Abstract
The current challenge, now that two plant genomes have been sequenced, is to assign a function to the increasing number of predicted genes. In Arabidopsis, approximately 55% of genes can be assigned a putative function, however, less than 8% of these have been assigned a function by direct experimental evidence. To identify these functions, many genes will have to undergo comprehensive analyses, which will include the production of chimeric transgenes for constitutive or inducible ectopic expression, for antisense or dominant negative expression, for subcellular localization studies, for promoter analysis, and for gene complementation studies. The production of such transgenes is often hampered by laborious conventional cloning technology that relies on restriction digestion and ligation. With the aim of providing tools for high throughput gene analysis, we have produced a Gateway-compatible Agrobacterium sp. binary vector system that facilitates fast and reliable DNA cloning. This collection of vectors is freely available, for noncommercial purposes, and can be used for the ectopic expression of genes either constitutively or inducibly. The vectors can be used for the expression of protein fusions to the Aequorea victoria green fluorescent protein and to the β-glucuronidase protein so that the subcellular localization of a protein can be identified. They can also be used to generate promoter-reporter constructs and to facilitate efficient cloning of genomic DNA fragments for complementation experiments. All vectors were derived from pCambia T-DNA cloning vectors, with the exception of a chemically inducible vector, for Agrobacterium sp.-mediated transformation of a wide range of plant species.
The genomic analysis of plants has progressed rapidly in recent years with the sequencing of the Arabidopsis and rice genomes. The Arabidopsis genome was the first of these reference plant organisms to be sequenced and has revealed that close to 30,000 genes are all that are necessary for a plant to function (Arabidopsis Genome Initiative, 2000). The challenge at present, however, is to determine how these genes function at the cellular and organismal level and how genetic pathways evolve to produce the diversity that has allowed plants to reproduce and colonize diverse habitats. This functional analysis can be approached in a number of ways, most of which rely on the expression of transgenes to manipulate biological processes in transgenic plants. These approaches include phenotypic studies, made by generating gain-of-function or loss-of-function mutants. Gain-of-function can be achieved by placing a gene under the transcriptional control of a constitutive promoter (Wilson et al., 1996; Schaffer et al., 1998). Such ectopic expression of a gene may provide a phenotype that helps to elucidate its function. For example, the ectopic expression of a gene that causes a switch in cell fate helps to elucidate the role this gene may play in development.
Constitutive misexpression of genes, although useful in the first instance, may, however, mask tissue specific effects or may lead to lethality or sterility. To overcome these problems, several methods have been employed that control the expression temporally. Such methods include regulating gene expression using a heat shock promoter (Holtorf et al., 1995) or using a chemically induced expression system (Gatz et al., 1992; Weinmann et al., 1994; Böhner et al., 1999; Martinez et al., 1999; Bruce et al., 2000; Zuo et al., 2000). These approaches allow the activity of a gene to be studied at a specific point in the life cycle of a plant. Conversely, loss-of-function mutants can be produced using antisense constructs or dominant negative constructs and the phenotype of the plants analyzed in the absence of activity of that gene, this again may suggest its functional role. Gene expression studies that identify the expression pattern of a gene are further approaches that help elucidate gene function (Gawantka et al., 1998). In this type of analysis, evidence of the likely spatial and temporal domains of expression of a gene can be revealed. Promoter-reporter constructs are frequently used to provide supporting evidence of the functional role of genes by identifying the likely spatial and temporal domains of the expression of a gene (Batni et al., 1996; Curtis et al., 1997).
A further revealing approach to study gene function is to examine the subcellular localization of the corresponding protein. Here, studies are frequently achieved using chimeric gene fusion constructs with reporter genes. The location of the reporter protein in a subcellular compartment, as directed by the unknown fused protein, often provides additional supporting evidence for the function of the gene (von Arnim et al., 1997; Mayer et al., 1998).
Although all of these approaches are effective methods of identifying gene function, the production of the constructs is laborious and is often hampered by inappropriately positioned restriction sites that make the production of transgene constructs a "bottle neck" in plant gene functional analysis. This is particularly true when cloning large genomic DNA fragments or creating a large number of clones in a functional genomics project.
