Roy et al. identify a critical role for PDX1 in maintaining acinar cell identity, thus resisting the formation of PanIN-derived pancreatic ductal adenocarcinoma. Upon neoplastic transformation, the role of PDX1 changes from tumor-suppressive to oncogenic.
Keywords: pancreatic cancer, pancreatitis, EMT, dedifferentiation
Abstract
Aberrant activation of embryonic signaling pathways is frequent in pancreatic ductal adenocarcinoma (PDA), making developmental regulators therapeutically attractive. Here we demonstrate diverse functions for pancreatic and duodenal homeobox 1 (PDX1), a transcription factor indispensable for pancreas development, in the progression from normal exocrine cells to metastatic PDA. We identify a critical role for PDX1 in maintaining acinar cell identity, thus resisting the formation of pancreatic intraepithelial neoplasia (PanIN)-derived PDA. Upon neoplastic transformation, the role of PDX1 changes from tumor-suppressive to oncogenic. Interestingly, subsets of malignant cells lose PDX1 expression while undergoing epithelial-to-mesenchymal transition (EMT), and PDX1 loss is associated with poor outcome. This stage-specific functionality arises from profound shifts in PDX1 chromatin occupancy from acinar cells to PDA. In summary, we report distinct roles of PDX1 at different stages of PDA, suggesting that therapeutic approaches against this potential target need to account for its changing functions at different stages of carcinogenesis. These findings provide insight into the complexity of PDA pathogenesis and advocate a rigorous investigation of therapeutically tractable targets at distinct phases of PDA development and progression.
Pancreatic ductal adenocarcinoma (PDA) is the third leading cause of cancer-related mortality in the United States and is slated to become the second by the end of this decade (Rahib et al. 2014). Despite recent therapeutic advances, the 5-year survival remains an abysmal 8% (Siegel et al. 2016). Consequently, understanding the molecular mechanisms of disease evolution and progression is imperative for developing novel therapeutic strategies.
PDA is thought to progress through a series of precursor lesions termed pancreatic intraepithelial neoplasia (PanIN) (Hruban et al. 2000). Consistent with >90% of PDA containing KRAS oncogenic mutations (Bailey et al. 2016), mutant KRAS appears to be responsible for PDA initiation. Mouse models expressing oncogenic Kras throughout the pancreatic parenchyma faithfully recapitulate the human disease with PanIN formation and progression to adenocarcinoma (Aguirre et al. 2003; Hingorani et al. 2003, 2005). Although its duct-like morphology suggests a ductal epithelial origin, recent data demonstrate that duct cells are mostly refractory to Kras-induced transformation, while PanIN lesions readily arise from mature acinar cells (De La O et al. 2008; Habbe et al. 2008; Kopp et al. 2012). The apparent morphologic transition of acinar cells to duct cells mimics a process associated with pancreatic wound healing, known as acinar-to-ductal metaplasia (ADM). Following injury, in conditions such as chronic pancreatitis, acinar cells transdifferentiate to form metaplastic ducts, gaining progenitor-like properties (Miyamoto et al. 2003). Once the insult has subsided, it is hypothesized that metaplastic ducts proliferate and differentiate back into acinar cells, allowing for repopulation of the exocrine pancreas. Oncogenic KRAS can initiate ADM but hijacks the healing process by blocking acinar redifferentiation and promoting a transition from metaplasia to PanIN (Morris et al. 2010).
