Abstract
Many processes are deregulated in melanoma cells and one of those is protein production. While much is known about protein synthesis in cancer cells, effective ways of therapeutically targeting this process remains an understudied area of research. A process that is upregulated in melanoma compared to normal melanocytes is proline biosynthesis, which has been linked to both oncogene and tumor suppressor pathways, suggesting an important convergent point for therapeutic intervention. Therefore, a RNA interference (RNAi) screen of a kinase library was undertaken, identifying aldehyde dehydrogenase 18 family, member A1 (ALDH18A1) as a critically important gene in regulating melanoma cell growth through proline biosynthesis. Inhibition of ALDH18A1, the gene encoding pyrroline-5-carboxylate synthase (P5CS), significantly decreased cultured melanoma cell viability and tumor growth. Knockdown of P5CS using siRNA had no effect on apoptosis, autophagy, or the cell cycle but cell doubling time increased dramatically suggesting that there was a general slowdown in cellular metabolism. Mechanistically, targeting ALDH18A1 activated the serine/threonine protein kinase GCN2 (general control nonderepressible 2) to inhibit protein synthesis, which could be reversed with proline supplementation. Thus, targeting ALDH18A1 in melanoma can be used to disrupt proline biosynthesis to limit cell metabolism thereby increasing the cellular doubling time mediated through the GCN2 pathway.
Keywords: ALDH18A1, P5CS, proline, GCN2, melanoma
Introduction
Melanoma is the deadliest form of skin cancer leading to 75% of skin cancer deaths (1), and mortality rates continue to rise (2). Constitutive activation of the MAPK signaling cascade is one important hallmark of melanoma whereby this pathway leads to increased cell growth and proliferation. Promising drugs have emerged for targeting members of this pathway (3, 4), but recurrent resistant disease develops in nearly every case (5). While recent melanoma research has focused on the MAPK pathway, other processes driving melanoma development might be equally important for therapeutic intervention. Production of protein is key to melanoma cell growth, and this process is deregulated to maximize the output of proteins for cellular survival. However, effective approaches to target this process for therapeutic application remain underdeveloped.
Amino acid production can play a key role in cancer cell survival by regulating cellular metabolism. The non-essential amino acid glutamine is an example of an amino acid important to cancer development (6). Melanoma cells have higher levels of de novo proline biosynthesis than normal melanocytes (7, 8). However, the importance of this process and pathways regulating its effect are understudied. Serine is another non-essential amino acid that was recently shown to be important for p53-null colorectal cancer cell proliferation (9); so it could be inferred that proline might play an equally important role in melanoma cell survival (10).
Proline is a nonessential amino acid that is a necessary component of proteins synthesized during mRNA translation (11). Mechanisms to regulate mRNA translation when the cell senses low levels of particular amino acids have been reported (12). The best known sensor of amino acid levels is the mTOR pathway, which is activated by the presence of amino acids, causing the phosphorylated activation of p70S6K and phosphorylated inactivation of 4EBP1 (12). Depletion of certain amino acids, particularly leucine, inactivates the mTOR pathway, preventing phosphorylation of the aforementioned targets, inhibiting initiation factor complex eIF4F activity, resulting in decreased initiation of mRNA translation (13).
General control nonderepressible 2 (GCN2) is another sensor of amino acid levels regulating mRNA translation through the ternary complex initiation factor eIF2 (14). Four kinases regulate translation initiation through eIF2, however only GCN2 responds to amino acid availability. GCN2 senses low amino acid levels by recognizing, and becoming activated by, uncharged tRNAs (15). Translation initiation can proceed when the guanine nucleotide exchange factor (GEF) eIF2B replenishes GDP on eIF2 with GTP (14). The phosphorylation of the alpha subunit of eIF2 by GCN2 at Ser52 in humans inhibits mRNA translation initiation by causing eIF2 to irreversibly bind to the eIF2B complex, preventing its GEF function (16). Thus, the mTOR and GCN2 pathways play an essential role in cancer cells by monitoring that adequate amino acid levels are present to aid tumor cell survival.
Aldehyde dehydrogenase 18 family, member A1 (ALDH18A1) is a gene that encodes Δ1-pyrroline-5-carboxylate synthase (P5CS) that is key to proline and ornithine production (17). P5CS is an ATP- and NAD(P)H-dependent mitochondrial enzyme that catalyzes the conversion of L-glutamate to pyrroline-5-carboxylate (P5C) (18). In addition to P5CS, proline dehydrogenase (PRODH), pyrroline-5-carboxylate dehydrogenase (P5CDH), and pyrroline-5-carboxylate reductase (PYCR) are essential components of this proline biosynthesis pathway. Enzymes of this pathway have been linked to MYC-driven oncogenesis and the tumor suppressor p53, which may help to regulate cancer development. PRODH, which synthesizes P5C from proline, can serve as a tumor suppressor and its transcription can be controlled by p53 (19, 20). ALDH4A1, the gene encoding P5CDH, synthesizing glutamate from P5C, has also been identified as a p53-inducible gene (21). Furthermore, PRODH, P5CS, and PYCR also have roles in MYC driven oncogenesis. MYC induces ALDH18A1 in T cells (22), inhibits PRODH expression, and increases P5CS and PYCR expression in Burkitt lymphoma and prostate cancer (23). PYCR mediates the conversion of P5C to proline and has been shown to be upregulated in melanoma (8). Collectively, these reports suggest that tumor suppressors such as p53 can maintain proline degradation while oncogenes such as MYC can promote synthesis.
Proline biosynthesis, which requires P5CS enzyme activity, is increased in melanoma cells (8), and is necessary for protein production. This study shows that targeting proline synthesis can be used to inhibit melanoma growth. siRNA-mediated knockdown of ALDH18A1 to decrease P5CS protein levels inhibited tumor development by 60 to 99% and decreased melanoma cell viability up to 90%, which could be partially restored by proline supplementation. Targeting P5CS proline-dependently increased GCN2 and eIF2α phosphorylation, but did not inactivate mTOR as a mechanism to decrease protein synthesis. Thus, the therapeutic targeting of P5CS may be an effective means to inhibit melanoma growth by disrupting the proline biosynthesis pathway and amino acid sensing through GCN2.
Materials and Methods
Cell line and culture conditions
Normal human FF2441 and Neonatal Fibroblasts (provided by the laboratory of Dr. Craig Myers, Penn State College of Medicine, Hershey, PA) were maintained in DMEM (Thermo HyClone, Logan, UT, USA) supplemented with 10% FBS (Thermo Hyclone) and 1X GlutaMAX (Life Technologies, Carlsbad, CA). Cancer lines UACC 903 (provided by Mark Nelson, University of Arizona, Tucson, AZ), 1205 Lu (provided by Dr. Meenhard Herlyn, Wistar Institute, Philadelphia, PA), A375M (CRL-1619; ATCC, Manassas, VA), and C8161 Cl.9 (provided by Dr. Danny Welch, University of Kansas, Kansas City, KS) were grown in DMEM with 10% FBS and 1X GlutaMAX. WM115 and WM278.1 cell lines (provided by Dr. Herlyn, Wistar Institute, Philadelphia, PA) were grown in media #2 (MCDB media supplemented with heat-inactivated FBS, GlutaMAX, and insulin) (24) except for LC-MS studies where they were cultured in DMEM with 10% FBS and 1X GlutaMAX. Normal FOM103 melanocytes (provided by Dr. Herlyn, Wistar Institute, Philadelphia, PA) were grown in DermaLife media (Lifeline Cell Technology, Walkersville, MD, USA) containing LifeFactors DermaLife M (Lifeline Cell Technology). Cells were incubated in a humidified incubator at 37°C and 5% CO2 atmosphere and periodically examined for cell microscopic phenotype, genetic biomarkers, and growth potential in culture and in mice to confirm identity of cell lines. Melanoma cell line mutation statuses listed in Figure 1D were obtained from the following sources: The Wistar Institute (http://www.wistar.org/lab/meenhard-herlyn-dvm-dsc/page/melanoma-cell-lines-0) and references (25–29).