Here, we describe a complete plant vector set that aims to remove this "bottle neck," providing a reliable and effective method for the rapid directional cloning of genes and their promoters. This set of vectors is freely available for noncommercial purposes and provides a comprehensive plant molecular genetic tool kit that permits a variety of genetic manipulations from subcellular localization to inducible ectopic gene expression. These vectors facilitate high throughput DNA analysis and characterization of gene products by incorporating stop codons (in all three reading frames) adjacent to the 3′ end of a Gateway (Hartley et al., 2000) cassette (where appropriate), so that genes lacking a native stop codon and flanked by att recombination sites can be freely transferred to all vectors within the series, whether the intention is to misexpress a gene or to make fusions with either β-glucuronidase (GUS) or green fluorescent protein (GFP). This capability is not available in the currently described plant Gateway vectors (Karimi et al., 2002), nor is the facility to induce gene expression.
In plants, subcellular localization using GUS or GFP fusion proteins has been widely adopted to investigate protein targeting. However, often the limitations in the production of such fusion proteins has been tailoring the gene to fit the GUS or GFP fusion vector. An advantage of using this vector set is that any gene, minus its native stop codon, cloned into a donor vector (and thus flanked by attP sites) can be conservatively transferred in one step, in the correct orientation, at high efficiency, into any vector in the series that have been made in all three reading frames. A further advantage is that these vectors contain the 8-bp restriction recognition sites for AscI and PacI. These sites flank the att recombination cassettes so that positive identification of new recombination clones, in which new DNA has replaced the att recombination cassette, can be achieved efficiently.
RESULTS AND DISCUSSION
We have constructed a variety of Gateway-compatible binary T-DNA destination vectors for a wide range of different applications in plants (Fig. 1). Details of these vectors can be found on the Web site (http://www.unizh.ch/botinst/Devo_Website/curtisvector/), which provides the complete DNA sequence and restriction maps of all constructs. The Web site will be updated by adding new constructs and relevant information, as they become available. With the exception of pMDC7, the backbone of all Gateway-compatible destination vectors is derived from the pCambia series of binary vectors for Agrobacterium sp.-mediated plant transformation (http://www.cambia.org/). The pMDC7 vector is derived from PER8 (Zuo et al., 2000). The Gateway recombination site for introduction of a DNA fragment of interest was placed towards the right border of the T-DNA in the pCambia vectors. Most of the T-DNA destination vectors described contain the hygromycin phosphotransferase plant-selectable marker gene. This selectable marker was chosen so that these vectors would be compatible with a large number of insertion lines that are kanamycin resistant, for example Ds insertions (http://genetrap.cshl.org/ and http://enhancertraps.bio.upenn.edu/EnhancerTraps.html), the SALK T-DNA insertions (http://signal.salk.edu/tabout.html), or the Arabidopsis knockout facility Madison (http://www.biotech.wisc.edu/Arabidopsis/), or that are herbicide resistant, for example SLAT lines (http://nasc.nott.ac.uk/info/slat_info1.html; Tissier et al., 1999) or the Institut National de la Recherche Agronomique lines, which are both kanamycin and herbicide resistant (http://www.Arabidopsis.org/abrc/inra.html; Bechtold et al., 1993). Vectors for complementation, however, either contain the neomycin phosphotransferase II, the hygromycin phosphotransferase, or the bialaphos acetyltransferase gene, which confer resistance to kanamycin, hygromycin, and glufosinate ammonium, respectively. All three selectable markers are under the transcriptional regulation of the cauliflower mosaic virus (CaMV) 35S promoter and nos terminator (Odell et al., 1985) and are adjacent to the left border of the T-DNA. For a comprehensive description of vector construction, refer to the supplementary materials.
