Abstract
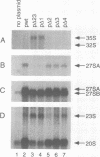
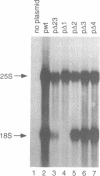
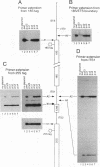
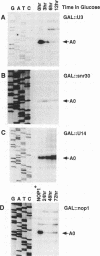
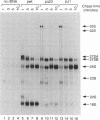
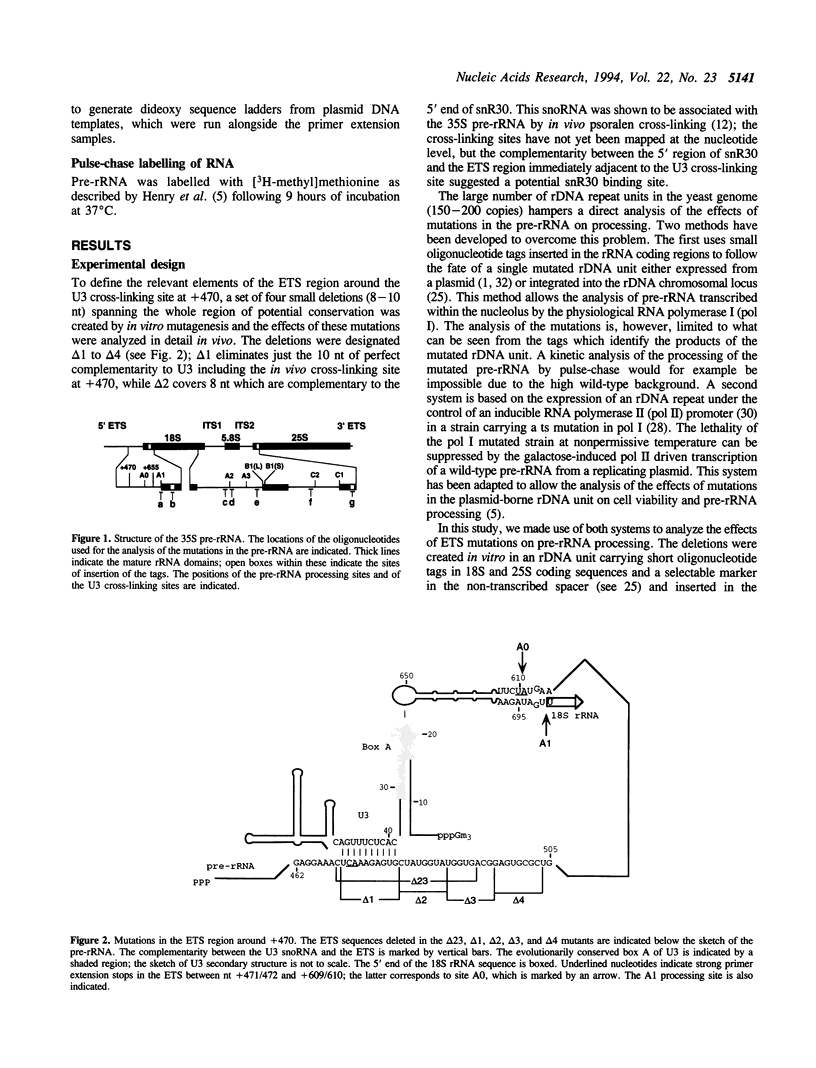
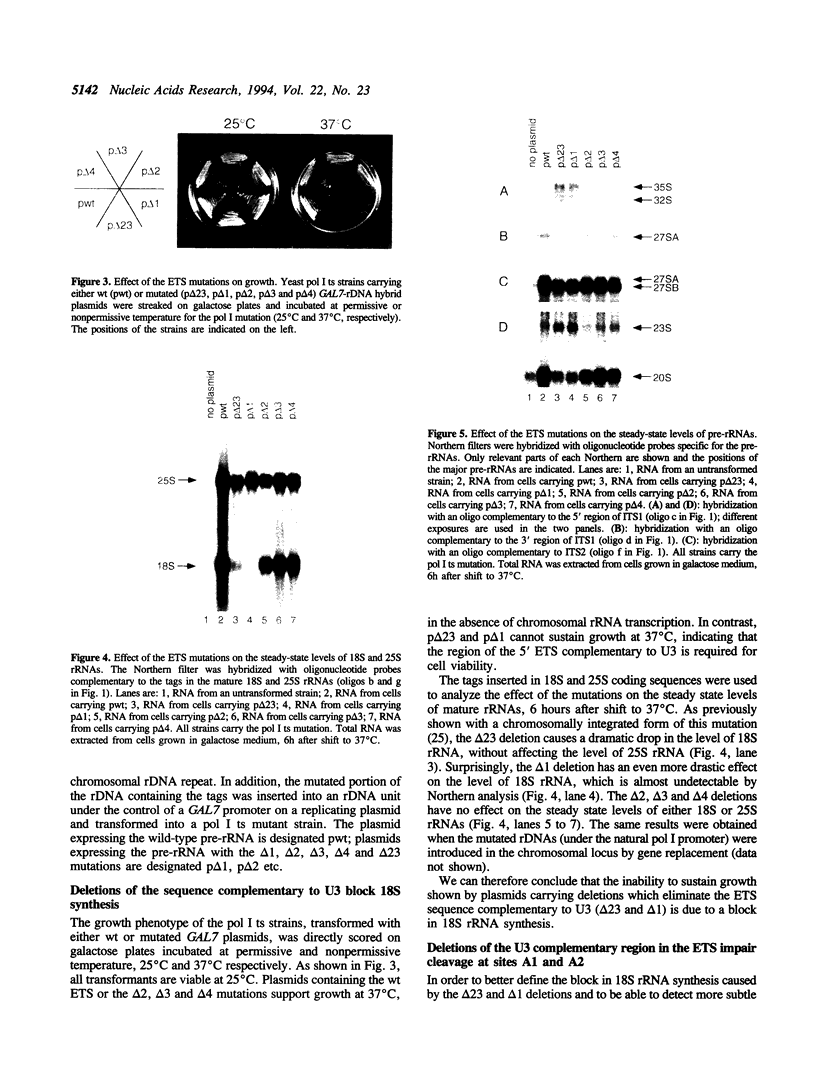
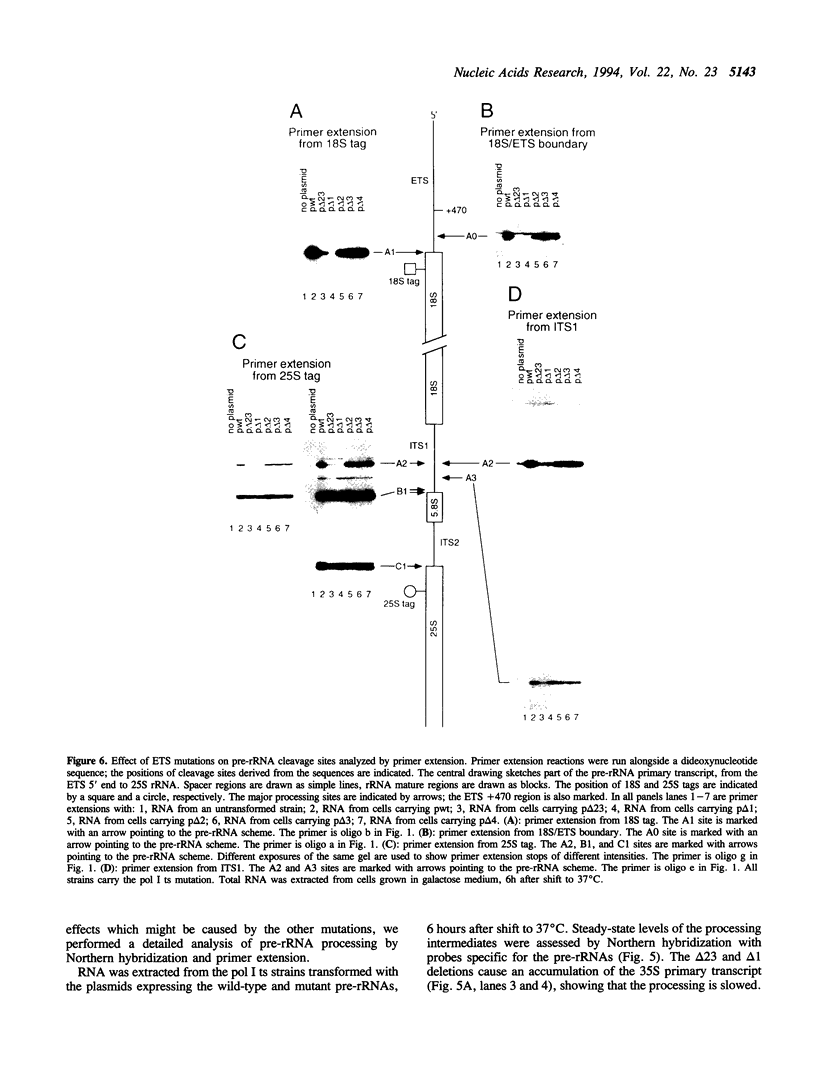
The small nucleolar RNA U3 is essential for viability in yeast. We have previously shown that U3 can be cross-linked in vivo to the pre-rRNA in the 5' external transcribed spacer (ETS), at +470. This ETS region contains 10 nucleotides of perfect complementarity to U3. In a genetic background where the mutated rDNA is the only transcribed rDNA repeat, the deletion of the 10 nt complementary to U3 is lethal. Cells lacking the U3 complementary sequence in pre-rRNA fail to accumulate 18S rRNA: pre-rRNA processing is inhibited at sites A0 in the 5' ETS, A1 at the 5' end of 18S rRNA and A2 in ITS1. We show here that effects on processing at site A0 are specific for U3 and its associated proteins and are not seen on depletion of other snoRNP components. The deletion of the sequence complementary to U3 in the ETS therefore mimics all the known effects of the depletion of U3 in trans. This indicates that we have identified an essential U3 binding site on pre-rRNA, required in cis for the maturation of 18S rRNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992 Apr;11(4):1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N., Kass S., Sollner-Webb B. Sequence organization and RNA structural motifs directing the mouse primary rRNA-processing event. Mol Cell Biol. 1991 Jan;11(1):458–467. doi: 10.1128/mcb.11.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M. J., Maxwell E. S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993 Apr;18(4):131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- Furlong J. C., Forbes J., Robertson M., Maden B. E. The external transcribed spacer and preceding region of Xenopus borealis rDNA: comparison with the corresponding region of Xenopus laevis rDNA. Nucleic Acids Res. 1983 Dec 10;11(23):8183–8196. doi: 10.1093/nar/11.23.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J. P., Lehtonen H., Caizergues-Ferrer M., Amalric F., Tollervey D., Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992 Feb;11(2):673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y., Wood H., Morrissey J. P., Petfalski E., Kearsey S., Tollervey D. The 5' end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994 May 15;13(10):2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. M., Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991 Dec;10(13):4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Tollervey D., Hurt E. C. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 1993 Jun;12(6):2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987 Aug;7(8):2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Sollner-Webb B. The first pre-rRNA-processing event occurs in a large complex: analysis by gel retardation, sedimentation, and UV cross-linking. Mol Cell Biol. 1990 Sep;10(9):4920–4931. doi: 10.1128/mcb.10.9.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J. A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990 Mar 23;60(6):897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Landt O., Grunert H. P., Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990 Nov 30;96(1):125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Mitchell P., Petfalski E., Séraphin B., Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994 Jun 15;8(12):1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Maser R. L., Calvet J. P. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5' external transcribed spacer of pre-ribosomal-RNA. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6523–6527. doi: 10.1073/pnas.86.17.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan S., Bachellerie J. P. Structure and organization of mouse U3B RNA functional genes. J Biol Chem. 1988 Dec 25;263(36):19461–19467. [PubMed] [Google Scholar]
- Morrissey J. P., Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993 Apr;13(4):2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougey E. B., O'Reilly M., Osheim Y., Miller O. L., Jr, Beyer A., Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993 Aug;7(8):1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- Mougey E. B., Pape L. K., Sollner-Webb B. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5' external transcribed spacer of Xenopus precursor rRNA. Mol Cell Biol. 1993 Oct;13(10):5990–5998. doi: 10.1128/mcb.13.10.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W., Boon K., van der Sande C. A., van Heerikhuizen H., Planta R. J. Functional analysis of transcribed spacers of yeast ribosomal DNA. EMBO J. 1990 Dec;9(12):3989–3996. doi: 10.1002/j.1460-2075.1990.tb07620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W., Venema J., van der Linden G., van Heerikhuizen H., Klootwijk J., Planta R. J. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989 Feb;9(2):551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Dodd J., Carles C., Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol. 1993 Jan;13(1):114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K. A., Steitz J. A. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987 Aug;7(8):2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer L. D., Miller O. L., Jr Electron microscopic study of Saccharomyces cerevisiae rDNA chromatin replication. Mol Cell Biol. 1986 Apr;6(4):1148–1157. doi: 10.1128/mcb.6.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R., Gerbi S. A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990 Jul;9(7):2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B. Novel intron-encoded small nucleolar RNAs. Cell. 1993 Nov 5;75(3):403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- Stroke I. L., Weiner A. M. The 5' end of U3 snRNA can be crosslinked in vivo to the external transcribed spacer of rat ribosomal RNA precursors. J Mol Biol. 1989 Dec 5;210(3):497–512. doi: 10.1016/0022-2836(89)90126-5. [DOI] [PubMed] [Google Scholar]
- Ségault V., Mougin A., Grégoire A., Banroques J., Branlant C. An experimental study of Saccharomyces cerevisiae U3 snRNA conformation in solution. Nucleic Acids Res. 1992 Jul 11;20(13):3443–3451. doi: 10.1093/nar/20.13.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M. P., Dahlberg J. E. Retention and 5' cap trimethylation of U3 snRNA in the nucleus. Science. 1994 May 13;264(5161):959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987 Dec 20;6(13):4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991 Mar;10(3):573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh L. C., Lee J. C. Structure analysis of the 5' external transcribed spacer of the precursor ribosomal RNA from Saccharomyces cerevisiae. J Mol Biol. 1992 Dec 5;228(3):827–839. doi: 10.1016/0022-2836(92)90867-j. [DOI] [PubMed] [Google Scholar]
- van Nues R. W., Rientjes J. M., van der Sande C. A., Zerp S. F., Sluiter C., Venema J., Planta R. J., Raué H. A. Separate structural elements within internal transcribed spacer 1 of Saccharomyces cerevisiae precursor ribosomal RNA direct the formation of 17S and 26S rRNA. Nucleic Acids Res. 1994 Mar 25;22(6):912–919. doi: 10.1093/nar/22.6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sande C. A., Kwa M., van Nues R. W., van Heerikhuizen H., Raué H. A., Planta R. J. Functional analysis of internal transcribed spacer 2 of Saccharomyces cerevisiae ribosomal DNA. J Mol Biol. 1992 Feb 20;223(4):899–910. doi: 10.1016/0022-2836(92)90251-e. [DOI] [PubMed] [Google Scholar]