Fig. 1. P5CS protein levels vary but proline levels are elevated in melanomas.
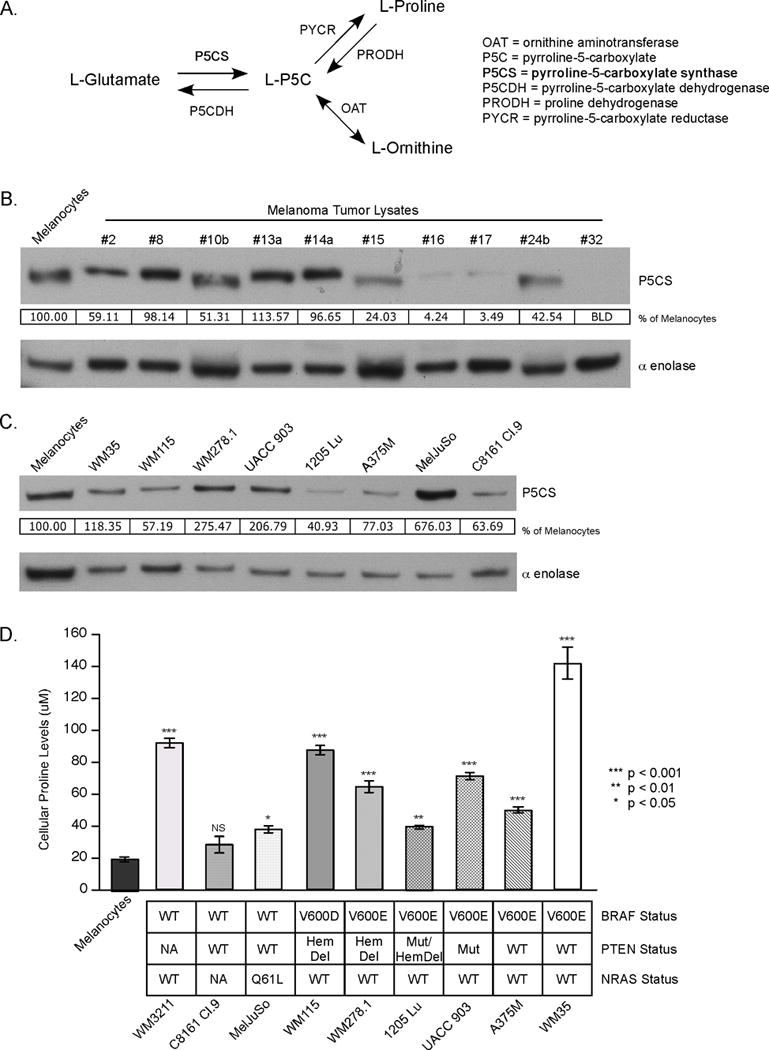
(A) Diagram of the biosynthetic pathway involving the ALDH18A1 enzyme P5CS; P5CS, Δ1-pyrroline-5-carboxylate synthase; P5C, pyrroline-5-carboxylate; P5CDH, pyrroline-5-carboxylate dehydrogenase; PYCR, pyrroline-5-carboxylate reductase; PRODH, proline dehydrogenase; OAT, ornithine aminotransferase. (B, C) Western blots comparing P5CS protein levels of melanocytes (FOM103) versus melanoma tumor lysates (B) and melanocytes versus melanoma cell lines (C). Protein band intensity was quantified using ImageJ software and P5CS values normalized to the alpha enolase protein loading control and expressed as a percentage of melanocyte control. (D) Intracellular proline levels measured by LC-MS/MS. Absolute cellular concentration was quantitated by comparison to a norvaline internal control (N=4). Bars; average, ±SEM. NS, not significant. Cell lines were stratified according to BRAF, PTEN, and NRAS mutational status. WT, wildtype. Hem Del, hemizygous deletion. Mut, mutation. NA, no data available.
siRNA transfection studies
Silencer Select siRNA (Life Technologies) were used in preliminary screening. Duplexed Stealth siRNA (Life Technologies) were used in all subsequent transfection experiments. Sequences are:
siScrambled – 5′ UCU CAC GUG ACA CGU UCG GAG AAU U-3′
siALDH18A1 #2 – 5′ ACU ACU UCA AUG CAU AAU UCC AGG U 3′
siALDH18A1 #3 – 5′ ACA CGG AUG UCA UCG UCA CAG AGG A 3′
Transfections to collect protein lysates were performed by incubating 100 pmoles siRNA with 7.5 uL RNAiMAX (Life Technologies) for every 1×106 cells according to manufacturers’ recommendations. Transfected cells were incubated at 37°C in a humidified, 5% CO2 atmosphere, incubator for specified length of time. For cells incubated in proline supplemented media, 1.25 mM L-proline (Sigma) (for UACC 903, A375M, and FF2441) or 2.5 mM L-proline (for 1205 Lu) was supplemented during transfection and maintained until lysate collection.
Determination of cell viability
Cell viability was measured 5 days after transfection using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega, Madison, WI). A Cell Proliferation BrdU ELISA (Roche, Indianapolis, IN) was used to quantify cell proliferation. Five days after transfection, absorbance was measured at 490 nm for the MTS and 370 nm for the BrdU ELISA, using Soft Max Pro software.
To test the effects of amino acid supplementation on cell viability, 1.25 mM or 2.5 mM L-proline, L-ornithine (Sigma), and L-citrulline (Sigma) were added to media. To determine the optimal concentrations of amino acid to use, each was tested individually with concentrations ranging from 0.15625 mM to 20 mM. Optimized amino acid concentrations were used for supplementation experiments.
Cell viability assessed by doubling time was measured via the sulforhodamine B (SRB) colorimetric assay (30). Cells transfected as above were fixed every day for 5 days followed by staining with SRB (S9012, Sigma) and OD measured. A doubling time calculator from http://www.doubling-time.com was used to estimate growth rate and doubling time using values from days 1 through 4, constituting the exponential growth phase of cells.
Similar to the SRB assay, a Hoechst DNA stain assay was performed to assess cell viability and estimate doubling time via total DNA content (31). Cells transfected as above were fixed and permeabilized every day for 5 days followed by staining with Hoechst 33258 (Life Technologies). Doubling time and growth rate were estimated as with the SRB assay.
Western blotting
Lysates were prepared and electrophoresed on gels as described previously (32). Membranes were probed with primary antibodies following each of the supplier’s recommendations: AKT (#4685), pAKT (S473) (#4060), ATG5 (#2630), caspase-3 (#9662), Cleaved PARP (D214) (#9541), p-eIF2α (S51) (#3398), ERK1/2 (#9102), pERK1/2 (T202/Y204) (#9101), GCN2 (#3302), LC3B (#2775), PRAS40 (#2610), pPRAS40 (T246) (#2997), and p62 (#5114) from Cell Signaling Technology (Danvers, MA), and p21 (sc-756), p27 (sc-528), Cyclin D1 (sc-718), α enolase (sc-7455), VDAC1 (sc-8828), and ERK2 (sc-1647) from Santa Cruz Biotechnology (Dallas, TX, USA). The P5CS primary antibody, ALDH18A1 (NBP1-83324), was purchased from Novus Biologicals (Littleton, CO, USA) and pGCN2 (T899) (ab75836) from Abcam. eIF2α and eIF2Bε antibodies were kind gifts from Dr. Scot Kimball. Secondary antibodies goat anti-rabbit IgG-HRP (sc-2004), goat anti-mouse IgG-HRP (sc-2005), and donkey anti-goat IgG-HRP (sc-2020) were purchased from Santa Cruz Biotechnology. The immunoblots were developed using ECL Western Blotting Substrate (Thermo Scientific) or Supersignal West Femto Chemiluminescent Substrate (Thermo Scientific).
