Abstract
Premise of the study:
We tested PCR amplification of 91 low-copy nuclear gene loci in taxa from Sapindales using primers developed for Bursera simaruba (Burseraceae).
Methods and Results:
Cross-amplification of these markers among 10 taxa tested was related to their phylogenetic distance from B. simaruba. On average, each Sapindalean taxon yielded product for 53 gene regions (range: 16–90). Arabidopsis thaliana (Brassicales), by contrast, yielded product for two. Single representatives of Anacardiaceae and Rutacaeae yielded 34 and 26 products, respectively. Twenty-six primer pairs worked for all Burseraceae species tested if highly divergent Aucoumea klaineana is excluded, and eight of these amplified product in every Sapindalean taxon.
Conclusions:
Our study demonstrates that customized primers for Bursera can amplify product in a range of Sapindalean taxa. This collection of primer pairs, therefore, is a valuable addition to the toolkit for nuclear phylogenomic analyses of Sapindales and warrants further investigation.
Keywords: Anacardiaceae, Burseraceae, low-copy nuclear genes, microfluidic PCR, Rutaceae
Low-copy nuclear gene regions offer increased phylogenetic utility for species- and population-level studies of plants as compared to chloroplast and nuclear ribosomal markers (Zimmer and Wen, 2012), yet sampling these regions remains challenging due to the dearth of universal primers and barriers to sequencing whole or partial nuclear genomes from multiple individuals. Consequently, assessing the phylogenetic limits of custom-designed target sequences or primers for low-copy nuclear gene regions is critical to fully realizing their broader impacts for advancing plant systematics. We report the results of a cross-amplification study incorporating primers for 91 low-copy nuclear gene loci created by Gostel et al. (2015) for species-level phylogenetics of Malagasy Commiphora Jacq. (Burseraceae). Primers for these markers were developed using genomic resources from two rosid orders by mapping sequence data from a transcriptome of Bursera simaruba (L.) Sarg. (Burseraceae; Sapindales) (Matasci et al., 2014) to 950 putative low- or single-copy nuclear gene loci of Arabidopsis thaliana (L.) Heynh. (Brassicaceae; Brassicales) (Duarte et al., 2010). Gostel et al. (2015) further optimized the primers for microfluidic PCR-based target enrichment, a method that allows simultaneous and cost-effective amplification of multiple loci (Blow, 2009; Uribe-Convers et al., 2016).
We tested cross-amplification of these primers using 10 taxa that have varying phylogenetic distances from B. simaruba within Sapindales and included A. thaliana as the outermost limit of the survey. Sapindales is a widespread group that includes ca. 6700 species within nine families (Angiosperm Phylogeny Group, 2016) (Fig. 1). Molecular phylogenies of this order often lack sufficient phylogenetic support along their backbone as well as at the species level (e.g., Fine et al., 2014; Grudinski et al., 2014), thus our understanding of Sapindalean systematics could benefit from an expanded phylogenetic toolkit such as that provided by the Gostel et al. (2015) primers.
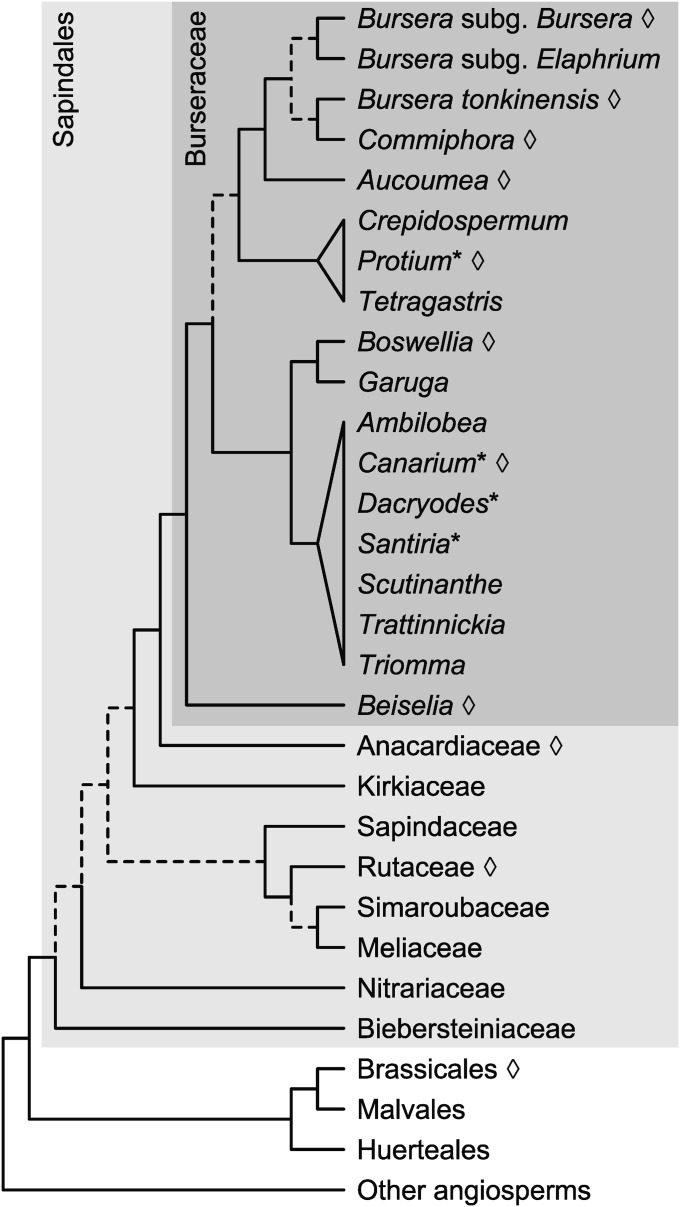
Fig. 1.