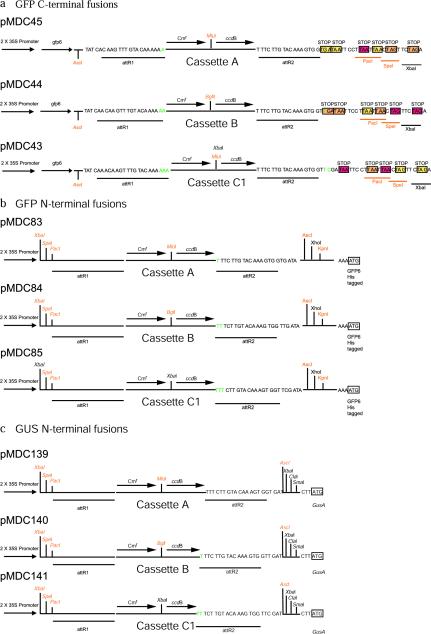
Figure 1.
A schematic illustrating the structure of the Gateway-compatible cloning vectors, showing the recombination sites flanked by AscI and PacI eight-nucleotide recognition sites. A, pMDC32, constitutive expression vector, harboring a dual 35S promoter, and pMDC30, a heat shock-inducible vector; B, pMDC7, derived from PER8, an estrogen-inducible vector. C, pMDC45, pMDC44, and pMDC43, for the construction of C-terminal GFP6 fusions. D, pMDC83, pMDC84, and pMDC85, for the construction of N-terminal GFP6his-tagged fusions. E, pMDC139, pMDC140, and pMDC141, for the construction of N-terminal GUS fusions. F, pMDC107, pMDC111, and pMDC110, for the construction of promoter-reporter (native promoter-gene fusion) GFP6 vectors. G, pMDC162, pMDC163, and pMDC164, for the construction of promoter-reporter (native promoter-gene fusion) GUS vectors. H, pMDC99, pMDC100, and pMDC123, for complementation of mutants with genomic fragments.
Destination Vectors for Constitutive Ectopic Gene Expression
To aid the production of constructs for the ectopic expression of genes in plants, the Gateway cassette B (see Fig. 2a) was placed adjacent to the dual 35S CaMV promoter in the destination vector pMDC32 (Fig. 1a). This promoter was chosen because it is highly active in most transgenic plant cells. The uidA gene (Jefferson et al., 1987) from an entry clone was inserted into the destination vector pMDC32 to produce an expression clone. The T0 and T1 generations of Arabidopsis ecotype Landsberg erecta transformed with this expression clone were tested to confirm that the Gateway-compatible construct was active in planta. These transgenic plant lines showed strong constitutive GUS activity in all of the expected plant tissue types associated with the expression of the 35S CaMV promoter (Odell et al., 1985). These plants also demonstrated that the att recombination sites do not inhibit transgene activity or interfere with enhancer activity (see supplementary data). This destination vector can, in addition to ectopic expression, be used to facilitate the rapid construction of antisense expression clones, although this latter application has not been tested.
Figure 2.
Nucleotide sequence adjacent to each Gateway cassette showing the reading frame for fusions with GFP (a and b) and GUS (c). These sequences also show the stop codons in the vectors where the attR2 site is followed by the PacI site but not in vectors where the attR2 site is followed by the AscI site.
Destination Vectors for Inducible Gene Expression
Heat-Inducible Vector
Although constitutive ectopic expression of genes provides a powerful tool for gene functional studies, the resulting ubiquitous expression may lead to lethality or may mask tissue-specific effects. We therefore produced a heat shock-inducible Gateway construct. The Gateway cassette B was placed downstream of a heat shock promoter (the Gmhsp17.3B promoter fragment from the SHS3252 plasmid) in the destination vector pMDC30 (Fig. 1a). Again, the uidA gene from an entry clone was used with the pMDC30 destination vector to produce an expression clone. T0 and T1 generations of Arabidopsis Landsberg erecta transformants were tested to confirm that the Gateway-compatible construct was heat inducible in planta (see supplementary data). In these plants, the transgene is strongly induced in newly dividing cells, particularly in developing leaves and in the primary and lateral root tips. Control plants showed no expression in the absence of heat treatment. Heat shock induction of gene activity is an effective method of providing temporal control of ectopic transgene expression, however, the physical stress of induction at 37°C may induce the expression of other genes in the plant, masking the effects of the gene under investigation. In our hands, no obvious detrimental effects on development were observed.