Animal studies
2×106 UACC 903 and 1205 Lu cell lines were transfected with 200 pmol siRNA to ALDH18A1 or siScrambled control using RNAiMAX. Sterile, nuclease free DEPC H2O (EMD Chemicals, Gibbstown, NJ, USA) was used as the Buffer control. 72 hours after transfection, 1×106 cells in 0.2 mL DMEM with 10% FBS and 1X GlutaMAX were subcutaneously injected into left and right flanks of 4–6 week old athymic female nude mice. Starting at day 6, tumors were measured every other day, using calipers to determine length, width, and depth to calculate tumor volume.
Protein synthesis analysis
Two or 3 days post transfection, cells were starved of methionine by washing plates with PBS then incubated with methionine-free DMEM for 1 hour. Respective plates were maintained with 1.25 or 2.5 mM proline and media changed after 2 days. Cystine and methionine-free DMEM (Life Technologies, 21013-024) supplemented with 48 mg/L L-cystine (#2470, CalBioChem) was used for methionine starvation. For cells incubated in proline, methionine-free DMEM was supplemented with 2.5 mM proline. The Click-iT Protein Reaction assay was performed as described previously (32).
Results
Melanoma cells have higher intracellular proline levels compared to melanocytes
To identify genes important in melanoma cell survival, a siRNA screen of over 700 kinases was undertaken in which cell growth inhibition following protein knockdown in UACC 903 and 1205 Lu melanoma cell lines was examined. The gene aldehyde dehydrogenase 18 family, member A1 (ALDH18A1) was identified as a target decreasing melanoma cell viability following protein knockdown (Fig. S1). ALDH18A1 encodes Δ1-pyrroline-5-carboxylate synthase (P5CS), an enzyme found in the proline biosynthesis pathway (Fig. 1A). It converts glutamate to P5C, which is then converted to proline by pyrroline-5-carboxylate reductase (PYCR). P5CS protein was present in the majority of melanoma tumor lysates and protein levels were found to be equivalent to or less than those occurring in melanocytes (Fig. 1B). Likewise, P5CS protein was expressed in all melanoma cell lines tested, and levels varied compared to melanocytes resulting in no discernable trend between melanoma cells and melanocytes (Fig. 1C). Next, intracellular proline levels in melanoma cell lines were examined. There was up to an 8-fold increase in intracellular proline levels compared to melanocytes in all but the C8161 Cl.9 cell line (Fig. 1D). Intracellular glutamate levels, upstream of the proline biosynthesis pathway, were not significantly altered in 6 of 9 melanoma cell lines tested (Fig. S2A), which served as a control to show that the increase in proline was not due to an increase in upstream metabolites. Intracellular levels of arginine and citrulline, two amino acids downstream of P5CS, were also significantly increased in the majority of melanoma cell lines compared to melanocytes (Figs. S2B and S2C). Collectively, these results suggest that metabolites downstream, but not upstream, of P5CS were increased in melanoma.
siRNA-mediated knockdown of P5CS protein levels decreased intracellular proline to reduce melanoma cell viability
Two independent siRNA targeting different regions of ALDH18A1 mRNA were transfected into multiple melanoma cell lines and normal fibroblast cells. ALDH18A1 siRNA transfection decreased P5CS protein levels by 65 to 97% in UACC 903, 1205 Lu, and A375M melanoma cell lines and normal FF2441 cells (Fig. 2A). Knockdown of ALDH18A1 decreased cell viability in melanoma cell lines by up to 90% but had a statistically insignificant decrease of 20% or less in normal human fibroblasts (Fig. 2B). A BrdU ELISA was performed to corroborate the MTS results demonstrating a 63 to 99% decrease in BrdU incorporation suggesting that the MTS assay findings were not an artifact from targeting of a mitochondrial aldehyde dehydrogenase (Fig. 2C).
Fig. 2. siRNA-mediated knockdown of P5CS protein levels decreased intracellular proline levels and reduced cultured melanoma cell viability.
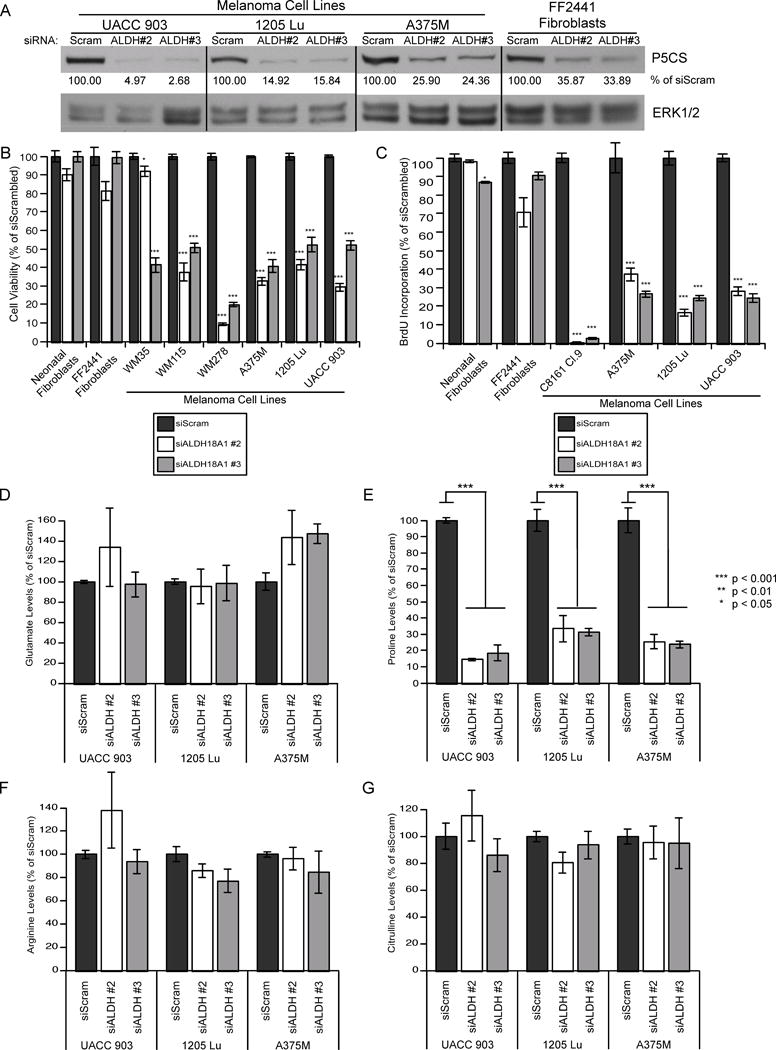
(A) siRNA targeting ALDH18A1 or a scrambled control were transfected into UACC 903, 1205 Lu, and A375M melanoma cell lines and FF2441 fibroblast cells. Protein expression was assessed via western blotting. ERK1/2 was used as a protein loading control. Protein band intensity was quantified using ImageJ software and P5CS values normalized to the ERK1/2 control and expressed as a percentage of siScrambled control. (B, C) siRNA targeting ALDH18A1 or a scrambled control were transfected into fibroblast or melanoma cell lines and cell viability measured by MTS assay (B) and cell proliferation assessed by a BrdU ELISA (C) (N≥3). Bars; average, ±SEM. (D–G) UACC 903, 1205 Lu, and A375M melanoma cells were transfected with siRNA targeting ALDH18A1 or a scrambled control. LC-MS/MS was used to quantify intracellular levels of glutamate (D), proline (E), arginine (F), and citrulline (G). Values were normalized to a norvaline internal control and expressed as a percentage of the siScrambled control (N=4). Bars; average, ±SEM.