Phylogeny of Sapindalean lineages condensed from Wang et al. (2009), Weeks et al. (2014), and Muellner-Riehl et al. (2016); nodes having low or conflicting support are indicated by dashed branches. Lineages sampled by the current study are noted by open diamonds. Generalized generic phylogeny of Burseraceae does not depict Rosselia or Pseudodacryodes, which have not been included in any molecular phylogenetic analysis; paraphyletic genera are indicated by asterisks.
METHODS AND RESULTS
Taxonomic sampling and molecular methods
Appendix 1 contains accession information for the 11 taxa sampled; Fig. 1 displays their phylogenetic relationships. Bursera simaruba (Bursera Jacq. ex L. subgenus Bursera) and C. grandifolia Engl. were included as positive controls; prior work has shown that all or most of the custom-designed primers amplify PCR product in these two species (Gostel et al., 2015). For experimental taxa, we included B. tonkinensis Guillaumin, which is sister to Commiphora (Weeks and Simpson, 2007), as well as Aucoumea Pierre, the monotypic genus sister to Bursera and Commiphora (Weeks et al., 2014). One species from each of Boswellia Roxb. ex Colebr., Canarium L., and Protium Burm. f. were included, as well as Beiselia Forman, the monotypic genus sister to all other Burseraceae (Weeks et al., 2014). We included one species of Anacardiaceae, the family that is sister to Burseraceae (Weeks et al., 2014), and one species of Rutaceae, which represents the Sapindalean clade sister to Burseraceae–Anacardiaceae–Kirkiaceae (Muellner-Riehl et al., 2016). Arabidopsis thaliana (Brassicales) was included because its genomic resources were used in primer design and can test the applicability of these primers to other closely related rosid lineages (Wang et al., 2009).
Whole genomic DNA was extracted from taxa using the FastPrep FastDNA Spin Kit (Bio101 Systems, La Jolla, California, USA) or the cetyltrimethylammonium bromide (CTAB) method (Weeks et al., 2005). Primer development for the 91 markers is detailed by Gostel et al. (2015); primer sequences are listed in Table 1. Markers were amplified via PCR in 15-μL reactions including: 0.15 μL of forward and reverse primers (50 μM), 0.75 μL spermidine (4 mM), 7.5 μL GoTaq Green Master Mix (Promega Corporation, Madison, Wisconsin, USA), 5.6 μL nuclease-free water, and 1 μL genomic DNA (0.1–25.8 ng/μL). Markers that failed to amplify for B. simaruba and C. grandifolia were then trialed using reaction chemistry based on that recommended for microfluidic PCR-based target enrichment including: 0.15 μL of forward and reverse primers (50 μM); FastStart High Fidelity PCR System reagents (Roche Diagnostics, Mannheim, Germany), composed of 1.5 μL FastStart High Fidelity Reaction Buffer without MgCl2 (10× concentration), 2.7 μL MgCl2 (25 mM), 0.75 μL DMSO, 1.2 μL Nucleotide Mix (10 mM), 0.15 μL FastStart High Fidelity Enzyme Blend (5 U/μL); 0.75 μL Loading Reagent (Fluidigm Corporation, San Francisco, California, USA); 6.8 μL nuclease-free water; and 1 μL genomic DNA.
Table 1.
Primer pair sequences and validation results by taxon.
| GenBank accession no.b | |||||||||||||||
| Locus IDa | Primer sequences (5′−3′)a | B. simaruba | C. grandifolia | Amplicon length range among all taxa | Arabidopsis thaliana | Aucoumea klaineana | Beiselia mexicana | Boswellia neglecta | Bursera simaruba | Bursera tonkinensis | Canarium pilosum | Commiphora grandifolia | Phellodendron amurense | Protium guianense | Schinus fasciculatus |
