DMRs were shared between CADs and SADs (P value ~0, hypergeometric test). This is consistent with the notion that the two components are derived from a common precursor. Interestingly, they shared many more hypermethylated DMRs (1582) than hypomethylated DMRs (145).
Fig. 6.
Definition, genomic feature enrichment, and GO analysis of CADs and SADs. (A) Definition and numbers of CADs, SADs, and shared DMRs. (B) Enrichment analysis of hypermethylated (top) and hypomethylated (bottom) DMRs located in different genomic regions, annotated by 15 chromHMM states. (C) Functional annotation of genes near different categories of DMRs using the GREAT GO tool.
We next sought to functionally annotate CADs and SADs. First, we asked if they distributed differently in the context of the reference human epigenome. The Roadmap Epigenomics Project [40] has produced an epigenomic annotation of the human genome across 127 tissue/cell types by integrating multiple types of histone modification datasets using chromHMM [41] and partitioning the genome into 15 chromatin states (e.g., enhancers, transcribed regions, quiescent regions). We found that both CADs and SADs were enriched for regulatory elements, including transcription start sites and bivalent enhancers (Figure 6B), and hypermethylated CADs and SADs in general were enriched slightly more than hypomethylated ones. Second, we asked if genes associated with CADs and SADs were enriched for similar or different functions. Toward this goal, we applied the GREAT tool [26] to our DMR lists. Interestingly, both CADs and SADs were associated with developmental genes, especially those involved in embryonic development and cell fate specification. In addition, CADs and SADs each enriched for terms related to their specific lineage (Figure 6C). For hypermethylated CADs, we found enrichment of genes related to epithelial development, including epithelial–mesenchymal cell signaling and respiratory system development; for hypermethylated SADs, we found enrichment of genes related to mesenchyme development, including lung-associated mesenchyme development and coronary artery development (Figure 6C). This result is consistent with the cancer cell identity crisis hypothesis, whereby, during tumorigenesis, cancer cells use epigenetic mechanisms including DNA methylation to silence genes that are specific to the tissue or cell types from which they derive [7], [42]. The DNA methylation differences between the carcinoma component and sarcoma component might indicate that they had a different developmental history during which different developmental genes were targeted for silencing.
Of 342 genes with CADs located in their promoters, elastic microfibril interface located protein (EMILIN1), neurofilament medium peptide (NEFM), and C-type lectin domain family 14 member A (CLEC14A) showed significant hypermethylation and decreased expression in endometrial carcinoma and carcinomatous components of UCS (Supplementary Figure 2, A and B). In contrast, SADs were linked to 261 genes. Plakophilin 3 (PKP3); family with sequence similarity 83, member F (FAM83F); and T-complex 11 (TCP11) were differentially methylated and expressed in two different cancer types (Supplementary Figure 2, C and D).