P5CS is involved in the conversion of glutamate to P5C, which is then converted to proline or ornithine (Fig. 1A). When P5CS protein levels were knocked down, no significant change in upstream intracellular glutamate levels occurred (Fig. 2D). In contrast, downstream of P5CS, intracellular proline levels significantly decreased by 66 to 85% upon P5CS protein knockdown (Fig. 2E). Intracellular ornithine levels of all samples were below the level of detection. Downstream of P5CS, ornithine is used for arginine and citrulline synthesis, however intracellular levels of arginine or citrulline were not significantly altered after ALDH18A1 knockdown (Figs. 2F and 2G). Collectively, these results suggest that P5CS protein knockdown can decrease intracellular proline production resulting in decreased melanoma cell viability, which was then tested by proline supplementation.
Addition of proline following knockdown of P5CS protein levels can partially restore cell viability
To demonstrate that decreased proline levels affected the proliferative potential of melanoma cells, proline was added to the cell culture media during and after siRNA transfection. Proline supplementation was able to partially or completely restore cell viability following ALDH18A1 knockdown in UACC 903, 1205 Lu, and A375M melanoma cell lines (Fig. 3), except for siALDH18A1 #2 in 1205 Lu cells. The ineffectiveness of recovery in 1205 Lu after siALDH18A1 #2 transfection was predicted to be due to a delay of 1205 Lu cells use of extracellular proline to restore protein synthesis. Similarly, ornithine supplementation partially or fully restored cell viability after ALDH18A1 knockdown (Figs. S3A–C). In contrast, supplementation with citrulline did not restore cell viability (Fig. S3D). Since proline supplementation led to a partial restoration of the proliferative potential of all except siALDH18A1 #2 in 1205 Lu cells, it appeared that decreased melanoma cell viability was in part due to a depletion of intracellular proline and/or ornithine.
Fig. 3. Proline supplementation following P5CS knockdown partially restored cell viability.
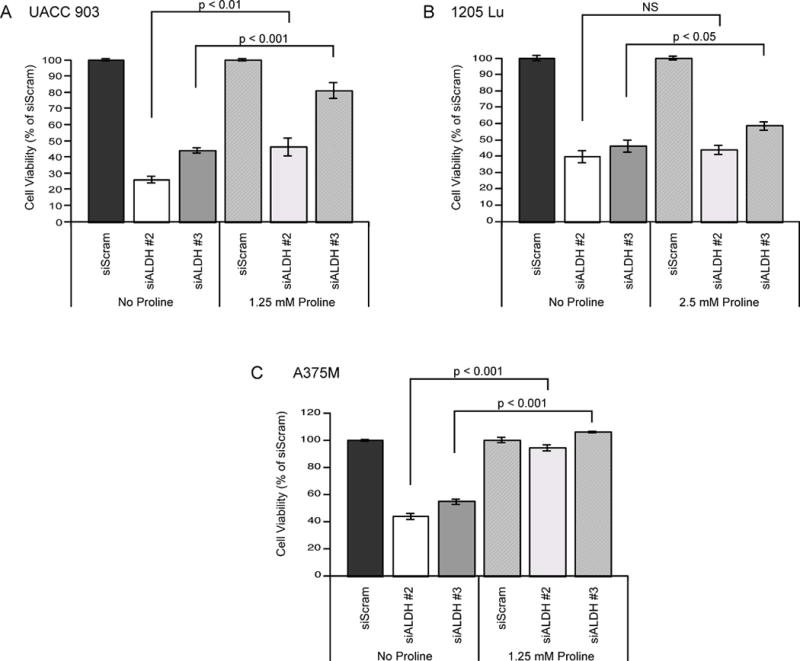
(A–C) ALDH18A1 and scrambled siRNA were transfected into UACC 903 (A), 1205 Lu (B), and A375M (C) melanoma cell lines. Transfected cells were grown for 5 days with or without proline supplementation and cell viability assessed via MTS assay (N≥4). Bars; average, ±SEM.
siRNA-mediated inhibition of ALDH18A1 decreased melanoma tumor development
To assess the effect of siRNA-mediated protein knockdown on tumor development, persistent decreased protein levels must be observed by western blotting for 6 to 8 days in cultured cells per published approaches (33). siRNA knockdown of ALDH18A1 decreased P5CS protein levels for at least 8 days in UACC 903 and 1205 Lu melanoma cells (Fig. 4A). Three days after transfection with siRNA targeting ALDH18A1, UACC 903 or 1205 Lu cells were subcutaneously injected into nude mice, which is the standardized approach for these types of assays (33). Developing tumors were measured on alternate days and tumor development was inhibited by 75 to 99% in UACC 903 cells after 28 days (Fig. 4B) and by 60% in 1205 Lu cells after 24 days (Fig. 4C). P5CS knockdown in tumor lysates were confirmed out to day 11 and 13 post injection (Fig. S4). This result suggests that disruption of the proline biosynthesis pathway can impair melanoma tumor development.
Fig. 4. siRNA-mediated knockdown of P5CS protein levels decreased melanoma tumor development.
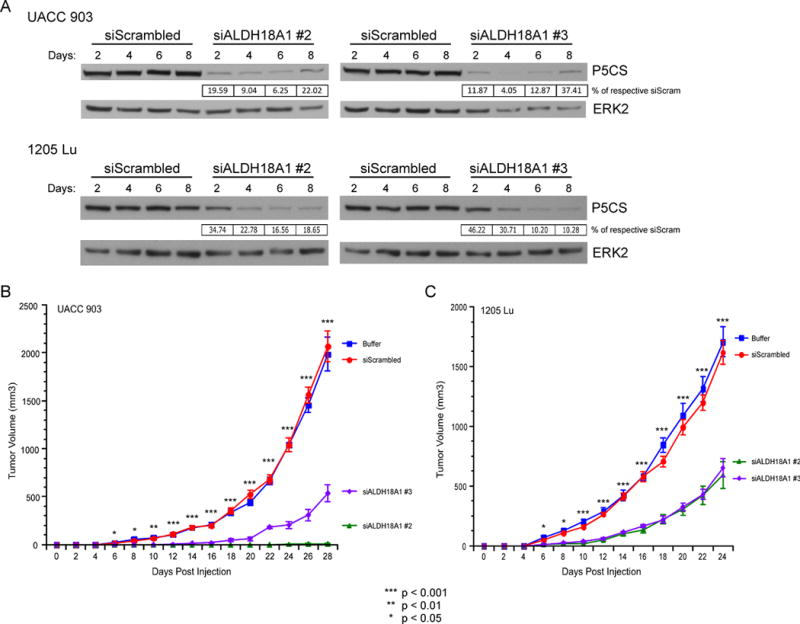
(A) UACC 903 or 1205 Lu melanoma cells were transfected with siRNA targeting ALDH18A1 or a scrambled control. Western blotting of lysates collected between days 2 and 8 shows decreased P5CS protein levels. Blots were quantitated by ImageJ, normalized to the ERK2 protein loading control, and expressed as a percentage of the respective siScrambled blot intensity. (B, C) UACC 903 (B) or 1205 Lu (C) cells were transfected with each respective siRNA or buffer control and incubated for 3 days prior to subcutaneous injection into nude mice. Tumor volumes were measured on alternate days (N=6). Points; mean, ± SEM (statistics, One-way ANOVA followed by Tukey’s test for multiple comparisons).
Mechanistically, knockdown of P5CS protein levels increased cell doubling time
The mechanism by which knockdown of P5CS protein levels inhibit melanoma development is unknown. To dissect the underlying mechanism, assays assessing protein content using an SRB assay or measuring DNA content with Hoechst staining were performed every day up to 5 days following siRNA-mediated knockdown of P5CS protein levels to determine if growth rate was affected. Growth curves using either approach led to a 60 to 90% reduction in growth by day 5 after siALDH18A1 transfection (Fig. 5A). Calculation of doubling time from both growth curves demonstrated that ALDH18A1 knockdown significantly decreased melanoma cell growth rate by 56 to 96% (Fig. 5B). These data suggest that targeting ALDH18A1 decreased cell viability by impairing cell growth leading to significant increases in cell doubling times.