| AT3G54460c | F: GGACACACCCTTGGCTCTAG | KX767982 | KX767983 | 270–290 | X | X | X | X | X | X | X | X | |||
| R: CTCCCATGACTTTTGGTTCTGTC | |||||||||||||||
| AT2G04620 | F: TCCACCATATTTTGAGTGAGAGGAA | KX76792 | KX767929 | 420–520 | X | X | X | X | X | X | X | X | X | ||
| R: AATGGGAGTGGGAATGAGAATGTG | |||||||||||||||
| AT4G37510c | F: TTCATTTTGAGACCTCCATTAGATGAC | KX768000 | 280 | X | X | X | X | X | X | X | X | X | |||
| R: GCTTAGCCGGATTATCGTCTCC | |||||||||||||||
| AT3G22660c | F: AGATGAAGATGTGAAATTGGTTGAACC | KX767974 | KX767975 | 450 | X | X | X | X | X | X | X | ||||
| R: TTTCTGCTTAGCTCTCTCTTTCATCT | |||||||||||||||
| AT1G21840c | F: TGTTGGAGAAGTTGAAGAGAGAGG | 630–640 | X | X | X | X | X | X | X | X | |||||
| R: CACCATTTATCCCAACCTCCTGAA | |||||||||||||||
| AT2G04740d | F: CAAACTCCAAAAACCCTAAACCGG | KX767930 | KX767931 | 460–590 | X | X | X | X | X | X | X | ||||
| R: TCAAAAGCCTTCAAAAGCTTCCTC | |||||||||||||||
| AT4G14605d | F: CTTCTCACTCATAGCAGGCAGAAG | KX767986 | KX767987 | 510–580 | X | X | X | X | X | X | X | ||||
| R: CTTCTTCACAGCCTTATCAAAGTCA | |||||||||||||||
| AT4G19900c | F: GTTCTCTGAGACGATTGAGCTTGA | KX767990 | KX767991 | 350–420 | X | X | X | X | X | X | X | X | |||
| R: CTTGTAGAGAGCAGCAAGTCGG | |||||||||||||||
| AT4G29590 | F: GAGCAATTCCCCTTCAAAGAGGAR: GTGCTTGTATCCTTTTGGGTAATGG | KX767994, KX767996 | KX767995, KX767997 | 490 | X | X | X | X | X | X | |||||
| AT5G04910 | F: TAAGAGTCCAACAGAGCATGAGTG | KX768005 | KX768006 | 260 | X | X | X | X | |||||||
| R: TAAAAGAATGATGTCACTCAGCTTCG | |||||||||||||||
| AT3G15110e | F: CTCACTGGTGCCATATCTGTCTTC | 1560 | X | ||||||||||||
| R: ATTCTCTGTACCTTTGCTTCTGGA | |||||||||||||||
| AT1G18060d | F: AACAAGAAAGGTTGCAGTAGAGGA | KX767902 | KX767903 | 740–930 | X | X | X | X | X | X | |||||
| R: GCTTGGCTCTCTGTCATCTTTTTG | |||||||||||||||
| AT2G03667c | F: CTAGTTGGCTCTGGTGCTGATG | KX767926 | KX767927 | 590 | X | X | X | X | X | X | X | ||||
| R: CACAAAGGAATATCAAGCAAAGTCCT | |||||||||||||||
| AT2G40760 | F: GGTGTATCATCTGGAAGGGGG | KX768007 | KX768008 | 400 | X | X | X | X | X | X | X | ||||
| R: CGCTCTCGCCCTCTCTTTC | |||||||||||||||
| AT2G20790c,g | F: CCAATTGTCAATGGTCTCTGAAGATG | KX767940 | KX767941 | 320–350 | X | X | X | X | X | X | X | X | X | X | |
| R: CCATGGTGCAAATTTAACTGTTCC | |||||||||||||||
| AT2G36740 | F: AGTCCACAAGAACTGCAGTGAT | 640–810 | X | X | X | X | X | X | |||||||
| R: CATCCTTTGAGAAATACCGTATCTGT | |||||||||||||||
| AT3G01380d | F: AATCATCATAATAGGGGCAGCCGR: CCAAGAAATATAGAAGTTAGTCGGGAC | KX767958, KX767960 | KX767959, KX767961 | 530–930 | X | X | X | X | X | X | X | ||||
| AT3G10400 | F: AGAAGAAAAAGACTAACAGTGACAGC | KX767966 | KX767967 | 340 | X | X | X | X | X | ||||||
| R: CGGTCTTTGAGCACGCTGA | |||||||||||||||
| AT1G59990d | F: GCTACTTGGTTCCTTTAATTGATAAGCR: TGACACCACGAATAAAATCCAAGC | KX767908, KX767910 | KX767909, KX767911 | 450–510 | X | X | X | X | X | X | X | X | |||
| AT2G22370Bc | F: AACCCACATGGACTGTTAAACATG | KX767944 | KX767945 | 610–780 | X | X | X | X | X | X | X | X | |||
| R: CATCAGACATAAGAGATGCAGCAG | |||||||||||||||
| AT1G31780c,g | F: CTTGTCCTTGGGTTACTTGATCCAR: TTGTGGGTCTCAATGATTTCAAGC | KX767904, KX767906 | KX767905, KX767907 | 520–570 | X | X | X | X | X | X | X | X | X | X | |
| AT1G31780 (INT)c,g | F: CTTGTCCTTGGGTTACTTGATCCA | 380–832 | X | Xh | X | X | X | X | X | X | X | ||||
| R: GGACCCAAAGTGTACTACAGAGAG | |||||||||||||||
| AT2G27760 | F: GAACCTTAAACCCTAACAATGGAGAA | 930 | X | ||||||||||||