Methylation-Associated Ectopic Expression of MIR200 in Two Cancer Types
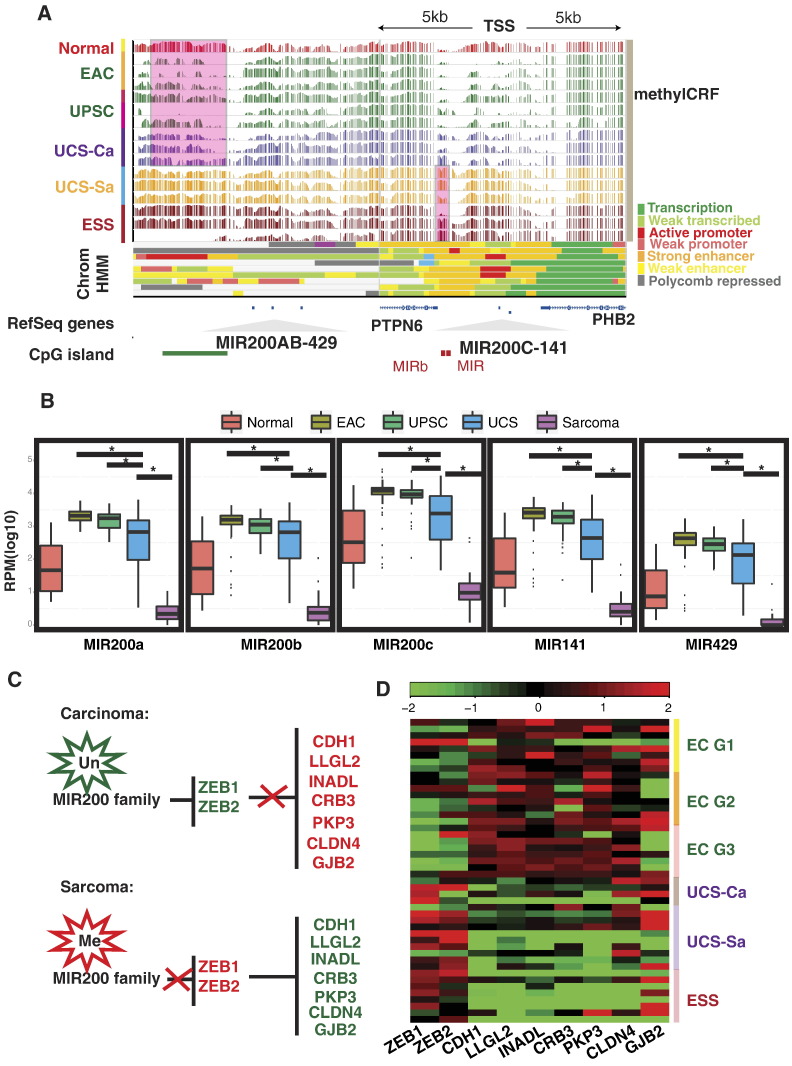
Our DMR analysis revealed a novel connection between epigenetic regulation of miRNAs and the distinction between the sarcomatous and carcinomatous components of UCS. This was highlighted by several DMRs targeting the MIR200 family (MIR200a, MIR200b, MIR200c, MIR141, MIR429) in UCS [43]. We found that CADs were located in MIR200b-MIR200a-MIR429’s promoter region (4 kb upstream of MIR200b), whereas in MIR200c-MIR141, SADs were localized in enhancer regions as predicted by chromHMM [41] (Figure 7A). Accordingly, expression levels of MIR200 family members showed significant differences between pure carcinoma samples and UCS samples (presumably due to a large difference between carcinoma and sarcoma) (Figure 7B). Although TCGA did not have expression data on ESS samples, expression levels of MIR200 of other sarcomas (37 dedifferentiated liposarcomas, 5 undifferentiated pleomorphic sarcomas, 2 myxofibrosarcomas, 50 leiomyosarcomas) were consistent with what we observed in the sarcoma component of UCS (Figure 7B).
Fig. 7.
Differences in MIR200 methylation between carcinoma and sarcoma. (A) Hypomethylation of promoters of MIR200A-MIR200B-MIR429 and hypermethylation of enhancers annotated by chromHMM state of MIR200C-MIR141 between two cancer types. Methylation scores of different uterine cancers are displayed. The chromHMM tracks of nine ENCODE cell lines (H1 ESC, GM12878, K562, HepG2, HUVEC, HMEC, HSMM, NHEK, and NHL) are displayed. (B) Differential expression of MIR200 family in different cancer types using TCGA expression data. (C and D) Proposed model for methylation-induced aberrant expression changes of MIR200 family and downstream pathway. EC represents endometrial cancers of grade 1, 2, or 3. UCSs may be either homologous or heterologous. Homologous UCSs are those whose sarcomatous component is nonspecific or corresponds to normal uterine tissue types such as an ESS or leiomyosarcoma. Heterologous UCSs are those containing an element of a specific nonuterine sarcoma such as rhabdomyosarcoma, chondrosarcoma, or osteosarcoma.
