Abstract
Ochrobactrum anthropi SUBG007 was isolated from the fruit of Prunus dulcis in Rajkot (22.30°N, 70.78°E), Gujarat, India. Here we present the 4.37 Mb genome sequence strain SUBG007, which may provide the genetic information for the application in environment pollution degradation and agriculture field. The strain also posses many genes cluster which involved in production of important secondary metabolites. The nucleotide sequence of this genome was deposited into NCBI GenBank under the accession LUAY00000000.
Keywords: O. anthropi, Plant pathogen, Xenobiotic compounds, Secondary metabolites
Specifications
| Organism | Ochrobactrum anthropi |
| Strain | SUBG007 |
| Sequencer | Ion Torrent PGM |
| Data format | Processed |
| Experimental factor | Microbial strain |
| Experimental features | Whole genome sequencing of O. anthropi SUBG007 |
| Sample source location | Rajkot, Gujarat, India |
1. Direct link to deposited data
https://www.ncbi.nlm.nih.gov/nuccore/LUAY00000000
Ochrobactrum anthropi is a gram-negative, aerobic, non-fermentative bacterium from the family Brucellaceae belonging to alphaproteobacterial, order Rhizobiales [1]. This organism strains are broadly distributed from diverse habitats including soil, plant and their rhizosphere, environment, waste water, animals and humans [2], [3]. O. anthropi is known to produce important enzymes that degrade the pesticides, petroleum waste and pollutant [4]. The complete genome sequence of O. anthropi strain SUBG007 is reported here and will provide genomic information on the specific properties of this strain.
2. Experimental design, materials and method
O. anthropi SUBG007 strain was isolated from infected fruit of Prunus dulcis. Genomic DNA was extracted from the strain Ochrobactrum anthropi SUBG007 according to [5]. Whole-genome shotgun sequencing of O. anthropi SUBG007 was performed using a high throughput ion torrent personal genome machine with ion torrent server (torrent suite v3.2), and with 50 × coverage was obtained. De novo assembly was performed using Velvet assembler (v. 1.2.10). The annotation of the genome was performed using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) (http://www.ncbi.nim.nih.gov/genome/annotation_prok/) utilizing GeneMark, Glimmer, and tRNAscan-SE tools [6] and functional annotation was carried out using the Rapid Annotations using subsystems Technology (RAST) server with the seed database [7]. Preliminary reference-based annotation using PATRIC was carried out to identify conserved pathways [8].
3. Data description
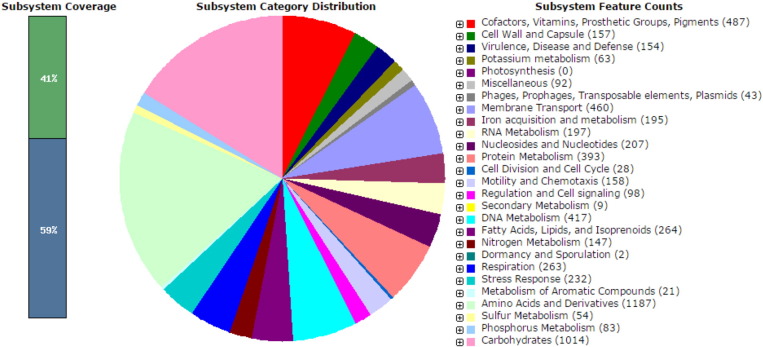
The total length of genome was estimated to be 4.37595 bp and contained 21 tRNA, 7rRNA and 5 ncRNA. Fig. 1 presents an overview of the count of each subsystem feature and the subsystem coverage of RAST server annotation. According to RAST annotated, O. santhropi having very important Type I and Type IV secretion system (T4SS), which are essential for the pathogenicity. Type IV systems are multicomponent, transenvelope complexes that translocate proteins and nucleoprotein complexes from donor cells to recipient cells in processes related to bacterial conjugation.
Fig. 1.
Subsystem distribution of O. anthropi strain SUBG007 (based on RAST annotation server).
The annotation revealed that genome of strain SUBG007 contains a large number of genes responsible for the degradation of xenobiotic compounds. The xenobiotic compound are involved gamma-Hexachlorocyclohexane, 1,1,1-Trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) degradation, benzoate degradation via hydroxylation, bisphenol, biphenyl, toluene and xylene, 1-2-methylnaphthalene, tetrachloroethene, naphthalene, anthracene, 1,4 Dichlorobenzene, ethylbenzene, styrene and caprolactam. The genome SUBG007 was contained the gene cluster related with the biosynthesis of some secondary metabolites like pucimycin, novobiocin, vancomycin, penicillin, puroycin, cephalosporium, flavonoid, phenyl propanoid and isoquinoline alkaloid. Interestingly genome also posses the key genes involved, IAA secretion and nitrogen metabolism genes.
Availability of the genome sequence SUBG007 provides opportunities for biotechnological exploitation of genome features related to the biosynthetic studies of some important secondary metabolite and provide promising application in bioremediation. This sequence will lead to a better understanding of its evolution and the development of biotechnological applications and the mechanisms of pathogenicity.
4. Nucleotide sequence accession number
The SUBG007 draft genome sequence has been deposited at GenBank under the accession LUAY00000000.
Acknowledgements
The authors are thankful to Department of Biosciences, Saurashtra University and Plant Biotechnology & Genetic Engineering Programme, Gujarat State Government for lab facility, India.
Contributor Information
Kiran S. Chudasama, Email: kiranchudasama@gmail.com.
Vrinda S. Thaker, Email: thakervs@gmail.com.
References
- 1.Scholz H.C., Pfeffer M., Witte A., Neubauer H., Al Dahouk S., Wernery U., Tomaso H. Specific detection and differentiation of Ochrobactrum anthropi, Ochrobactrum intermedium and Brucella spp. by a multi-primer PCR that targets the recA gene. J. Med. Microbiol. 2008;57:64–71. doi: 10.1099/jmm.0.47507-0. [DOI] [PubMed] [Google Scholar]
- 2.Bathe S., Achouak W., Hartmann A., Heulin T., Schloter M., Lebuhn M. Genetic and phenotypic microdiversity of Ochrobactrum spp. FEMS Microbiol. Ecol. 2005;56:272–280. doi: 10.1111/j.1574-6941.2005.00029.x. [DOI] [PubMed] [Google Scholar]
- 3.Ozdemir D., Soppacacl Z., Sahin I., Bicik Z., Sencan I. Ochrobactrum anthropi endocarditis and septic shock in a patient with no prosthetic valve or rheumatic heart disease: case repot and review of the literature Jpn. J. Infect. Dis. 2006;59:264–265. [PubMed] [Google Scholar]
- 4.Ramasamy S., Mathiyalagan P., Chandran P. Characterization and optimization of EPS producing and diesel oil-degrading Ochrobactrum anthropi MP3 isolated from refinery waste water. Pet. Sci. 2014;11:439–445. [Google Scholar]
- 5.Chudasama K.S., Thaker V.S. Biological control of phytopathogenic bacteria Pantoea agglomerans and Erwinia Chrysanthemi using 100 essential oils. Arch. Phytopathol. Plant Protect. 2014;47:2221–2232. [Google Scholar]
- 6.Disz T., Akhter S., Cuevas D., Olson R., Overbeek R., Vonstein V., Stevens R., Edwards R.A. Accessing the SEED genome databases via web services API: tools for programmers. BMC Bioinf. 2013;11:319. doi: 10.1186/1471-2105-11-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wattam A.R., Abraham D., Dalay O., Disz T.L., Driscoll T., Gabbard J.L. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:581–591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]