| R: GGCGGTTCCGTGACCATAT | |||||||||||||||
| AT2G27760 (INT) | F: GAACCTTAAACCCTAACAATGGAGAA | 160–470 | X | X | X | X | X | X | X | X | X | ||||
| R: CGAAATTCCTTAGCAGTGAACTCC | |||||||||||||||
| AT1G63160 (INT) | F: GACGCTGTATCTAGGCTCCAAG | 220–640 | X | X | X | X | X | ||||||||
| R: AAAATGTTGCATGTGAAGTTTGGC | |||||||||||||||
| AT1G63160 | F: GACGCTGTATCTAGGCTCCAAG | 1070–1490 | X | X | X | X | X | X | |||||||
| R: CACCATGAGACGGCCAAGTAT | |||||||||||||||
| AT1G65030 | F: CGGTTTTCTGTAACTCGGTACAG | KX767912 | KX767913 | 340 | X | X | X | X | |||||||
| R: CGGGGGAAAGAGAGGTTTTGG | |||||||||||||||
| AT5G52180 | F: CTCCGAGAATTTGGTTGGAAATGT | KX768003 | KX768004 | 460 | X | X | X | X | |||||||
| R: CATACAGAAAGCCGCGTCGATA | |||||||||||||||
| AT2G44760d | F: CAGCATGGAATACGTTTGCTAGTA | KX767954 | KX767955 | 530–900 | X | X | X | X | X | X | X | X | X | ||
| R: TATCAACTGGACCCCTGGAATAAG | |||||||||||||||
| AT2G05320Ac | F: TGCCAAGTGAAACAGATATTTGCT | KX767934 | KX767935 | 440 | X | X | X | X | X | X | X | X | |||
| R: TCTCCAAACAGTCTGGTTAAAGGA | |||||||||||||||
| AT4G31770c | F: GCGGTGAGAAATGAGAATGACATG | KX767998 | KX767999 | 580–780 | X | X | X | X | X | X | X | X | |||
| R: AACAAGTTCCTTCCAATTCCCAAA | |||||||||||||||
| AT2G20330c,g | F: TCATTGAAGGTTTGGGATTTACGC | KX767938 | KX767939 | 610–750 | X | X | X | X | X | X | X | X | X | X | X |
| R: ACGACTTGGCTGATCTCTGAATAA | |||||||||||||||
| AT1G66080 | F: CCTCTTCTCTTCCATAGTGTTGCT | 900 | X | ||||||||||||
| R: CCCACAAAACGACTGCATAAAGTT | |||||||||||||||
| AT2G05170Bc | F: GCACAGTACATTAACACCATTGGT | 430–480 | X | X | X | X | X | X | X | X | X | ||||
| R: TGGCTTGTGGTCTATGAGAATCTT | |||||||||||||||
| AT1G65070 | F: CCTAATACTGGAGGGAAAACTGCT | KX767914 | KX767915 | 510–600 | X | X | X | X | |||||||
| R: CAGTACTTCCCCAGAGAATTCGAA | |||||||||||||||
| AT5G67220 | F: CGGTTAAAAATGCTCTCAGGATCC | 690 | X | ||||||||||||
| R: CATCTGCCGAATGAGTAACCTTCT | |||||||||||||||
| AT2G17265c,d,g | F: TTATGGGAGGTTTCGTTTTGATTCG | KX767936 | KX767937 | 470–1690 | X | X | X | X | Xh | Xh | X | X | X | ||
| R: CTAGCACCAACTCTATCCAACCTC | |||||||||||||||
| AT2G46890Bd | F: TTCTTTGCTGTCTACCTCTCTCAG | 570–780 | X | X | X | X | X | X | |||||||
| R: CGATGTCGTCTTCTGATATAGCCT | |||||||||||||||
| AT2G31890Bc,g | F: CTCTCCAGTGCTCAGTTTTAACAG | KX767946 | KX767947 | 410 | X | X | X | X | X | X | X | X | X | ||
| R: CTTGAGAAATGTGTTGGTCCATCA | |||||||||||||||
| AT2G46100c | F: TTTAAAGGACTTCGCCGTTTCAAA | KX767956 | KX767957 | 310–370 | X | X | X | X | X | X | X | X | X | ||
| R: GGCAGAAAGAATAGGCCTCCAG | |||||||||||||||
| AT3G26580c,g | F: AGGTGAACGGTGTGGATTATGATGR: GTGACGGTTATTTGCCTCGTAAG | KX767976, KX767978 | KX767977, KX767979 | 660–920 | X | X | X | X | X | X | X | X | X | X | |
| AT2G44660B | F: GTTTTTGCAGGAAGGGATGGATTT | KX767952 | KX767953 | 590–1130 | X | X | X | X | X | X | |||||
| R: TGAAGGTTGTTGCTGGAGTTATCT | |||||||||||||||
| AT2G44660B (INT)c | F: GTTTTTGCAGGAAGGGATGGATTT | KX767952 | KX767953 | 520–900 | X | X | X | X | X | X | X | ||||
| R: TGCCTGAATCTTGAACCCTAGTTT | |||||||||||||||
| AT3G49730 | F: CCGAAACTGGAGATGGCTTTG | 140 | X | ||||||||||||
| R: AATCAACTCAGGCCTTTCTTTTCTC | |||||||||||||||
| AT2G44660Af | F: ATCGTATCACAGCACAGACTTTGA | KX767950 | KX767951 | 790 | X | X | |||||||||
| R: GCAAAAACAAAACCACCCATCAAA | |||||||||||||||
| AT2G21710c | F: TTTCCTCCTTTACTAACATACAGCCTR: CTTGTCTGCAACCTTCTGATTGAA | KX767942 (5′ only) | KX767943 (5′ only) | 1040–1360 | X | X | X | X | X | X | X | X | |||