Because DNA hypermethylation predominantly results in loss of gene expression, we hypothesize that increased DNA methylation leads to loss of MIR200 family expression, which would repress genes in the EMT pathway, including ZEB1 and ZEB2, known targets of the MIR200 family [44]. This was supported by altered gene expression in the MIR200 EMT pathway in different cancer types (Figure 7C). To provide additional evidence that MIR200 expression was responsible for different cancer phenotypes, we found that the expression of MIR429, another gene product of the MIR200a-MIR200b-MIR429 pre-MIRNA transcript, was altered in a variety of cancer types, including kidney, breast, colon, lung, and uterine cancers (Supplementary Figure 3). Taken together, these results suggest that the methylation pattern of MIR200 family members is an important epigenetic biomarker to distinguish carcinomatous and sarcomatous components of UCS.
Discussion
Recent studies have demonstrated the importance of DNA methylation in regulating gene expression, and there is considerable evidence that DNA methylation alteration plays important roles in cancer. In this study, we generated the first complete DNA methylome maps of UCS and characterized global and local DNA methylation abnormalities in this disease.
We discovered 3029 DMRs in UCS and validated a subset in a larger and independent cohort of TCGA tumors. These DMRs had consistent methylation differences between cancer and normal tissues. We identified 79 tumor suppressor genes with methylated promoters. Some of the candidate genes we identified have been investigated previously in UCSs. UCSs with high WT1 and high estrogen receptor β expression were shown to have decreased survival, supporting a role for WT1 as a biomarker in this tumor type [45]. However, Coosemans et al. found only 1 of 12 UCSs with nuclear expression of WT1 [46]. Our study thus helps reconcile some of the conflicts in the field and could establish candidate genes for further mechanistic studies and biomarker analysis.
Our study is also the first to combine LCM and modern DNA methylomics to study the carcinomatous and sarcomatous components of UCSs. We identified candidate genes, including PCDH cell adhesion genes, exhibiting component-specific methylation patterns. Our study provides a valuable resource for future investigation to determine the function and importance of the component-specific DMRs in cancer development and progression.
Among other candidate genes with a differential methylation pattern between the carcinomatous and sarcomatous components, we noted that EMILIN1 and CLEC4A harbored carcinoma-associated DMRs in their promoters and exhibited decreased expression in carcinoma. EMILIN1 maintains vascular elastin components and cell–cell interactions [47], whereas CLEC4A is a tumor endothelial marker [48]. According to the protein–protein interaction network [49], CLEC4A and EMILIN1 interacted with each other and both promoted tumor angiogenesis. These findings highlight the possibility that carcinosarcoma may induce specific gene expression patterns to facilitate its interaction with the microenvironment.
Our study also joins a recent effort to investigate the DNA methylation and expression pattern of MIR200 family across different cancer types and normal tissues [43]. We identified component-specific DNA methylation abnormalities of MIR200 in UCS-Ca and in UCS-Sa, as well as in other endometrial tumor types. The expression of MIR200 family members in carcinoma and the apparent loss of expression of MIR200 in sarcoma predict that MIR200 plays a profound role in determining different cancer cell phenotypes within the same tumor.
Recent findings provide clues to the mechanism of genome-wide epigenetic alterations in UCSs. Compared to pure carcinomas, UCS cases were found to have an increase in histone H2A/H2B mutations; furthermore, a majority of UCSs showed amplification of chromosome arm 6p, harboring the HIST1H histone gene cluster, and 1q22, harboring the HIST2H gene cluster [16]. Functional studies showed that at least some of these histone gene mutations caused epithelial to mesenchymal transition in cell culture. Altered histone protein sequences could alter the accessibility of chromatin to methyltransferases as well as to mechanisms of active demethylation [50]. It is not immediately obvious how this mechanism would confer specificity for particular regions to be targeted for differential methylation; indeed, the nonspecificity of the process could account for the variable phenotypes observed in UCSs.