Estrogen-Inducible Vector
An alternative to heat shock induction is the use of chemical induction. An estrogen-inducible ectopic gene expression vector, PER8, was described by Zuo et al. (2000). This system shows efficient induction with no toxic effects in transgenic plants. The PER8 vector was kindly provided to us by Prof. Nam-Hai Chua and was made Gateway compatible using cassette B, to produce the destination vector pMDC7 (Fig. 1b). The uidA reporter gene was inserted down-stream of the lexA-binding domain and used to transform Arabidopsis Landsberg erecta plants. T0, T1, and T2 generation transformants were tested to confirm that the att recombination sites did not inhibit the estradiol-inducible expression of the reporter gene in vivo (see supplementary data). In these plants, the transgene is strongly induced ubiquitously, particularly in the roots, which were in direct contact with the inducer in the media. Control plants showed no expression in the absence of estradiol treatment.
The WUSCHEL cDNA was used to confirm that a gene without its native stop codon, when inserted into these vectors, would be expressed without interference from the additional nucleotides at the 3′ end that included the att recombination site and vector sequence prior to an inframe stop codon. WUSCHEL was chosen in conjunction with the modified PER8 vector (pMDC7) because the expression of this gene after 17-β-estradiol induction was well characterized (Zuo et al., 2002). WUSCHEL cDNA was amplified from plasmid A1-A (Mayer, 1998; a gift from Thomas Laux) without its native stop codon. This amplified product was used to produce an Entry clone. The WUSCHEL cDNA was inserted downstream of the lexA-binding site in pMDC7. In this vector, the WUSCHEL cDNA relies on one of the stop codons in the vector to provide a translational stop. The gene now has 36 additional nucleotides, excluding the stop codon at its 3′ end encoding 12 additional amino acids. T1 generation Arabidopsis Landsberg erecta transformants were tested to confirm that these additional nucleotides did not change the estradiol-inducible expression of WUSCHEL in vivo that was previously described by Zuo et al. (2002; Fig. 3). In these plants, plant hormone independent somatic embryo formation was observed only in the presence of the inducer, confirming that the incorporation of these additional nucleotides and use of a stop codon within the vector sequence has no deterimental effect on this randomly selected gene. Control plants showed no expression in the absence of estradiol treatment.
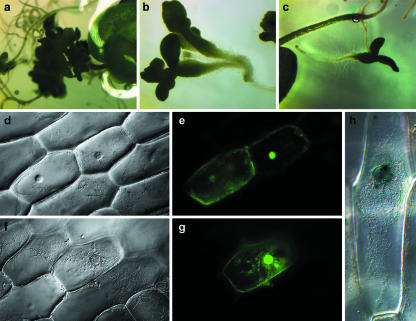
Figure 3.
17-β-Estradiol induced WUSCHEL expression (a-c) in 31-d-old seedlings of three independent transformants showing germinating somatic embryos growing at the primary and lateral root tips. Noninduced seedlings show no somatic embryo development (data not shown). The same WUSCHEL gene from the same entry clone was used to make C- and N-terminal fusions with GFP and N-terminal fusions with GUS. d, f, and h show light microscope images, and e and g show fluorescent microscope images. e, Fluorescent microscope image of pMDC114 expression in bombarded onion epidermal cells (WUSCHEL cDNA, fused to C-terminus of GFP, in pMDC43). g, Fluorescent microscope image of pMDC116 expression in bombarded onion epidermal cells (WUSCHEL cDNA, fused to N-terminus of GFP, in pMDC84). h, Light microscope image of pMDC153 expression in bombarded epidermal onion cells (WUSCHEL cDNA, fused to N-terminus of GUS, in pMDC141). All show that the marker protein was localized to the nucleus.
Destination Vectors for Analysis of Subcellular Localization of Proteins
To investigate the subcellular localization of particular proteins, a further series of T-DNA destination vectors was constructed to allow C- or N-terminal protein fusions with GFP or GUS.
The vectors pMDC43, pMDC44, and pMDC45 were produced for GFP C-terminal fusions (Fig. 1c), and the vectors pMDC83, pMDC84, and pMDC85 were produced for N-terminal fusions (Fig. 1d). These vectors provide Gateway cassettes in three reading frames (Fig. 2, a and b). Gateway-compatible N-terminal fusions with GUS can also be produced using vectors pMDC139, pMDC140, and pMDC141 (Fig. 1e), again in the three reading frames (Fig. 2c).