| AT2G21710 (INT)d | F: TTTCCTCCTTTACTAACATACAGCCT | 750–860 | X | X | X | X | X | X | X | ||||||
| R: GCTGCATCCCAAGAGCTCTGG | |||||||||||||||
| AT2G22370Ac | F: ATGTTGAGGCCCTTGAGATTCTTC | 980–1320 | X | X | X | X | X | X | X | X | |||||
| R: TAGGTGCTGTTACTTCAACCAGTT | |||||||||||||||
| AT1G77930Ad | F: ACCCTAATTCTGTTCTGCGATTTG | KX767924 | KX767925 | 580–740 | X | X | X | X | X | ||||||
| R: GAGCAGTTCATAAGCAGCTTGAAT | |||||||||||||||
| AT1G77930A (INT) | F: ACCCTAATTCTGTTCTGCGATTTG | 410–460 | X | X | X | X | X | X | X | ||||||
| R: GCATCCCTCTCTAACTCTGCAATT | |||||||||||||||
| AT5G02250d | F: CACTTATCCCATGTTTCCAGAGAAC | 1240–1680 | X | X | X | X | X | X | |||||||
| R: GGATCTGCCTGGTTTTCAACATAT | |||||||||||||||
| AT2G31440 | F: GTATGGAGGGCTTTTCTTCCTTTG | 1000–1350 | X | X | |||||||||||
| R: ATTCCTGCAGCAAGATGAACTACA | |||||||||||||||
| AT1G77550Ad | F: TGTGAGCTTTTCTATATTGTGGCC | KX767920 | KX767921 | 740–860 | X | X | X | X | X | ||||||
| R: TGATGCTTCATGACCAGACAAGA | |||||||||||||||
| AT3G15290e | F: GATGTTGTAGTCGAGGCTATTGTG | 1090 | X | ||||||||||||
| R: ATCTGCAAGTTCTAAAGGACCCAT | |||||||||||||||
| AT5G11980 | F: TTCAACCATGCATCCCAAATTACC | N/A | |||||||||||||
| R: GACAGAGATCCGCCTTCAGTTATC | |||||||||||||||
| AT5G14580e | F: TATACGTATTGGCAGAATTTCCGG | 1030–1750 | X | X | |||||||||||
| R: TCTTGTGCAATCTTATCTAAGGCCT | |||||||||||||||
| AT2G31840d | F: AGTGATTGATTGGTGTCCTGATGT | 480–1220 | X | X | X | X | X | X | |||||||
| R: CATCTTGGTGAAGGTAGCCTACAG | |||||||||||||||
| AT5G57655 | F: TTGGTTATGCTCAGTGTAATCCGA | 340–1340 | Xh | Xh | X | Xh | Xh | X | |||||||
| R: CTACAGTGCAGATTGGAAAAGCAT | |||||||||||||||
| AT2G47760 | F: CAGCATGGAATACGTTTGCTAGTA | 620–1480 | X | X | |||||||||||
| R: TATCAACTGGACCCCTGGAATAAG | |||||||||||||||
| AT3G29130d | F: TTTGCCGAGGTTTCTGGTGATTR: AAGTACTTCTCTTGTTGATTCATCCG | KX767980, KX767981 | 980–1720 | X | Xh | X | |||||||||
| AT3G13200e | F: AACTCATCGGCTTTTTCCTCTCT | 1970 | X | ||||||||||||
| R: GAATCATCAGCATCTACATTGCGT | |||||||||||||||
| AT4G33030d | F: GATGGTGTCTTTGGTACTGCTTTG | 770–1340 | X | X | X | X | |||||||||
| R: CCAAGAAACAGTGGGCATTATCTG | |||||||||||||||
| AT1G73180d | F: AACTCCTGCCAGTGTCCAAATATA | 810–1000 | X | X | X | X | X | X | X | ||||||
| R: AGAATGCCATATCACCAGGTAAGT | |||||||||||||||
| AT2G31890A | F: AGATTGGAGGGGAGCTACTTTATT | KX767948 | KX767949 | 450–620 | X | X | X | X | X | X | X | X | |||
| R: CCTCCCTATACTGCTCTGAAATCC | |||||||||||||||
| AT3G46220d | F: CAATTGAGGAATGAAATGGTGGCT | 330–570 | X | X | X | X | |||||||||
| R: TCCATTTCTTGCAAAAGCTTCTGT | |||||||||||||||
| AT2G05120c,g | F: TGTCAAAGCTCTGGTCTCATGAAA | KX767932 | KX767933 | 370–570 | X | X | X | X | X | X | X | X | X | ||
| R: CGAAGGAAGAACTGAAGCATCTAG | |||||||||||||||
| AT1G73740d | F: TTGATATTGGGAGGCTCTTTGGG | 870–1230 | Xh | X | Xh | X | Xh | ||||||||
| R: CACCAGCTCTTGAAACAACGAG | |||||||||||||||
| AT4G31790d | F: ATTTGGTTGTTCGAGCCAAGAAAA | 1620–2180 | X | X | X | X | |||||||||
| R: GTCCAAAATCAACCATCTGCAGTT | |||||||||||||||
| AT5G10460 | F: TGGTCATCATTAGCAATTCTTCACG | 1320–1800 | X | X | X | X | X | ||||||||
| R: GCTTCTTCAACATTCTCCACAACT | |||||||||||||||
| AT4G26980d | F: CTGCTAGTGGTGTTTCTGAATTGG | 940–1170 | X | X | X | X | X | ||||||||
| R: ACTTCTCAGCCATTGACAACTCAT | |||||||||||||||
| AT5G48790d | F: GAGGATTTTGGTTTCACTGAAAAGG | 680–1250 | X | X | X | X | X | X | X | ||||||
| R: TCGCGACCTTTAAAATTGTGAATGT | |||||||||||||||
| AT5G15680A | F: TTCTCATTCAAACATCATTGGGCC | KX768001 | KX768002 | 560 | X | X | X | X | X | X | X | X | |||