The present study has several limitations. One set of limitations revolves around sample size. Frozen UCS specimens suitable for microdissection were few in our biosample bank once criteria for sample size, tumor viability, and admixture of carcinoma and sarcoma elements were applied. Three samples were microdissected, making it possible to identify recurrent epigenetic features. A larger sample size might make it possible to discover a larger set of features and confirm their recurrent nature.
The purity of material dissected by LCM is difficult to benchmark. Each LCM specimen is expected to be highly enriched in the desired component, but some admixture of another tumor component, benign stroma, or other elements may be present.
For practical reasons, we studied only two components per UCS sample: carcinomatous components histologically resembling high-grade serous cancer, and homologous sarcomatous components. It would be of interest to study multiple components (e.g., rhabdomyosarcomatous and leiomyosarcomatous elements from a single sample) to determine the epigenetic events that allow for heterologous differentiation and to determine the stability of epigenetic alterations across varying sarcoma and carcinoma types within a single tumor. However, the combinatorial complexity of the analysis escalates rapidly if a wider variety of histologies is incorporated.
We have used methylation data and expression data from TCGA samples as a validation and extension set. We did not have sufficient material to derive gene mutation or expression data from our microdissected UCS samples, which could have permitted more in-depth analyses.
Another caveat is that our conclusions depend upon the controls that we have selected. We have analyzed the epigenetic profile of UCS components in comparison with nontumor endometrium, with pure carcinomas other than UCSs (i.e., endometrioid and serous), and with ESS. The common types of endometrial carcinoma are a natural comparison set because UCS is thought to be fundamentally a type 2 carcinoma exhibiting divergent sarcomatous differentiation [51], [52], [53]. It is less obvious whether the sarcomatous component of UCS is best compared to ESS, to uterine leiomyosarcoma (as has been done in some prior studies [24]), or to some other type of sarcoma. ESS is a common type of sarcoma occurring in the uterus, and UCS often has sarcomatous components closely resembling ESS; however, ESS is characterized by a specific translocation [54], which may drive its biology. Ultimately, the premise of comparing UCS to any pure uterine sarcoma represents an implicit assumption that this comparison is meaningful.
In conclusion, in this study, we have used next-generation DNA methylation profiling to establish an epigenomic map of UCS. There are recurrent epigenetic differences between the carcinomatous and sarcomatous components, in keeping with the phenotypic differences between them. The preponderance of hypermethylated DMRs in UCS could suggest a role for hypomethylating agents in clinical practice. Repression of MIR200 family members may be responsible for adoption of the sarcomatous phenotype and could also represent a therapeutic avenue. Future studies should focus on replicating these results in larger cohorts and on testing mechanistic hypotheses based upon them. It would be of interest to extend this methodology to other tumors with biphasic histology, such as metaplastic carcinomas of breast, sarcomatoid carcinoma of the head and neck, and dedifferentiated sarcomas.
The following are the supplementary data related to this article.
Sample information of Roadmap Project.
Supplementary table: Globale DNA methylation changes.
Additional analysis of UCS DMRs in context of TCGA data. (A) Overlap of DMRs with TCGA Infinium probes. (B) Expression of DNMT in UCS. (C) Methylation changes of TSGs in TCGA UCS cohorts. (D) GO analysis of UCS common DMRs by GREAT.
Examples of CADs and SADs. (A and C) Epigenome Browser views of promoters with hypermethylated changes in carcinoma (3 EAC, 3 UPSC, and UCS carcinoma components) and sarcoma (3 UCS sarcoma components and 3 ESSs). (B and D) Expression level of candidate genes representing CAD and SAD using public array data (GSE32507). EC represents endometrial cancers of grade 1, 2, or 3. UCS may be either homologous or heterologous bulk carcinosarcomas without separating two components. Homologous UCSs are those whose sarcomatous component is nonspecific or corresponds to normal uterine tissue types. Heterologous UCSs are those containing an element of a specific non-uterine sarcoma such as rhabdomyosarcoma, chondrosarcoma, or osteosarcoma.