Fig. 5. siRNA-mediated knockdown of P5CS protein levels decreased cell growth rate and protein synthesis.
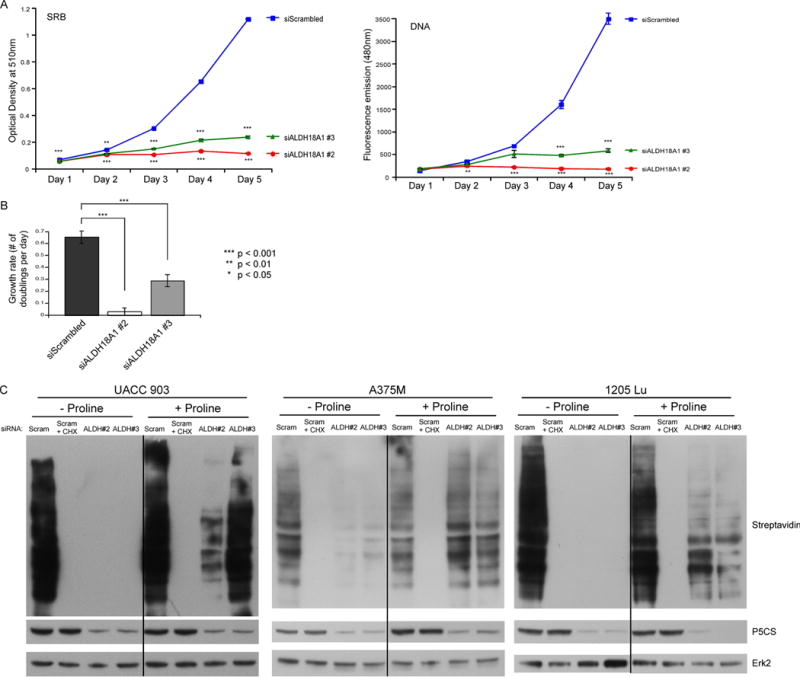
(A) UACC 903 cells were transfected with siRNA targeting ALDH18A1 or a scrambled control. Cell viability was measured by an SRB assay or Hoechst stain DNA assay for consecutive days (N=6). Points; average, ±SEM. *p<0.05, **p<0.01, ***p<0.001 (B) Optical density (O.D.) values from the SRB and DNA assays representing the exponential growth phase of cells were input into a doubling time calculator from http://www.doubling-time.com and the calculated growth rate values were plotted (N=5). Bars; average, ±SEM. (C) UACC 903, A375M, and 1205 Lu cells transfected with siRNA targeting ALDH18A1 or a scrambled control were subjected to methionine starvation followed by azidohomoalanine (AHA) incubation at 3 (UACC 903), 2 (A375M), or 8 (1205 Lu) days post transfection. Cycloheximide (CHX) served as a positive assay control. Biotin-alkyne was attached to newly synthesized proteins containing AHA before performing western blotting. Membranes were probed with streptavidin-HRP to identify the extent of new protein synthesis and P5CS primary antibody used to confirm protein knockdown. ERK2 primary antibody serves as a control for equal protein loading.
siRNA knockdown of ALDH18A1 did not modulate typical processes such as apoptosis, cell cycle arrest, or autophagy to increase doubling time but instead decreased protein synthesis
The next goal was to identify the mechanism by which P5CS protein knockdown increased cell doubling time and decreased tumor growth. To establish the underlying mechanism, apoptosis and autophagy as well as expression levels of cell cycle regulators and MAPK pathway signaling proteins were examined in cultured cells following ALDH18A1 knockdown (Fig. S5). Levels of proteins serving as indicators of apoptosis or as measures of cell cycle changes (Fig. S5A), autophagy (Fig. S5B), and MAPK signaling (Fig. S5C) were unchanged following P5CS protein knockdown. Since multiple papers have shown that the proline biosynthesis pathway modulates apoptosis or autophagy through reactive oxygen species (ROS) generation, ROS levels were examined next. Amplex Red and H2DCFDA assays were performed on UACC 903 siALDH18A1-transfected cells and no consistent changes in ROS activity were observed (Fig. S6). Next, propidium iodide was used to measure DNA levels in siALDH18A1-transfected cells to determine whether any changes occurred in the cell cycle. ALDH18A1 knockdown did not arrest cells in any cell cycle phase (Fig. S7A). Since no changes in apoptosis, autophagy, or cell cycle arrest were observed, the possibility of impaired cellular metabolism was examined next. Glycolytic activity was measured using a Seahorse Flux Analyzer. Glycolytic activity significantly decreased upon ALDH18A1 knockdown in UACC 903 cells in a proline-dependent manner (Fig. S7B), but this was not observed in 1205 Lu melanoma cells. Collectively, these results suggest that the most common causes of decreased growth rate involving apoptosis, cell cycle arrest, or autophagy are unaffected by ALDH18A1 knockdown in melanoma, suggesting cellular metabolic activity is a possible cause of the decreased growth rate. While effects on metabolism may be cell line dependent, one commonality may be its effect on protein synthesis.
To determine whether ALDH18A1 knockdown inhibited protein synthesis to increase cell doubling time, the methionine analog azidohomoalanine (AHA) was used to measure the extent of new protein synthesis (34). Knockdown of ALDH18A1 decreased protein synthesis in UACC 903, A375M, and 1205 Lu melanoma cell lines (Fig. 5C). To demonstrate protein synthesis was affecting the proliferative potential and doubling time of melanoma cells, proline was added to determine if it could reverse the effect. Protein synthesis was partially restored by supplementation of proline to cell media in UACC 903 cells by day 3 (Fig. 5C) and 1205 Lu cells by day 8 (Fig. 5C) and completely restored in A375M cells by day 2 (Fig. 5C). Timing of protein synthesis recovery was predicted to be variable due to differences in activity of proline metabolism and regulation of translation initiation between the different cell lines. For 1205 Lu cells, proline supplementation did not affect protein synthesis by day 5 (Fig. S8) but by day 8 protein synthesis levels were restored (Fig. 5C), suggesting delayed recovery for this cell line. Collectively, these data suggest that impairment of proline biosynthesis decreased protein synthesis, consequently affecting the proliferative potential of melanoma cells leading to an increased cell doubling time.
GCN2 and eIF2α were activated, but mTOR was not inactivated, upon P5CS knockdown in melanoma cells
Amino acid availability sensing by a cell can occur through recognition of uncharged aminoacyl-tRNAs by GCN2 (15) (Fig. 6A), which can modulate mRNA translation. A decrease in amino acids increases levels of uncharged tRNA, which causes the autophosphorylation of GCN2. It in turn phosphorylates eIF2α, which blocks eIF2B to shut down protein synthesis (16). To determine whether the GCN2 pathway was activated following knockdown of P5CS protein levels, protein levels of key pathway members regulating this process, such as GCN2 and eIF2α, and the protein’s phosphorylation states were examined in the melanoma cell lines and compared to the response in normal fibroblasts. Since activation of the GCN2 pathway is dependent on the timing of amino acid depletion following transfection, lysates were collected after 48 and 96 hours for analysis. At 48 hours, P5CS knockdown increased GCN2 phosphorylation at T899 in 1205 Lu and UACC 903 melanoma cell lines by ~500% and ~300%, respectively, and eIF2α phosphorylation at S52 in 1205 Lu and A375M cell lines by 500% and ~200%, respectively (Figs. 6B and 6C). By 96 hours, total eIF2α protein levels were elevated in UACC 903 and 1205 Lu cells by approximately 200%. Furthermore, by 96 hours, phospho-eIF2α protein levels were elevated in UACC 903 cells by ~200% and in 1205 Lu cells by ~400%. Both total and phospho-eIF2α were unchanged in A375M. Importantly, at neither 48 nor 96 hours post transfection did P5CS knockdown increase GCN2 or eIF2α phosphorylation in normal human fibroblasts FF2441 (Fig. 6C), suggesting the effect observed was specific to the melanoma cells.