Pancreatic and duodenal homeobox 1 (PDX1) is a transcription factor essential for pancreas development (Jonsson et al. 1994; Offield et al. 1996). Initially expressed in the pancreatic anlage, it is required for differentiation of all pancreatic cell lineages (Jonsson et al. 1994; Offield et al. 1996; Holland et al. 2002; Hale et al. 2005; Gannon et al. 2008). PDX1 expression persists at high levels in β cells, where it is required for efficient insulin gene transcription (Ohlsson et al. 1993; Ahlgren et al. 1998; Holland et al. 2002), but is maintained at lower levels in exocrine cells (Guz et al. 1995; Wu et al. 1997) where its function has not been thoroughly investigated. Pdx1 is up-regulated in the adult pancreas in ADM induced by constitutive overexpression of TGFα (Song et al. 1999). Likewise, expression of oncogenic KRAS in the pancreatic parenchyma leads to neoplasia with increased expression of PDX1 (Hingorani et al. 2003). PDX1 up-regulation in these models suggests its possible role in metaplasia and neoplasia. Indeed, PDX1 has been proposed as an oncogene, as its overexpression in PDA cell lines increases proliferation, invasiveness, and growth in soft agar (Liu et al. 2008). However, a more recent large-scale study suggests that PDX1 loss is associated with a more aggressive subtype of PDA (Bailey et al. 2016). With these seemingly contradictory findings in mind, we set out to clarify the roles of PDX1 in the adult exocrine pancreas and its associated diseases, examining organ homeostasis, pancreatitis, tumorigenesis, and PDA progression using an acinar cell-specific Pdx1 conditional knockout mouse and RNAi approaches.
Results
Pdx1 maintains acinar cell identity
To analyze PDX1 expression at the early stages of pancreatic neoplasia, we used the Ptf1aCre/+;KrasLSL-G12D/+;Trp53f/+ (KPC) mouse model of PDA. These animals harbor PanINs by 4–6 wk of age that progress to well-differentiated PDA by 18–24 wk of age. We observed uniformly strong PDX1 expression in ADM and PanIN lesions (Supplemental Fig. S1A) and low-level expression in acinar cells in both adult murine and human pancreata (Supplemental Fig. S1A–C). Thus, increased PDX1 expression correlates with transformation. Pdx1 expression in embryonic acinar cells has been noted (Offield et al. 1996), but its low-level expression in adult acinar cells has been largely dismissed. Considering prior work pointing to acinar cells as progenitors for PanIN-derived PDA, we were interested in whether PDX1 performs critical functions in acinar cells.
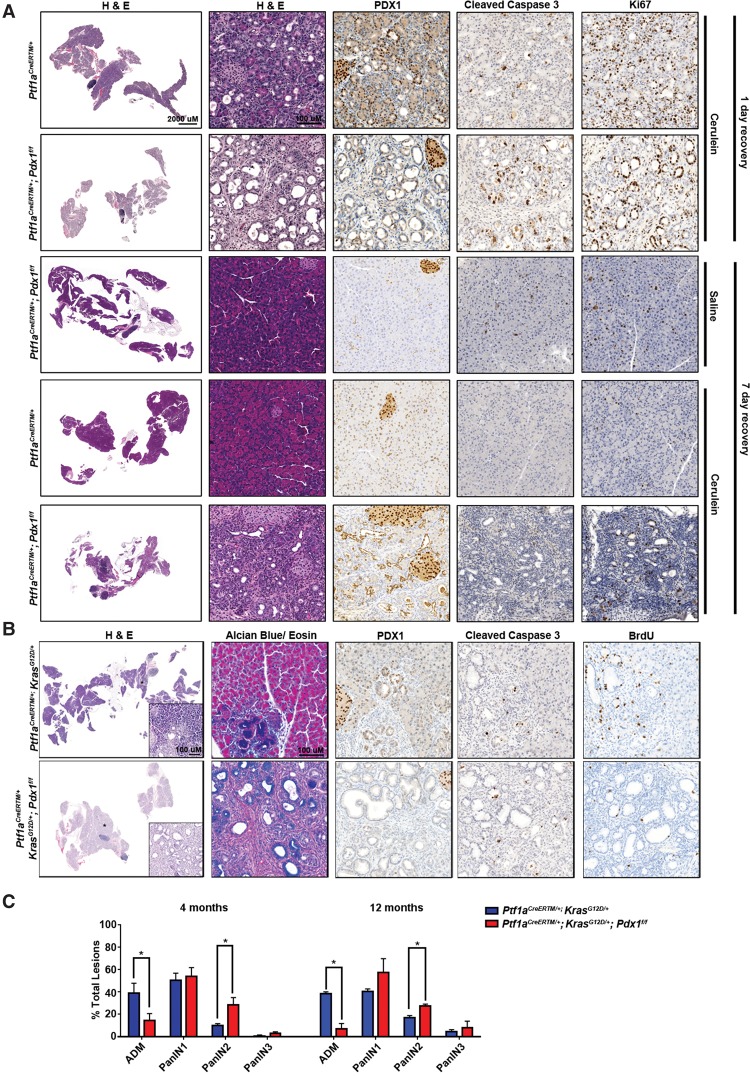
To address Pdx1 function in acinar cells, we used Ptf1aCreERTM/+;Pdx1f/f animals to ablate Pdx1 specifically from adult acinar cells via tamoxifen administration (Pan et al. 2013). Eight-week-old to 12-wk-old Ptf1aCreERTM/+ (control) and Ptf1aCreERTM/+;Pdx1f/f animals were tamoxifen-treated to induce recombination and sacrificed 2 wk later. Pancreata from control animals appeared histologically normal, with PDX1 expression high in islet and low in acinar, centro-acinar, and ductal cells (Supplemental Fig. S2A). In insulin-producing β cells, PDX1 maintains cell function, and its depletion erodes their