Differential expression of MIR429 in different carcinomas and sarcomas using public array data.
Supplementary Tables
Acknowledgements
This work was supported by American Cancer Society Research Scholar grant RSG-14-049-01-DMC (to T.W.), National Institutes of Health P50 CA134254 (to D.G.M., subawards to T.W. and I.S.H.), and Shanghai Sailing Program 16YF1414900 (to J.L.). T. W. is also supported by National Institutes of Health grants R01HG007354, R01HG007175, R01ES024992, U01CA200060, and U24ES026699.
Footnotes
This work was supported by American Cancer Society Research Scholar grant RSG-14-049-01-DMC (to T.W.), National Institutes of HealthP50 CA134254 (to D.G.M., subawards to T.W. and I.S.H.), and Shanghai Sailing Program16YF1414900 (to J.L.). T. W. is also supported by National Institutes of Health grants R01HG007354, R01HG007175, R01ES024992, U01CA200060, and U24ES026699.
The authors declare no financial conflicts of interest.
References
- 1.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 4.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochem Cell Biol. 2005;83:296–321. doi: 10.1139/o05-036. [DOI] [PubMed] [Google Scholar]
- 6.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, Xing X, Li J, Lowdon RF, Zhou Y, Lin N, Zhang B, Sundaram V, Chiappinelli KB, Hagemann IS. Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers. BMC Genomics. 2014;15:868. doi: 10.1186/1471-2164-15-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saglam O, Husain S, Toruner G. AKT, EGFR, C-ErbB-2, and C-kit expression in uterine carcinosarcoma. Int J Gynecol Pathol. 2013;32:493–500. doi: 10.1097/PGP.0b013e31827fedef. [DOI] [PubMed] [Google Scholar]
- 9.Berton-Rigaud D, Devouassoux-Shisheboran M, Ledermann JA, Leitao MM, Powell MA, Poveda A, Beale P, Glasspool RM, Creutzberg CL, Harter P. Gynecologic Cancer InterGroup (GCIG) consensus review for uterine and ovarian carcinosarcoma. Int J Gynecol Cancer. 2014;24:S55–S60. doi: 10.1097/IGC.0000000000000228. [DOI] [PubMed] [Google Scholar]
- 10.Denschlag D, Thiel FC, Ackermann S, Harter P, Juhasz-Boess I, Mallmann P, Strauss HG, Ulrich U, Horn LC, Schmidt D. Sarcoma of the uterus. Guideline of the DGGG (S2k-level, AWMF Registry No. 015/074, August 2015) Geburtshilfe Frauenheilkd. 2015;75:1028–1042. doi: 10.1055/s-0035-1558120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Weinberg RA. Epithelial–mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Samarnthai N, Hall K, Yeh IT. Molecular profiling of endometrial malignancies. Obstet Gynecol Int. 2010;2010:162363. doi: 10.1155/2010/162363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada H, Enomoto T, Fujita M, Yoshino K, Nakashima R, Kurachi H, Haba T, Wakasa K, Shroyer KR, Tsujimoto M. Molecular evidence that most but not all carcinosarcomas of the uterus are combination tumors. Cancer Res. 1997;57:5379–5385. [PubMed] [Google Scholar]
- 14.Jones S, Stransky N, McCord CL, Cerami E, Lagowski J, Kelly D, Angiuoli SV, Sausen M, Kann L, Shukla M. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat Commun. 2014;5:5006. doi: 10.1038/ncomms6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McConechy MK, Hoang LN, Chui MH, Senz J, Yang W, Rozenberg N, Mackenzie R, McAlpine JN, Huntsman DG, Clarke BA. In-depth molecular profiling of the biphasic components of uterine carcinosarcomas. J Pathol Clin Res. 2015;1:173–185. doi: 10.1002/cjp2.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S, Bellone S, Lopez S, Thakral D, Schwab C, English DP, Black J, Cocco E, Choi J, Zammataro L. Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial–mesenchymal transition. Proc Natl Acad Sci U S A. 2016;113:12238–12243. doi: 10.1073/pnas.1614120113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Zhang B, Xing X, Wang T. Combining MeDIP-seq and MRE-seq to investigate genome-wide CpG methylation. Methods. 2015;72:29–40. doi: 10.1016/j.ymeth.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [Article published online before print in May 2002] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiyoda T, Tsuda H, Tanaka H, Kataoka F, Nomura H, Nishimura S, Takano M, Susumu N, Saya H, Aoki D. Expression profiles of carcinosarcoma of the uterine corpus-are these similar to carcinoma or sarcoma? Genes Chromosomes Cancer. 2012;51:229–239. doi: 10.1002/gcc.20947. [DOI] [PubMed] [Google Scholar]
- 25.Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens M, Cheng JB, Li D, Xie M, Hong C, Maire CL, Ligon KL, Hirst M, Marra MA, Costello JF. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 2013;23:1541–1553. doi: 10.1101/gr.152231.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B, Zhou Y, Lin N, Lowdon RF, Hong C, Nagarajan RP, Cheng JB, Li D, Stevens M, Lee HJ. Functional DNA methylation differences between tissues, cell types, and across individuals discovered using the M&M algorithm. Genome Res. 2013;23:1522–1540. doi: 10.1101/gr.156539.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sproul D, Kitchen RR, Nestor CE, Dixon JM, Sims AH, Harrison DJ, Ramsahoye BH, Meehan RR. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol. 2012;13:R84. doi: 10.1186/gb-2012-13-10-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Faveri LE, Hurst CD, Platt FM, Taylor CF, Roulson JA, Sanchez-Carbayo M, Knowles MA, Chapman EJ. Putative tumour suppressor gene necdin is hypermethylated and mutated in human cancer. Br J Cancer. 2013;108:1368–1377. doi: 10.1038/bjc.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimada H, Hashimoto Y, Nakada A, Shigeno K, Nakamura T. Accelerated generation of human induced pluripotent stem cells with retroviral transduction and chemical inhibitors under physiological hypoxia. Biochem Biophys Res Commun. 2012;417:659–664. doi: 10.1016/j.bbrc.2011.11.111. [DOI] [PubMed] [Google Scholar]
- 34.Simmons CD, Pabona JM, Heard ME, Friedman TM, Spataro MT, Godley AL, Simmen FA, Burnett AF, Simmen RC. Kruppel-like factor 9 loss-of-expression in human endometrial carcinoma links altered expression of growth-regulatory genes with aberrant proliferative response to estrogen. Biol Reprod. 2011;85:378–385. doi: 10.1095/biolreprod.110.090654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK, Liloglou T. Hypomethylation of retrotransposable elements correlates with genomic instability in non–small cell lung cancer. Int J Cancer. 2009;124:81–87. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 36.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2016;164:1073. doi: 10.1016/j.cell.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimoto K, Suzuki AM, Dos Santos A, Desterke C, Collino A, Ghisletti S, Braun E, Bonetti A, Fort A, Qin XY. CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus-induced tumors. Genome Res. 2015;25:1812–1824. doi: 10.1101/gr.191031.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dallosso AR, Hancock AL, Szemes M, Moorwood K, Chilukamarri L, Tsai HH, Sarkar A, Barasch J, Vuononvirta R, Jones C. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' tumor. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz-Martin J, Diaz-Lopez A, Moreno-Bueno G, Castilla MA, Rosa-Rosa JM, Cano A, Palacios J. A core microRNA signature associated with inducers of the epithelial-to-mesenchymal transition. J Pathol. 2014;232:319–329. doi: 10.1002/path.4289. [DOI] [PubMed] [Google Scholar]