Fig. 6. siRNA-mediated knockdown of P5CS protein levels activated the GCN2 pathway in melanoma cells.
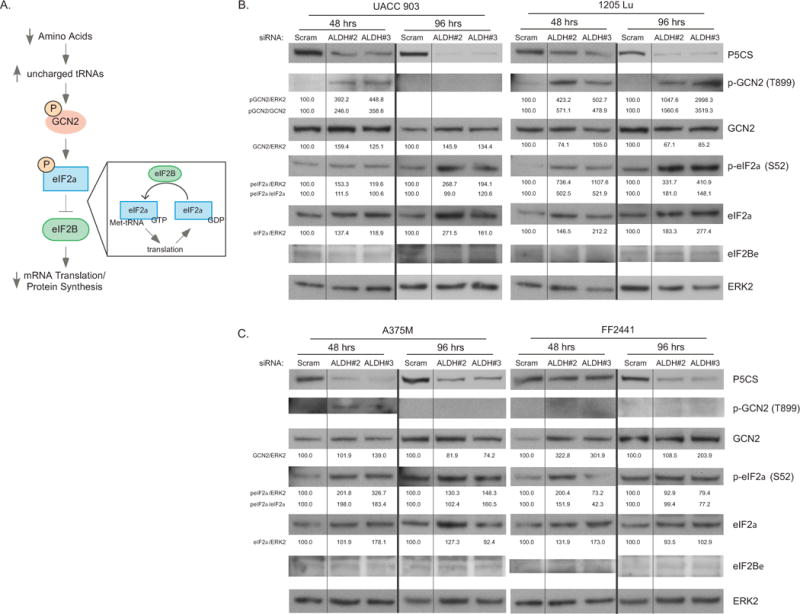
(A) Schematic flow diagram of GCN2 pathway activation. A decrease in amino acids causes an increase in respective uncharged tRNAs. GCN2 binds to uncharged tRNAs leading to GCN2 autophosphorylation. Activated GCN2 phosphorylates eIF2α, which then tightly binds to eIF2B preventing its guanine nucleotide exchange factor activity. This results in inhibition of mRNA translation initiation and protein production. (B, C) Melanoma cell lines UACC 903, 1205 Lu (B), and A375M (C) or the fibroblast cell line FF2441 (C) were transfected with siRNA targeting ALDH18A1 or a scrambled control siRNA. Western blots were probed for phosphorylated and total protein levels of GCN2 pathway members. Blots were quantitated by ImageJ, normalized to the ERK2 protein loading control, and expressed as a percentage of the siScrambled blot intensity.
mTOR is another nutrient sensor able to detect amino acid availability (13). To determine whether the mTOR pathway was altered by decreased proline levels mediated by P5CS knockdown, protein levels and phosphorylation states of downstream targets of mTOR were examined. mTOR directly phosphorylates and activates p70S6K (12) but knockdown of P5CS did not alter p70S6K phosphorylation (Fig. S9). mTOR also phosphorylates, to inactivate, 4EBP1 (12). Knockdown of P5CS decreased unphosphorylated alpha and beta 4EBP1 bands, and increased phosphorylated, active, gamma band (Fig. S9). Thus, p70S6K and 4EBP1 phosphorylation statuses act as controls to demonstrate the specificity of the effect of P5CS knockdown to mediate signaling through the GCN2 pathway. Collectively, these results suggest that disruption of P5CS activity leads to activation of the GCN2 pathway to inhibit mRNA translation, without inactivation of mTOR.
Proline supplementation of melanoma cells in which P5CS has been knocked down reversed effects on the GCN2 pathway
To demonstrate that the effects on the GCN2 pathway could be reversed by proline supplementation following P5CS knockdown, protein levels and phosphorylation states in cells grown in media supplemented with proline were examined. Proline supplementation of melanoma cells in which P5CS had been knocked down prevented phosphorylation of eIF2α in A375M or UACC 903 cells and prevented an increase in total eIF2α protein levels in UACC 903 (Fig. 7). While proline supplementation did not prevent increased total or phospho-eIF2α in 1205 Lu, it reduced the increase from approximately 500% to 190% by 48 hours (Fig. 7). Thus, proline supplementation reduced GCN2 pathway activation following P5CS protein knockdown.
Fig. 7. Proline supplementation during P5CS protein knockdown suppressed GCN2 activation.
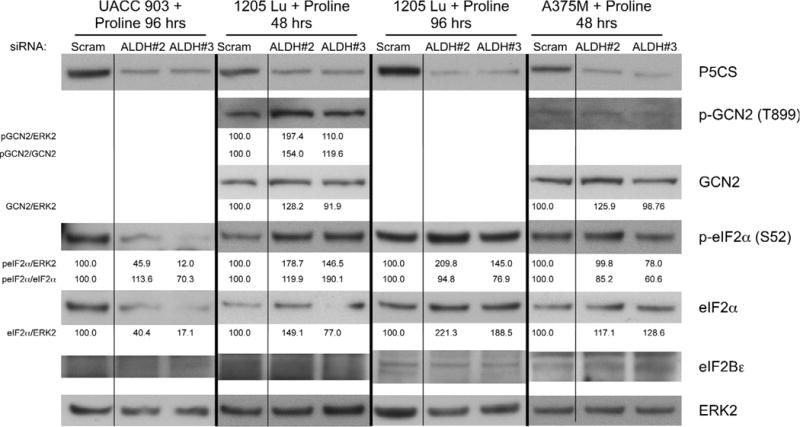
Melanoma cell lines UACC 903, 1205 Lu, and A375M were transfected with siRNA targeting ALDH18A1 or a scrambled control siRNA. Cells were maintained in 1.25 mM (UACC 903, A375M) or 2.5 mM (1205 Lu) proline media for 48 or 96 hours prior to collecting protein lysates. Western blotting compared phosphorylated and total protein levels. Protein band intensity was quantified using ImageJ software, normalized to ERK2 loading control, and expressed as a percentage of the scrambled siRNA blot intensity.
Discussion
High levels of protein synthesis are key to meet the high proliferative rates of cancer cells, suggesting that this process could be therapeutically targeted. This report demonstrates that disrupting proline biosynthesis can impair protein synthesis to inhibit melanoma tumor development. This is the first report to demonstrate that targeting P5CS, a protein necessary for proline biosynthesis, can significantly decrease melanoma tumor development. P5CS knockdown depleted intracellular proline levels to increase cell doubling time mediated by decreased protein synthesis. Although mTOR senses amino acid sufficiency and GCN2 responds to amino acid insufficiency, this study shows that only the GCN2 pathway responded to proline depletion in melanoma cells.
No trend in P5CS protein expression in melanoma cell lines or tumor lysates compared to melanocytes was observed in this study. This contradicts a previous report that found higher P5CS levels in melanomas (8). However, both studies reported increased intracellular proline levels in melanoma cells irrespective of mutational status. Although both studies focused on BRAF mutant cell lines, it is conceivable that a similar phenomenon occurs in wildtype BRAF cell lines, since these cell lines also contain elevated proline levels. Depletion of proline by P5CS knockdown did not cause cell death or cell cycle arrest, but slowed the growth rate of cells by increasing cell doubling times. Cell doubling time increased by at least 2.5 times because cells were simply growing more slowly. This is conceivable since protein synthesis is required for cell growth (35) and heavily relied upon for cancer cell survival (36). Therefore, short-term knockdown could reduce protein synthesis because of a reduced nutrient supply without inducing autophagy or apoptosis. Long-term knockdown of P5CS might activate an autophagic response in an attempt to replenish depleted amino acids (37) but this was not investigated in this study. Other groups have found that disruption of proline biosynthesis at other enzymatic steps of the pathway caused apoptosis and autophagy. For instance, overexpressing PRODH induced apoptosis through the mitochondrial and death receptor pathways (38, 39) and downregulated COX-2, EGFR, and Wnt/β-catenin pathways (19). Induction of apoptosis was dependent on ROS generation (39, 40); but no effect on ROS was observed in this study. A possible explanation is that targeting different enzymes in the proline biosynthesis pathway has variable effects on cells because each enzyme could uniquely affect its associated metabolites. Alternatively, there could be intrinsic differences of proline biosynthesis disruption in melanoma versus other cancer types.