differentiation status (Puri et al. 2014). Pdx1 ablation from acinar cells did not induce any overt exocrine abnormalities despite an increase in both apoptosis and proliferation (Supplemental Fig. S2A). We then treated cohorts of mice with cerulein, a cholecystokinin analog that stimulates transient ADM in wild-type pancreata. Adult Ptf1aCreERTM/+ and Ptf1aCreERTM/+;Pdx1f/f animals were treated with tamoxifen followed by cerulein to induce ADM. One day after cerulein treatment, Ptf1aCreERTM/+;Pdx1f/f mice presented with enhanced acinar dedifferentiation accompanied by increased apoptotic activity (Fig. 1A). Lesions in both wild-type and Pdx1f/f pancreata were proliferative, as shown by Ki67-positive staining. Metaplastic lesions resolved within 7 d after cerulein in control animals, but Ptf1aCreERTM/+;Pdx1f/f animals maintained widespread ADM and had a severe reduction in the pancreas to body mass ratio, demonstrating that ADM derived from acinar cells require PDX1 for redifferentiation (Fig. 1A; Supplemental Fig. S2C). Of note, the profound loss of acinar cells in the cerulein-treated Ptf1aCreERTM/+;Pdx1f/f pancreata 7 d after treatment makes the PDX1 expression in the normal ducts not affected by the Ptf1aCreERTM/+ driver especially evident (Fig. 1A). These results highlight that it is the acinar cell-derived metaplastic ducts that are uniquely capable of regenerating the damaged organ. Consistent with these in vivo data, acinar cell explants derived from Ptf1aCreERTM/+;Pdx1f/f mice treated with tamoxifen transdifferentiated to ductal structures more rapidly than Ptf1aCreERTM/+-derived explants in response to cerulein treatment (Supplemental Fig. S2B). Collectively, PDX1 appears to both maintain and re-establish acinar differentiation in injury models.
Figure 1.
Pdx1 loss accelerates inflammation and oncogene-induced ductal metaplasia. (A) Acinar cells lacking Pdx1 undergo more rapid and extensive dedifferentiation. Eight-week-old to 12-wk-old Ptf1aCreERTM/+ and Ptf1aCreERTM/+;Pdx1f/f mice were treated with tamoxifen. Three days following tamoxifen administration, animals were treated with saline or cerulein for 7 d and allowed to recover for 1 or 7 d. Representative H&E stainings at the indicated time points are shown. PDX1 immunohistochemistry was performed to confirm efficient Pdx1 ablation. Immunohistochemistry for cleaved caspase 3 and Ki67 measured apoptosis and cell proliferation, respectively. (B) Eight-week-old to 12-wk-old Ptf1aCreERTM/+;KrasLSL-G12D/+ and Ptf1aCreERTM/+;KrasLSL-G12D/+;Pdx1f/f mice were treated with tamoxifen and euthanized 6 wk later. Two hours prior to euthanasia, mice were injected intraperitoneally with BrdU. Representative H&E staining is shown. Alcian blue staining highlights the mucinous region, whereas PDX1 immunohistochemistry demonstrates efficient Pdx1 deletion. Immunohistochemistry for cleaved caspase 3 and BrdU measured apoptosis and cell proliferation, respectively. (C) Ductal lesions (ADM and PanIN 1–3) were counted and graded in 10 10× fields from three pancreata of each genotype and time point. Samples were blinded. Shown are the percentages of specific grades compared with the total counted.