| R: GAGGAATTGCATCAGATTCTCGTC | |||||||||||||||
| AT3G04650 | F: CAAATCGCTTGCTTGGTTCATCAR: CTGTGGCAGTTGGGATGTTTTC | KX767962, KX767964 | KX767963, KX767965 | 490–660 | X | X | X | X | X | X | X | ||||
| AT2G25570d | F: GACAACTCAAATACACATGCCAGG | 660–1160 | X | X | X | X | X | X | |||||||
| R: GTCCCTTCTCTGATGCCCTATG | |||||||||||||||
| AT2G31040d | F: AAGTACTGGGGTGGAGAAAAAGAG | 1230–1690 | X | X | X | X | X | X | |||||||
| R: CCAAGTGTGAGGATTTGCAACTTC | |||||||||||||||
| AT4G04955 | F: GAACAGATACGGTACAGAGCCAG | 440–1170 | X | X | |||||||||||
| R: TGCAGCTTTAGTCCCTGAAGG | |||||||||||||||
| AT3G21540c | F: GTTGCTATTAGTCCTGATGCCAAAR: AATGGTTCTTCAGTACGATCCCAA | KX767970, KX767972 | KX767971, KX767973 | 730–1020 | X | X | X | X | X | X | X | ||||
| AT2G05170Ac | F: GAAAGGAAATGTACCAGTGGAGGA | 650–880 | X | X | X | X | X | X | X | X | |||||
| R: TGAGAAGAATTGGTGGAGCTTCTT | |||||||||||||||
| AT2G28450f | F: TTTCTGAGATCATGCTTATAGTTCAGG | 1330 | X | X | |||||||||||
| R: CGCCCAATTGTTCCAGTACCA | |||||||||||||||
| AT3G07750d | F: GCTATATTTGTTGATTGCAGCCCT | 940–1330 | X | X | X | X | X | X | |||||||
| R: TGGTTGCTCATCGTTTAATGCATC | |||||||||||||||
| AT1G76450d | F: TGGTCCGGATTTTACAAGAATGGAR: CGTCCGCACAAAAATCAAAATAGG | KX767916, KX767918 | KX767917, KX767919 | 1330–1570 | X | X | X | ||||||||
| AT3G10530c,d | F: GGTCTTCTTGCATAATGAGCTGTT | 700–1960 | X | X | X | X | X | X | X | X | |||||
| R: TCAAATTTCCGCAAGTCCCAAATC | |||||||||||||||
| AT3G61620e | F: ATTTCCCCATTCAAATTCCACTCG | 1330 | X | ||||||||||||
| R: ACCTCATGCGATCTCCAGTACTAA | |||||||||||||||
| AT4G21770d | F: TGGAGCTGTTTATTATGCCCTTGT | KX767992 | KX767993 | 730–1130 | X | X | X | X | X | X | |||||
| R: TAGTCCTAGCTAACACAACACAGC | |||||||||||||||
| AT3G22990d | F: TTCTAGGTCCATCTCTTCAAGTGC | 900–1130 | X | X | X | X | X | X | X | ||||||
| R: TTATGATATTTGAGGCAGCAACGG | |||||||||||||||
| AT4G18810B | F: CCTAAAAGGTGATGGTCGAAGGTA | KX767988 | KX767989 | 540–640 | X | X | X | X | X | X | X | ||||
| R: TCTCCCTTCAACTTGAATGTGAGA | |||||||||||||||
| AT1G77550Bd | F: ACAGTTCAGAAGGCACATGGATC | KX767922 | KX767923 | 760–820 | X | X | X | X | |||||||
| R: CTGGTGTGTTCATGTGATTGAGTC | |||||||||||||||
| AT5G16690d | F: TGCTTCCTCTAGACAATTGTTCACT | 760–820 | X | X | X | ||||||||||
| R: TCCAAAGAAGCCGCTATGAATTG | |||||||||||||||
| AT4G00560 | F: CTGCTATGTCTATAAACGTTCCTTCCR: GTCCACCAACATTCAACAGTAACT | KX767984 (5′ only) | KX767985 (5′ only) | 900–1180 | X | X | X | X | X | ||||||
| AT3G17170d | F: GATGATGAACTATTTTTCCCTGAGGC | 630–900 | X | X | X | X | X | ||||||||
| R: TCTTGAACTTTCTCATTCACACTGC | |||||||||||||||
| AT3G14910c | F: GGAGCTATTTTATCAAAAGTTGTGCC | KX767968 | KX767969 | 360–840 | X | X | X | X | X | X | X | X | X | ||
| R: AAAGCAATATACGACCAAGAGAATCTG | |||||||||||||||
| Total no. of primers amplified/taxon | 2 | 16 | 47 | 68 | 90 | 53 | 71 | 72 | 26 | 54 | 34 | ||||
Note: INT = reverse primer is an internal primer for the locus.
Primer originally developed by Gostel et al. (2015).
GenBank accession numbers from loci used in phylogenetic analysis in Gostel et al. (2015). GenBank numbers were only created for loci of Bursera simaruba and Commiphora grandifolia that were used in the phylogenetic analysis in Gostel et al. (2015). Some loci have two GenBank numbers for a species because sequence reads did not cover the full length of the locus. The first GenBank number corresponds to the read from the 5′ end of the locus; the second GenBank number corresponds to the read from the 3′ end of the locus.