To confirm that these vectors were suitable for studying subcellular localization of plant proteins, we created fusions between the WUSCHEL protein, previously shown to be nuclear targeted (Mayer et al., 1998), and the GFP6 or GUS proteins. To illustrate the versatility of these vectors, the same WUSCHEL entry clone (described above, containing the WUSCHEL gene without its native stop codon) was used to generate GFP and GUS fusions with WUSCHEL.
These vectors were used in biolistic bombardment experiments on onion epidermal cells (Varagona et al., 1992), and the subcellular localization of the fusion proteins was investigated by light (GUS) and fluorescence (GFP) microscopy. The results demonstrated that the GFP and GUS proteins were targeted to the nucleus when fused to the WUSCHEL protein (Fig. 3). Thus the att recombination sites do not interfere with the ability of WUSCHEL to direct the subcellular localization of the GUS or GFP proteins.
Promoter-Reporter (or Native Promoter-Gene Fusion) Constructs
For promoter-reporter analysis with GFP6, the vectors pMDC107, pMDC110, and pMDC111 were constructed (Fig. 1f). These constructs were produced with Gateway cassettes in all three reading frames so that, in addition to promoter analysis, GFP6-gene fusions could be constructed so that the fused product is under the transcriptional control of the native promoter of the gene. Similarly, the vectors pMDC162, pMDC163, and pMDC164 were constructed for promoter-reporter analysis with GUS. These constructs were again made in all three reading frames to facilitate GUS-gene fusions for subcellular localization studies using an appropriate gene with its native promoter (Fig. 1g). To test the function of these vectors, the 35S promoter was amplified from plasmid pCambia 3300 (data not shown) so that it was flanked by attB1 and attB2 sites. This amplified product was used to make a 35S-promoter entry clone (construct available, data not shown). The 35S-promoter entry clone was used to generate a GUS expression clone (construct available; data not shown). Plants transformed with this construct showed constitutive GUS expression in transgenic plant tissues, consistent with the expression pattern of the 35S promoter (see supplementary data).
Constructs for Complementation Analysis of Mutant Plant Lines
For complementation analysis in mutant backgrounds, the T-DNA destination vectors pMDC99, pMDC100, and pMDC123 can be used to rapidly clone large fragments. These three vectors contain the Gateway cassette C1 and differ from each other only in the plant selectable markers that they contain. Vector pMDC99 confers hygromycin resistance, pMDC100 confers kanamycin resistance, and pMDC123 confers BASTA resistance. Fragments of Arabidopsis genomic DNA, up to 12 kb, have been cloned successfully between att recombination sites (Norbert Huck, personal communication) using Escherichia coli DH5α. It may be possible to clone larger fragments using bacterial strains such as Stbl2 (Invitrogen, Carlsbad, CA) that stabilize large genomic fragments.
In summary, we have produced a new set of Gateway-compatible plant expression vectors that enable efficient construction of transgenes for high throughput DNA analysis and characterization of gene products. These vectors provide a reliable method of cloning that reduces the usual multistep cloning approach to a single-step approach. This single-step approach is common to all vectors in the series so that a gene under investigation can be cloned using the same strategy into many vectors that have been designed to help elucidate gene function.
MATERIALS AND METHODS
Plasmid Construction
Standard gene cloning methods (Sambrook and Russell, 2001) were used to make the gene constructs. Detailed description of how the vectors were constructed can be found in the online version of this article under supplementary data at http://www.plantphysiol.org.
Plant Materials, Growth Conditions, and Plant Transformation
Arabidopsis Landsberg erecta plants were used for plant transformations using the floral dip method (Clough and Bent, 1998). Plants were grown under continuous white light at 22°C on Murashige and Skoog agar (1× Murashige and Skoog salts, 3% [w/v] Suc, and 0.8% [w/v] agar). Detailed description of methods used in experiments can be found in the online version of this article under supplementary data at http://www.plantphysiol.org.