Possible side effects to therapeutically targeting P5CS would need to be investigated. P5CS is a component of the proline biosynthesis pathway, and as such, is also needed for normal tissue functioning. mRNA levels of ALDH18A1 are expressed at high levels in the pancreas, kidney, testis, and ovary (17). Defects in P5CS can cause congenital disorders such as hypoprolinemia, hypoornithinemia, hypocitrullinemia, hypoargininemia and hyperammonemia, neurodegeneration, and connective tissue anomalies (18, 41, 42). It would be important to determine whether these phenotypes occur when targeting P5CS or whether the phenotypes are the result of early developmental problems. Targeting a therapy directly to tumors could alleviate possible side effects. In addition, since intracellular proline levels are elevated in melanomas, a therapeutic window may exist, whereby decreasing proline biosynthesis would impair cancer cell growth disproportionately to that of normal cells. A P5CS targeting drug should also not disrupt the functioning of other ALDH family members, which might be required for normal physiological processes.
Targeting P5CS could impair proline and ornithine synthesis since both uses P5C as a precursor. Proline and ornithine supplementation partially restored cell viability following P5CS knockdown. Ornithine can have multiple fates: being used in the urea cycle to remove ammonia (43), converted to proline through ornithine aminotransferase (OAT) (44), or converted to polyamines via ornithine decarboxylase (45). Ornithine supplementation might have the same effect as proline supplementation because ornithine can be converted to proline via OAT and PYCR. The ornithine supplementation effect did not appear to be mediated by its effect on the urea cycle because knockdown of P5CS did not decrease arginine or citrulline levels, both of which are members of the urea cycle. Furthermore, citrulline supplementation did not alter cell viability after P5CS knockdown compared to addition of proline and ornithine. The ornithine supplementation effect is also not from ornithine-specific effects on polyamines because supplementing media with various polyamines did not affect cell viability after P5CS knockdown (data not shown).
Proline supplementation only partially restored viability and protein synthesis of UACC 903 and 1205 Lu melanoma cells. In 1205 Lu cells proline supplementation did not restore protein synthesis at all until 8 days post transfection, possibly explaining why restoration of cell viability from proline in this cell line was minimal. This could have occurred because GCN2 was still activated. It is conceivable that 1205 Lu cells do not internalize extracellular proline as efficiently as the other cell lines. Alternatively, it is possible that this cell line will not internalize nonessential nutrients like proline until after nutrient stress, like GCN2 activation, ensues. It is also possible that knockdown of P5CS affects more processes than just protein synthesis.
Impairment of amino acid biosynthesis can have multiple effects in addition to protein production due to links between protein synthesis and other metabolic processes. For instance, proline biosynthesis not only regulates protein production but is also linked to glycolysis and the pentose phosphate pathway (PPP) through the proline cycle (10, 46, 47). Oxidation of NAD(P)H to NAD(P)+ by PYCR can be used by glycolysis or the PPP. Although glycolysis was responsive to proline availability in UACC 903 cells this did not occur in other cell lines, suggesting that while all melanoma cell lines activated the GCN2 pathway upon inhibition of proline biosynthesis, other unidentified cell line specific effects may explain this variability. Each cell line is unique and may differ in particular metabolic activities such as how closely proline biosynthesis is tied to glycolysis or the PPP.
Cellular responsiveness to proline synthesis inhibition could vary due to differences in the kinetics of cellular metabolism. Sensing amino acid levels through the amino acid response (AAR) can cause autophosphorylation of GCN2 at T899 whenever uncharged tRNAs are detected (15). This in turn activates GCN2 kinase activity allowing the phosphorylation of eIF2α (48). Normally, the guanine nucleotide exchange factor eIF2B exchanges GDP with GTP on eIF2α enabling protein synthesis (49). However, when eIF2α is phosphorylated at Ser52, it becomes a competitive inhibitor of eIF2B, preventing GEF activity and inhibiting mRNA translation (50, 51). Activation of this process depends on the timing of amino acid depletion. In this study, siRNA was used to deplete P5CS protein levels, which decreased proline biosynthesis, leading to depletion of intracellular proline levels. This process is therefore dependent on the half-life of P5CS, siRNA transfection efficiency, and proline turnover. Thus, timing of this sequential process will vary dependent on the cell line, which alters the timing of GCN2 pathway activation and protein synthesis inhibition.
GCN2 pathway activation requires eIF2α phosphorylation to enable inhibition of mRNA translation initiation and thus, protein synthesis inhibition. In this study, P5CS knockdown increased eIF2α phosphorylation and total eIF2α protein levels by 96 hours in UACC 903 and 1205 Lu cells. Even though absolute levels of phosphorylated eIF2α increased, relative proportion of eIF2α phosphorylation compared to total eIF2α protein levels did not change. Thus, cells may only need an increase in absolute phosphorylated eIF2α levels rather than a relative increase in eIF2α phosphorylation. Since total protein levels of eIF2Bε were unchanged upon P5CS knockdown, an absolute increase in phosphorylated eIF2α appears to be sufficient to bind and inhibit all eIF2B complexes. The increase in total eIF2α protein levels could be explained by binding of eIF2α to eIF2B causing increased protein stability.
GCN2 pathway activation and mTOR-dependent regulation of the eIF4F complex both regulate mRNA translation initiation, however, only the GCN2 pathway was affected when proline synthesis was disrupted in melanoma cells. mRNA translation initiation can be disrupted when 4EBP1 binds to eIF4E, a subunit of eIF4F (52). However, mTOR can override inhibition by phosphorylating and inactivating 4EBP1 (53). When 4EBP1 becomes phosphorylated it migrates to a higher molecular weight on polyacrylamide gels (54). Thus, the low band, α, is active 4EBP1 and the high band, γ, is inactive 4EBP1. P5CS knockdown decreased the active α 4EBP1 bands while increasing the γ 4EBP1 inactive bands, which is counterintuitive to what should be occurring with the decrease in protein synthesis. This likely can be explained by an increase in intracellular amino acids. With protein synthesis inhibited, an accumulation of amino acids would ensue. Indeed, protein synthesis inhibition using several protein synthesis inhibitors can activate mTOR signaling (55, 56). Thus, inhibition of mRNA translation initiation by GCN2 activation could cause the accumulation of amino acids, resulting in 4EBP1 phosphorylation.
In conclusion, this report suggests that targeting P5CS inhibits proline synthesis leading to decreased protein production mediated by the GCN2 pathway. Knockdown of P5CS decreased intracellular proline levels, leading to impaired tumor growth and increased cellular doubling time. P5CS knockdown activated the GCN2 pathway with a corresponding decrease in protein synthesis, both of which could be partially restored by proline supplementation. Thus, results of this study suggest that proline has a significant role in cancer development and reliance of cells on this amino acid can be exploited to therapeutically target melanoma cells.
Supplementary Material
Implications.
This study demonstrates that melanoma cells are sensitive to disruption of proline synthesis and provides a proof-of-concept that the proline synthesis pathway can be therapeutically targeted in melanoma tumors for tumor inhibitory efficacy.
Acknowledgments
The authors thank Dr. Scot Kimball for his valuable suggestions related to translation initiation regulation and for providing eIF2α and eIF2Bε antibodies for this study.