Universal Burseraceae primer (excluding Aucoumea).
Primer for which high-fidelity TAQ increased amplification success for Commiphora grandifolia.
Primer for which high-fidelity TAQ increased amplification success for Bursera simaruba.
Primer for which high-fidelity TAQ increased amplification success for Bursera simaruba and Commiphora grandifolia.
Universal Sapindales primer (excluding Aucoumea).
Faint double band observed.
The PCR thermocycler protocol followed that of Gostel et al. (2015) and included three alternating standard and Cot cycles (Mathieu-Daude et al., 1996), beginning with 2 min at 50°C, 20 min at 70°C, and 10 min at 95°C. The first set of 10 standard cycles included a denaturation step at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min. Two Cot cycles followed, including four steps consisting of 95°C for 15 s, 80°C for 30 s, 60°C for 30 s, and 72°C for 1 min. Standard and Cot cycles alternated two more times with eight, two, eight, and five cycles, respectively. After 35 cycles, samples were held at 4°C prior to being visually verified via agarose gel electrophoresis (1% agarose; 94 V for 40 min). Low DNA mass ladder (Invitrogen, Carlsbad, California, USA) was included in the first and last wells of each gel to guide length estimation of PCR products.
Marker amplification results
Table 1 contains amplification results for the low-copy nuclear loci, including the range of amplicon lengths for all taxa and GenBank numbers for markers sequenced by Gostel et al. (2015) for B. simaruba and C. grandifolia that had ≥15 sequence reads mapped. Table 2 summarizes marker amplification success for each taxon. Ninety primer pairs amplified product in B. simaruba and, on average, 54 primer pairs worked for other Burseraceae taxa. The low number of markers amplified in Aucoumea (16) was unexpected given its close relationship to Bursera. This result may have been caused by primer mismatch due to increased genetic change within this monotypic genus, as evidenced by its long branch within Burseraceae phylogeny (Weeks et al., 2014). In total, nine primer pairs worked for every Burseraceae taxon tested, and if Aucoumea is excluded as an outlier, the panel of family-universal primer pairs increases to 26. Thirty-four and 26 primer pairs generated product in Anacardiaceae and Rutaceae, respectively, while only two primer pairs worked in Arabidopsis. Comparing the Burseraceae panel to that of Anacardiaceae and Rutaceae reveals 16 and 12 successfully amplified regions in common, respectively, with eight shared among the three families. PCR chemistry may have suppressed amplification of markers, as high-fidelity PCR reagents were not used due to their high cost. Among the positive controls, high fidelity as compared to standard PCR reagents increased amplification success by 8% (Bursera, 83 to 90 primer pairs) and 85% (Commiphora, 39 to 72 primer pairs). Thus, our experimental results report a conservative baseline for the cross-amplification success of these primer pairs.
Table 2.
Number of primer pairs amplified of the 91 primer pairs tested for each of the 11 taxa.
| Species tested (Order; Family) | Primer pairs amplified/tested (%) |
| Arabidopsis thaliana (Brassicales; Brassicaceae) | 2/91 (0.02) |
| Aucoumea klaineana (Sapindales; Burseraceae) | 16/91 (17) |
| Beiselia mexicana (Sapindales; Burseraceae) | 47/91 (52) |
| Boswellia neglecta (Sapindales; Burseraceae) | 68/91 (75) |
| Bursera simaruba (Sapindales; Burseraceae) | 90/91 (99) |
| Bursera tonkinensis (Sapindales; Burseraceae) | 53/91 (58) |
| Canarium pilosum (Sapindales; Burseraceae) | 71/91 (78) |
| Commiphora grandifolia (Sapindales; Burseraceae) | 72/91 (79) |
| Phellodendron amurense (Sapindales; Rutaceae) | 26/91 (28) |
| Protium guianense (Sapindales; Burseraceae) | 54/91 (59) |
| Schinus fasciculatus (Sapindales; Anacardiaceae) | 34/91 (37) |
CONCLUSIONS
Our study demonstrates that 90 of 91 primer pairs for novel low-copy nuclear loci developed by Gostel et al. (2015) for B. simaruba successfully amplify product in a broad range of Sapindalean taxa and effectively expand the phylogenomic toolkit for this order. Twenty-six markers amplify all Burseraceae taxa (excluding Aucoumea) and eight amplify all Sapindalean groups tested. Our results present a new source for universal targets or primers for phylogenetic reconstruction of taxa within Sapindales. Future efforts will include sequencing amplicons to determine the number of phylogenetically informative characters for each locus.
Appendix 1.