Next we sought to determine whether Pdx1 contributes to Kras-induced transformation. To address this, we crossed KrasLSL-G12D/+ mice with Ptf1aCreERTM/+;Pdx1f/f animals. Eight-week-old to 12-wk-old Ptf1aCreERTM/+;KrasLSL-G12D/+;Pdx1f/f and Ptf1aCreERTM/+;KrasLSL-G12D/+ control animals were tamoxifen-treated and sacrificed 6 wk after treatment. As expected, controls expressing only KrasG12D in their acinar cells formed infrequent ADM lesions (von Figura et al. 2014a). In contrast, the additional ablation of Pdx1 showed widespread loss of acinar cells, accompanied by extensive formation of ductal lesions (Fig. 1B). Pdx1-null lesions showed histological and molecular characteristics of PanIN, such as highly acidic mucin content marked by positive Alcian blue staining (Fig. 1B), but were less proliferative, as shown by BrdU incorporation. Thus, the accelerated transformation in Pdx1-null animals appeared to be due to rapid erosion of acinar differentiation rather than expansion through increased cell proliferation. No differences were observed in apoptosis. In line with their accelerated initial transformation, pancreata from Pdx1-null mice had rapidly progressed past ADM to form high-grade PanIN (PanIN 2/3) at 4 and 12 mo of age (Fig. 1C) compared with age-matched controls. Collectively, our results point to a dynamic pattern of Pdx1 expression that maintains acinar cell differentiation during pancreatic damage and oncogenic Kras-driven transdifferentiation. Acinar redifferentiation can then be overridden by KrasG12D expression or blocked by Pdx1 ablation, with the two synergizing to enhance the overall amount of transformation (Supplemental Fig. S2D).
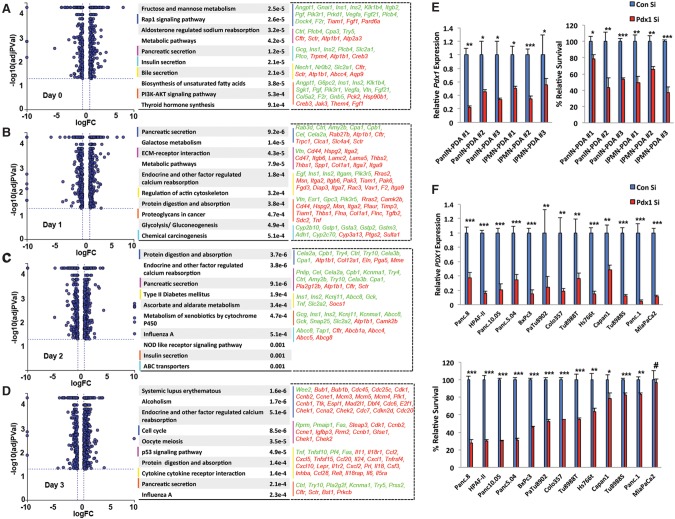
To gain mechanistic insight into how Pdx1 inhibits ADM, we performed genome-wide transcriptome profiling. Acinar cells were isolated from Ptf1aCreERTM/+ or Ptf1aCreERTM/+;Pdx1f/f animals treated with tamoxifen and incubated with TGFα to induce ADM. Cells were harvested at days 0, 1, 2, and 3 following TGFα treatment, and gene expression was analyzed by RNA sequencing (RNA-seq). At day 0, even before evident ductal metaplasia, Pdx1 ablation initiated destabilization of acinar identity (Fig. 2A). Pathway enrichment analysis revealed several acinar-specific genes to be down-regulated (e.g., Cpa3), whereas genes defining duct cells, such as Cftr, were up-regulated in Pdx1-null acinar cells, suggesting that Pdx1 may function as a “gatekeeper” of acinar identity (Fig. 2A; Supplemental Fig. S3). At days 1 and 2, when acinar cells underwent initial dedifferentiation, several genes belonging to pancreatic secretion pathways were differentially regulated in Pdx1-null acinar cells (Fig. 2B,C). Specifically, genes that define the mature acinar cell state, such as Amy2b, Cpa1, Cpb1, Cel, and Cela2a, were down-regulated in Pdx1-null acinar cells, pointing toward accelerated erosion of the mature acinar state. In addition, genes regulating ECM–receptor interactions, actin cytoskeleton, and proteoglycans were differentially expressed. Interestingly, at day 3, when the acinar ductal reprogramming process had concluded, differentially regulated genes belonged to the networks that support tumorigenesis, such as cell cycle, p53 signaling, and cytokine/receptor interactions (Fig. 2D). These findings shed light on how Pdx1 function promotes acinar identity in wild-type acinar cells and how it may regulate aspects of redifferentiation to resolve ADM. Importantly, longer-term loss of Pdx1 promotes proliferative signals that may complement oncogenic Kras in the transition from ADM toward PanIN and PDA.