AttB Primers, PCR, and Recombination Reaction for Introduction of Sequences into pDONR207
Primers with attB1 and attB2 sequences were purchased from Invitrogen. PCRs and in vitro BP clonase recombination reactions were carried out according to the manufacturer's instructions (Invitrogen). The product of recombination reactions (BP reactions) was used to transform competent Escherichia coli, strain DH5α using heat shock.
Recombination Reactions for Introduction of Sequences into Destination Vectors
LR clonase reactions to transfer DNA fragments from entry clones to destination vectors were carried out according to the manufacturer's instructions (Invitrogen). The product of recombination reactions (LR reactions) was used to transform competent E. coli strain DH5α using heat shock.
Distribution of Materials
All the vectors described in this publication will be made freely available and will be distributed from the University of Zurich for noncommercial research purposes (http://www.unizh.ch/botinst/Devo_Website/curtisvector/).
Supplementary Material
Acknowledgments
We thank Nam-Hai Chua (Rockefeller University, New York) for kindly providing the vector PER8, the Center for Application of Molecular Biology to International Agriculture for the pCambia vectors, David Jackson (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for the plasmid pSKp-mgfp6, and Thomas Laux (University of Freiburg, Germany) for the plasmid A1-A. We thank Valeria Gagliardini, Jana Schneider, and Brigitte Gabathuler for help with sequencing and Peter Kopf for technical assistance. We also thank Rita Gross-Hardt and Siân Curtis for critical reading of the manuscript and Jean-Jacques Pittet for his expert help with processing images found in the supplementary data.
The online version of this article contains Web-only data.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Batni S, Scalzetti L, Moody SA, Knox BE (1996) Characterization of the Xenopus rhodopsin gene. J Biol Chem 271: 3179-3186 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194-1199 [Google Scholar]
- Böhner S, Lenk I, Rieping M, Herold M, Gatz C (1999) Transcriptional activator TGV mediates dexamethasone-inducible and tatracycline-inactivatable gene expression. Plant J 19: 87-95 [DOI] [PubMed] [Google Scholar]
- Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B (2000) Expression profiling of the maize flavanoid pathway genes controlled by estrodiol-inducible transcription factors CRC and P. Plant Cell 12: 65-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Argobacterium-mediated transformation of Arabidopsis thaliana 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Rae AL, Rusu AG, Harrison SJ, Manners JM (1997) A peroxidase gene promoter induced by phytopathogens and methyl jasmonate in transgenic plants. Mol Plant-Microbe Interact 10: 326-338 [DOI] [PubMed] [Google Scholar]
- Gatz C, Frohberg C, Wendenburg R (1992) Stringent repression and homogeneous de-repression by tetracycline of modified CaMV35S promoter in intact transgenic tobacco plants. Plant J 2: 397-404 [DOI] [PubMed] [Google Scholar]
- Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, Blumenstock C, Niehrs C (1998) Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech Dev 77: 95-141 [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA (2000) DNA cloning using in-vitro site-specific recombination. Genome Res 10: 1788-1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtorf S, Apel K, Bohlmann H (1995) Comparison of different constitutive and inducible promoters for the overexpression of transgenes in Arabidopsis thaliana. Plant Mol Biol 29: 637-646 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193-195 [DOI] [PubMed] [Google Scholar]
- Martinez A, Sparks C, Hart CA, Thompson J, Jepson I (1999) Ecdysone agonist inducible transcription in transgenic tobacco plants. Plant J 19: 97-106 [DOI] [PubMed] [Google Scholar]
- Mayer KF (1998) Klonierung und Expressionsanalyse des Meristemgens WUSCHEL (WUS) von Arabidopsis thaliana. PhD thesis. Universität Tübingen, Tübingen, Germany
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805-815 [DOI] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 3: 13: 810-812 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1999) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219-1229 [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JD (1999) Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841-1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV (1992) Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 10: 1213-1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Osterlund MT, Kwok SF, Deng XW (1997) Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol 114: 779-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann P, Gossen M, Hillen W, Bujard H, Gatz C (1994) A chimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J 5: 559-569 [DOI] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G (1996) A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8: 659-671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Chau N-H (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in plants. Plant J 24: 265-273 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Frugis G, Chau N-H (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30: 349-359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.