Accession information for taxa used in this study, including voucher information, country of origin, and latitude and longitude coordinate data, if available, and DNA extraction method.
| Species | Voucher (Herbarium) | Country of origin | Geographic coordinates | DNA extraction methoda |
| Sapindales | ||||
| Burseraceae | ||||
| Aucoumea klaineana Pierre | Walters et al. 466 (MO) | Gabon | 00°07′12″S, 11°42′57″E | 1 |
| McPherson 16293 (MO) | Gabon | 00°27′S, 11°45′E | 1 | |
| Beiselia mexicana Forman | Pell s.n. (TEX) | Mexico | NA | 1, 2 |
| Boswellia neglecta S. Moore | Weeks 00-VII-29-1 (TEX) | Ethiopia | NA | 2 |
| Bursera simaruba (L.) Sarg. | Weeks 16-VI-16-01 (GMUF) | USA | NA | 1 |
| Goldman s.n. (BH) | USA | NA | 2 | |
| Bursera tonkinensis Guillamin | Daly et al. 13929 (NY) | Vietnam | 20°15′12.6″N, 105°43′2.5″E | 1 |
| Canarium pilosum A. W. Benn. | Bogler s.n. (TEX) | Malaysia | NA | 2 |
| Commiphora grandifolia Engl. | Gostel 121 (GMUF) | Madagascar | 23°39′19.64″S, 44°37′44.36″E | 1 |
| Weeks 10-I-09-10 (GMUF) | Madagascar | 12°14′16.14″S, 49°22′12.906″E | 1 | |
| Protium guianense (Aubl.) Marchand | Miller and Hauk 9391 (MO) | Suriname | 04°45′22″N, 056°52′30″W | 1 |
| Anacardiaceae | ||||
| Schinus fasciculatus (Griseb.) I. M. Johnst. | Silva-Luz 287 (NY) | Argentina | 24°52′05.4″S, 65°32′41.4″W | 1 |
| Rutaceae | ||||
| Phellodendron amurense Rupr. | Weeks 15-VII-13-01 (GMUF) | USA | 38°49′53.76″N, 77°18′32.04″W | 1 |
| Brassicales | ||||
| Brassicaceae | ||||
| Arabidopsis thaliana (L.) Heynh. | Gostel s.n. (GMUF) | USA | NA | 1 |
Note: NA = not available.
1 = FastDNA, 2 = CTAB.
LITERATURE CITED
- Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181: 1–20. [Google Scholar]
- Blow N. 2009. Microfluidics: The great divide. Nature Methods 6: 683–686. [Google Scholar]
- Duarte J. M., Wall P. K., Edger P. P., Landerr L. L., Ma H., Pires J. C., Leebens-Mack J., dePamphilis C. W. 2010. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis, and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evolutionary Biology 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine P. V. A., Zapata F., Daly D. C. 2014. Investigating processes of Neotropical rain forest tree diversification by examining the evolution and historical biogeography of the Protieae (Burseraceae). Evolution 68: 1988–2004. [DOI] [PubMed] [Google Scholar]
- Gostel M. R., Coy K. A., Weeks A. 2015. Microfluidic PCR-based target enrichment: A case study in two rapid radiations of Commiphora (Burseraceae) from Madagascar. Journal of Systematics and Evolution 53: 411–431. [Google Scholar]
- Grudinski M., Pannell C. M., Chase M. W., Ahmad J. A., Muellner-Riehl A. N. 2014. An evaluation of taxonomic concepts of the widespread plant genus Aglaia and its allies across Wallace’s Line (tribe Aglaieae, Meliaceae). Molecular Phylogenetics and Evolution 73: 65–76. [DOI] [PubMed] [Google Scholar]
- Matasci N., Hung L.-H., Yan Z., Carpenter E. J., Wickett N. J., Mirarab S., Nguyen N., et al. 2014. Data access for the 1,000 plants (1KP) project. GigaScience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu-Daude F., Welsh J., Vogt T., McClelland M. 1996. DNA rehybridization during PCR: The ‘Cot effect’ and its consequences. Nucleic Acids Research 24: 2080–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellner-Riehl A. N., Weeks A., Clayton J. W., Buerki S., Nauheimer L., Chiang Y.-C., Cody S., Pell S. K. 2016. Molecular phylogenetics and molecular clock dating of Sapindales based on plastid rbcL, atpB and trnL-trnF DNA sequences. Taxon 65: 1019–1036. [Google Scholar]
- Uribe-Convers S., Settles M. L., Tank D. C. 2016. A phylogenomic approach based on PCR target enrichment and high throughput sequencing: Resolving the diversity within the South American species of Bartsia L. (Orobanchaceae). PLoS ONE 11: e0148203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Moore M. J., Soltis P. S., Bell C. D., Brockington S. F., Alexandre R., Davis C. C., et al. 2009. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proceedings of the National Academies of Science, USA 106: 3853–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks A., Daly D. C., Simpson B. B. 2005. The phylogenetic history and biogeography of the frankincense and myrrh family (Burseraceae) based on nuclear and chloroplast sequence data. Molecular Phylogenetics and Evolution 35: 85–101. [DOI] [PubMed] [Google Scholar]
- Weeks A., Simpson B. B. 2007. Molecular phylogenetic analysis of Commiphora (Burseraceae) yields insight on the evolution and historical biogeography of an “impossible” genus. Molecular Phylogenetics and Evolution 42: 62–79. [DOI] [PubMed] [Google Scholar]
- Weeks A., Zapata F., Pell S. K., Daly D. C., Mitchell J. D., Fine P. V. A. 2014. To move or to evolve: Contrasting patterns of intercontinental connectivity and climatic niche evolution in “Terebinthaceae” (Anacardiaceae and Burseraceae). Frontiers in Genetics 5: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer E. A., Wen J. 2012. Using nuclear gene data for plant phylogenetics: Progress and prospects. Molecular Phylogenetics and Evolution 65: 774–785. [DOI] [PubMed] [Google Scholar]