Figure 2.
PDX1 regulates distinct sets of genes in acinar cells undergoing ADM. (A–D) Acinar cells were isolated from tamoxifen-treated Ptf1aCreERTM/+ (n = 3) and Ptf1aCreERTM/+;Pdx1f/f (n = 3) mice. Cells were treated with TGFα to induce in vitro ADM. At days 0, 1, 2, and 3, cells were harvested, and whole-genome transcriptome profiling was done by RNA-seq. The graphs at the left show differentially expressed genes between Pdx1 wild-type and Pdx1-null acinar cells at each time point. The tables at the right show pathway analyses of genes differentially regulated at least ±1.5-fold. The top 10 pathways from each data set are shown along with their respective P-values. Pathways of interest are color-coded, and genes that belong to those pathways are listed. Green indicates genes down-regulated in Pdx1-null cells, and red indicates up-regulated genes in Pdx1-null cells. (E, left) Quantitative PCR analysis of Pdx1 in mouse PDA-derived cell lines transfected with control or Pdx1 siRNA. (Right) Relative survival of cells following siRNA-mediated Pdx1 knockdown in mouse PDA-derived cell lines. (F, top) Quantitative PCR analysis of PDX1 in human PDA-derived cell lines transfected with control or PDX1 siRNA. (Bottom) Relative survival of cells following siRNA-mediated PDX1 knockdown in human PDA-derived tumor cell lines.
Opposing roles of Pdx1 in tumor maintenance
Pdx1 ablation promotes acinar dedifferentiation. However, prior work demonstrated a profound increase in PDX1 expression in neoplastic lesions, indicating that its function may change during cellular transdifferentiation and tumorigenesis. To test this, we attenuated Pdx1 expression in mouse PDA-derived cell lines. Two independent PanIN-derived PDA tumor lines were transfected with constructs expressing doxycycline (dox)-inducible Pdx1 targeted or control shRNAs. In both cell lines, dox treatment significantly reduced PDX1 expression compared with controls (Supplemental Fig. S4A, top). Pdx1 deficiency blocked cell growth, suggesting protumorigenic functions of PDX1 (Supplemental Fig. S4A, bottom), consistent with previous reports (Wu et al. 2014). As PDA can originate from neoplastic lesions other than PanIN, we asked next whether Pdx1 has analogous oncogenic activities in PDA emerging from PanIN or intraductal papillary mucinous neoplasia (IPMN). Six different primary tumor cell lines were analyzed, including three isolated from PanIN-derived PDA from Ptf1aCre/+;KrasLSL-G12D/+ mice and three cell lines established from IPMN-derived PDA from Ptf1aCre/+;KrasLSL-G12D/+;Brg1f/f mice. Reducing Pdx1 expression using siRNA in these PDA lines impaired cell survival (Fig. 2E), demonstrating that PDX1 maintains tumorigenicity of malignant cells irrespective of their origin (von Figura et al. 2014a).