Financial Support: This work was supported by The Foreman Foundation for Melanoma Research, Melanoma Research Foundation and NIH grants CA-127892, CA-136667, and CA-138634.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Tarver T. Cancer Facts & Figures 2012. American Cancer Society (ACS) Journal of Consumer Health on the Internet. 2012;16:366–7. [Google Scholar]
- 2.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 5.Kugel CH, 3rd, Aplin AE. Adaptive resistance to RAF inhibitors in melanoma. Pigment cell & melanoma research. 2014 doi: 10.1111/pcmr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science (New York, NY) 1955;122:501–14. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 7.Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, et al. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. The Journal of biological chemistry. 2011;286:42626–34. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Ingeniis J, Ratnikov B, Richardson AD, Scott DA, Aza-Blanc P, De SK, et al. Functional specialization in proline biosynthesis of melanoma. PLoS One. 2012;7:e45190. doi: 10.1371/journal.pone.0045190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–6. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phang JM, Liu W, Hancock C, Christian KJ. The proline regulatory axis and cancer. Front Oncol. 2012;2:60. doi: 10.3389/fonc.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belitz IH-D, Grosch IW. Food chemistry. Springer Berlin Heidelberg; 1999. Amino acids, peptides, proteins; pp. 8–91. [Google Scholar]
- 12.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. The Journal of biological chemistry. 1998;273:14484–94. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 13.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature reviews Molecular cell biology. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 14.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nature reviews Molecular cell biology. 2004;5:827–35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Molecular cell. 2000;6:269–79. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 16.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–56. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 17.Hu CA, Lin WW, Obie C, Valle D. Molecular enzymology of mammalian Delta1-pyrroline-5-carboxylate synthase. Alternative splice donor utilization generates isoforms with different sensitivity to ornithine inhibition. The Journal of biological chemistry. 1999;274:6754–62. doi: 10.1074/jbc.274.10.6754. [DOI] [PubMed] [Google Scholar]
- 18.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Borchert GL, Surazynski A, Phang JM. Proline oxidase, a p53-induced gene, targets COX-2/PGE2 signaling to induce apoptosis and inhibit tumor growth in colorectal cancers. Oncogene. 2008;27:6729–37. doi: 10.1038/onc.2008.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–5. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 21.Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49:134–40. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 22.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8983–8. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satyamoorthy K, DeJesus E, Linnenbach AJ, Kraj B, Kornreich DL, Rendle S, et al. Melanoma cell lines from different stages of progression and their biological and molecular analyses. Melanoma research. 1997;7(Suppl 2):S35–42. [PubMed] [Google Scholar]
- 25.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. The Journal of investigative dermatology. 2004;122:337–41. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg SA, Niglio SA, Salehomoum N, Chan JL, Jeong BS, Wen Y, et al. Targeting Glutamatergic Signaling and the PI3 Kinase Pathway to Halt Melanoma Progression. Transl Oncol. 2015;8:1–9. doi: 10.1016/j.tranon.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–22. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 28.Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer research. 2006;66:9483–91. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 29.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer research. 2011;71:2750–60. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–6. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 31.Schulz J, Dettlaff S, Fritzsche U, Harms U, Schiebel H, Derer W, et al. The amido black assay: a simple and quantitative multipurpose test of adhesion, proliferation, and cytotoxicity in microplate cultures of keratinocytes (HaCaT) and other cell types growing adherently or in suspension. J Immunol Methods. 1994;167:1–13. doi: 10.1016/0022-1759(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 32.Kardos GR, Dai MS, Robertson GP. Growth inhibitory effects of large subunit ribosomal proteins in melanoma. Pigment cell & melanoma research. 2014 doi: 10.1111/pcmr.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599EB-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer research. 2005;65:2412–21. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- 34.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9482–7. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baxter GC, Stanners CP. The effect of protein degradation on cellular growth characteristics. J Cell Physiol. 1978;96:139–45. doi: 10.1002/jcp.1040960202. [DOI] [PubMed] [Google Scholar]
- 36.White-Gilbertson S, Kurtz DT, Voelkel-Johnson C. The role of protein synthesis in cell cycling and cancer. Molecular oncology. 2009;3:402–8. doi: 10.1016/j.molonc.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. The Journal of biological chemistry. 2005;280:31582–6. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Borchert GL, Surazynski A, Hu CA, Phang JM. Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: the role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene. 2006;25:5640–7. doi: 10.1038/sj.onc.1209564. [DOI] [PubMed] [Google Scholar]
- 39.Maxwell SA, Rivera A. Proline oxidase induces apoptosis in tumor cells, and its expression is frequently absent or reduced in renal carcinomas. The Journal of biological chemistry. 2003;278:9784–9. doi: 10.1074/jbc.M210012200. [DOI] [PubMed] [Google Scholar]
- 40.Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, et al. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer research. 2001;61:1810–5. [PubMed] [Google Scholar]
- 41.Kamoun P, Aral B, Saudubray JM. A new inherited metabolic disease: delta1-pyrroline 5-carboxylate synthetase deficiency. Bull Acad Natl Med. 1998;182:131–7. discussion 8–9. [PubMed] [Google Scholar]
- 42.Baumgartner MR, Rabier D, Nassogne MC, Dufier JL, Padovani JP, Kamoun P, et al. Delta1-pyrroline-5-carboxylate synthase deficiency: neurodegeneration, cataracts and connective tissue manifestations combined with hyperammonaemia and reduced ornithine, citrulline, arginine and proline. Eur J Pediatr. 2005;164:31–6. doi: 10.1007/s00431-004-1545-3. [DOI] [PubMed] [Google Scholar]
- 43.Meijer AJ, Lamers WH, Chamuleau RA. Nitrogen metabolism and ornithine cycle function. Physiol Rev. 1990;70:701–48. doi: 10.1152/physrev.1990.70.3.701. [DOI] [PubMed] [Google Scholar]
- 44.Smith RJ, Phang JM. The importance of ornithine as a precursor for proline in mammalian cells. J Cell Physiol. 1979;98:475–81. doi: 10.1002/jcp.1040980306. [DOI] [PubMed] [Google Scholar]
- 45.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–90. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 46.Phang JM, Downing SJ, Yeh GC, Smith RJ, Williams JA. Stimulation of the hexose-monophosphate pentose pathway by delta 1-pyrroline-5-carboxylic acid in human fibroblasts. Biochemical and biophysical research communications. 1979;87:363–70. doi: 10.1016/0006-291x(79)91805-9. [DOI] [PubMed] [Google Scholar]
- 47.Hagedorn CH, Phang JM. Catalytic transfer of hydride ions from NADPH to oxygen by the interconversions of proline and delta 1-pyrroline-5-carboxylate. Arch Biochem Biophys. 1986;248:166–74. doi: 10.1016/0003-9861(86)90413-3. [DOI] [PubMed] [Google Scholar]
- 48.Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–62. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 49.Webb BL, Proud CG. Eukaryotic initiation factor 2B (eIF2B) Int J Biochem Cell Biol. 1997;29:1127–31. doi: 10.1016/s1357-2725(97)00039-3. [DOI] [PubMed] [Google Scholar]
- 50.Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Molecular and cellular biology. 2001;21:5018–30. doi: 10.1128/MCB.21.15.5018-5030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimball SR. Eukaryotic initiation factor eIF2. Int J Biochem Cell Biol. 1999;31:25–9. doi: 10.1016/s1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- 52.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–7. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 53.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science (New York, NY) 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 54.Lawrence JC, Jr, Abraham RT. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends in biochemical sciences. 1997;22:345–9. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- 55.Beugnet A, Tee AR, Taylor PM, Proud CG. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J. 2003;372:555–66. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe-Asano T, Kuma A, Mizushima N. Cycloheximide inhibits starvation-induced autophagy through mTORC1 activation. Biochemical and biophysical research communications. 2014;445:334–9. doi: 10.1016/j.bbrc.2014.01.180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