- 44.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guntupalli SR, Cao D, Shroff R, Gao F, Menias C, Stewart Massad L, Powell MA, Mutch DG, Thaker PH. Wilms' tumor 1 protein and estrogen receptor beta expression are associated with poor outcomes in uterine carcinosarcoma. Ann Surg Oncol. 2013;20:2373–2379. doi: 10.1245/s10434-012-2838-9. [DOI] [PubMed] [Google Scholar]
- 46.Coosemans A, Nik SA, Caluwaerts S, Lambin S, Verbist G, Van Bree R, Schelfhout V, de Jonge E, Dalle I, Jacomen G. Upregulation of Wilms' tumour gene 1 (WT1) in uterine sarcomas. Eur J Cancer. 2007;43:1630–1637. doi: 10.1016/j.ejca.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Zanetti M, Braghetta P, Sabatelli P, Mura I, Doliana R, Colombatti A, Volpin D, Bonaldo P, Bressan GM. EMILIN-1 deficiency induces elastogenesis and vascular cell defects. Mol Cell Biol. 2004;24:638–650. doi: 10.1128/MCB.24.2.638-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mura M, Swain RK, Zhuang X, Vorschmitt H, Reynolds G, Durant S, Beesley JF, Herbert JM, Sheldon H, Andre M. Identification and angiogenic role of the novel tumor endothelial marker CLEC14A. Oncogene. 2012;31:293–305. doi: 10.1038/onc.2011.233. [DOI] [PubMed] [Google Scholar]
- 49.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765–780. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 51.Sreenan JJ, Hart WR. Carcinosarcomas of the female genital tract. A pathologic study of 29 metastatic tumors: further evidence for the dominant role of the epithelial component and the conversion theory of histogenesis. Am J Surg Pathol. 1995;19:666–674. [PubMed] [Google Scholar]
- 52.Ferguson SE, Tornos C, Hummer A, Barakat RR, Soslow RA. Prognostic features of surgical stage I uterine carcinosarcoma. Am J Surg Pathol. 2007;31:1653–1661. doi: 10.1097/PAS.0b013e3181161ba3. [DOI] [PubMed] [Google Scholar]
- 53.Taylor NP, Zighelboim I, Huettner PC, Powell MA, Gibb RK, Rader JS, Mutch DG, Edmonston TB, Goodfellow PJ. DNA mismatch repair and TP53 defects are early events in uterine carcinosarcoma tumorigenesis. Mod Pathol. 2006;19:1333–1338. doi: 10.1038/modpathol.3800654. [DOI] [PubMed] [Google Scholar]
- 54.Lee CH, Nucci MR. Endometrial stromal sarcoma—the new genetic paradigm. Histopathology. 2015;67:1–19. doi: 10.1111/his.12594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample information of Roadmap Project.
Supplementary table: Globale DNA methylation changes.
Additional analysis of UCS DMRs in context of TCGA data. (A) Overlap of DMRs with TCGA Infinium probes. (B) Expression of DNMT in UCS. (C) Methylation changes of TSGs in TCGA UCS cohorts. (D) GO analysis of UCS common DMRs by GREAT.
Examples of CADs and SADs. (A and C) Epigenome Browser views of promoters with hypermethylated changes in carcinoma (3 EAC, 3 UPSC, and UCS carcinoma components) and sarcoma (3 UCS sarcoma components and 3 ESSs). (B and D) Expression level of candidate genes representing CAD and SAD using public array data (GSE32507). EC represents endometrial cancers of grade 1, 2, or 3. UCS may be either homologous or heterologous bulk carcinosarcomas without separating two components. Homologous UCSs are those whose sarcomatous component is nonspecific or corresponds to normal uterine tissue types. Heterologous UCSs are those containing an element of a specific non-uterine sarcoma such as rhabdomyosarcoma, chondrosarcoma, or osteosarcoma.
Differential expression of MIR429 in different carcinomas and sarcomas using public array data.
Supplementary Tables