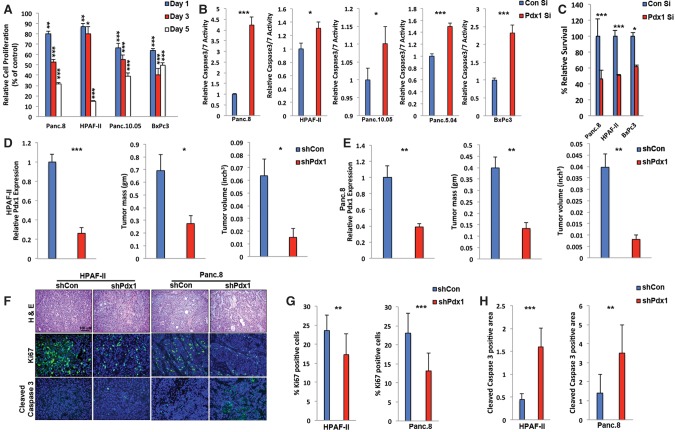
We next addressed whether PDX1 has similar tumor-promoting activities in human PDA. We compared PDX1 expression in several human PDA-derived tumor lines with HPDE, a human duct cell line that expresses low levels of PDX1, and human islets, which have high PDX1 expression. Most PDA cell lines expressed PDX1 at levels significantly higher than HPDEs but lower than islets (Supplemental Fig. S4B), emphasizing that moderate PDX1 up-regulation accompanies exocrine cell transformation. To assess whether PDX1 up-regulation influenced tumor cell survival, we reduced PDX1 expression using siPDX1 in 13 different human PDA lines. Similar to the mouse PDA lines and consistent with previous studies (Yu et al. 2016), decreasing PDX1 expression led to reduction in cellular growth or survival, albeit at variable levels (Fig. 2F). To elucidate the protumorigenic functions of PDX1 in greater detail, we performed cell proliferation and apoptosis assays. In four independent human PDA lines, siPDX1 reduced cell proliferation (Fig. 3A) and increased cleaved caspase 3/7 activity (Fig. 3B). We conclude that PDX1 has two apparent protumorigenic functions in established tumors: maintenance of cellular proliferation and inhibition of apoptosis.
Figure 3.
PDX1 maintains tumorigenicity of human PDA cells in vivo. (A) Relative cell proliferation of the indicated human PDA-derived cell lines at 1, 3, and 5 d following transfection with control or PDX1 siRNA. (B) Relative caspase 3/7 activity of the indicated human PDA-derived cell lines 24 h after transfection with control or PDX1 siRNA. (C) Relative survival of cells in three dimensions following siRNA-mediated PDX1 knockdown in human PDA-derived tumor cell lines. HPAF-II (D) and Panc.8 (E) cells expressing either control shRNA or shRNA targeting PDX1 were subcutaneously injected into NSG mice, and tumor growth was measured. Mice were sacrificed 4 wk after inoculation, and tumor sizes were measured. (F) H&E and costaining of Ki67, cleaved caspase 3, and DAPI of tumors obtained in D and E. Quantification of Ki67-positive cells (G) and the cleaved caspase 3-positive area (H) of tumors obtained in D and E.
Another important characteristic of tumor cells is their ability to grow in anchorage-independent conditions. Attenuating PDX1 expression in three-dimensional (3D) culture conditions resulted in reduced colony formation, indicating that PDX1 maintains the transformed phenotype of PDA cells (Fig. 3C). To examine PDX1 function in vivo, we stably transduced two human PDA lines with lentivirus expressing shControl or shPDX1 and transplanted them subcutaneously into immunocompromised NSG mice. Tumor size was significantly lower in shPDX1-expressing cells (Fig. 3D,E) due to reduced cell proliferation and increased apoptosis (Fig. 3F–H). Thus, in contrast to its role in resisting acinar transformation, PDX1 promotes tumorigenicity of PDA cells (Supplemental Fig. S4C).
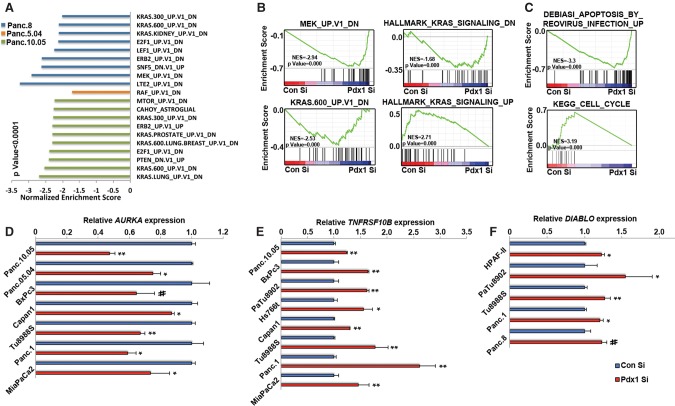
To gain mechanistic insight into how PDX1 loss reduces tumorigenicity, we performed genome-wide transcriptome analysis of four different human PDA lines transfected with either control or PDX1 siRNA (Supplemental Fig. S5A). Several genes were consistently differentially regulated between control and PDX1 siRNA-treated cells, but the extent to which gene expression was affected varied significantly. Gene set enrichment analysis (GSEA) on differentially expressed genes revealed several oncogenic pathways that were perturbed, the most prominent among them being the KRAS signaling pathway. In three out of four lines, genes down-regulated by oncogenic KRAS or its downstream effectors, RAF/MEK, were enriched in PDX1 siRNA-treated cells, suggesting that the KRAS signaling pathway is affected by PDX1 knockdown (Fig. 4A,B). Surprisingly, when the MAPK pathways downstream from KRAS were surveyed, we found that PDX1 down-regulation consistently resulted in higher pERK levels (Supplemental Fig. S5B,C). It is possible that up-regulation of pERK is a compensatory feedback mechanism that tumor cells activate in order to overcome the stress induced by PDX1 knockdown, which is accompanied by up-regulation of proapoptotic genes and attenuation of cell cycle genes associated with proliferation (Fig. 4C). We then focused on apoptosis and cell cycle pathways to identify key factors in these pathways that may be regulated by PDX1. Although the gene sets were heterogeneous between cell lines, several genes were up-regulated/down-regulated in at least three out of four cell lines analyzed. Among them was Aurora kinase A (AURKA), a serine/threonine protein kinase that promotes G2/M transition during the cell cycle. We further confirmed that PDX1 positively regulates AURKA expression by quantitative PCR, suggesting a defect in G2–M cell cycle transition that may lead to reduced proliferation of cells following PDX1 knockdown (Fig. 4D). Several apoptosis-regulating factors were also differentially expressed between control and PDX1 siRNA cells, including the proapoptotic factors Caspase 3, Diablo, FADD, FAS, TNFRSF10B, and TNFSF10 and anti-apoptotic protein XIAP (Supplemental Fig. S5D). Quantitative PCR showed that TNFRSF10B and Diablo were up-regulated in several PDA lines following PDX1 siRNA treatment, suggesting that PDX1 attenuation triggers a signaling cascade predisposing cells to undergo apoptosis (Fig. 4E,F). Thus, PDX1 enhances the tumorigenicity of PDA cells by controlling several critical pathways that regulate cell proliferation and apoptosis.
Figure 4.
PDX1 maintains a transcriptional landscape conducive to oncogenic Kras signaling. (A) GSEA of RNA-seq data from Supplemental Figure S5A. Genes differentially expressed at least ±1.5-fold between control siRNA and PDX1 siRNA-treated cells were compared with gene set-defining oncogenic signatures. (B) Kras signature enrichment plots in human PDA-derived cell lines transfected with control or PDX1 siRNA. (C) Cell cycle and apoptosis signature enrichment plots in human PDA-derived cell lines transfected with control or PDX1 siRNA-treated cells. Quantitative PCR analysis of AURKA (D), TNFRSF10B (E), and DIABLO (F) in human PDA-derived cell lines transfected with control or PDX1 siRNA.
Profound shifts in PDX1 chromatin occupancy in acinar cells vs. PDA
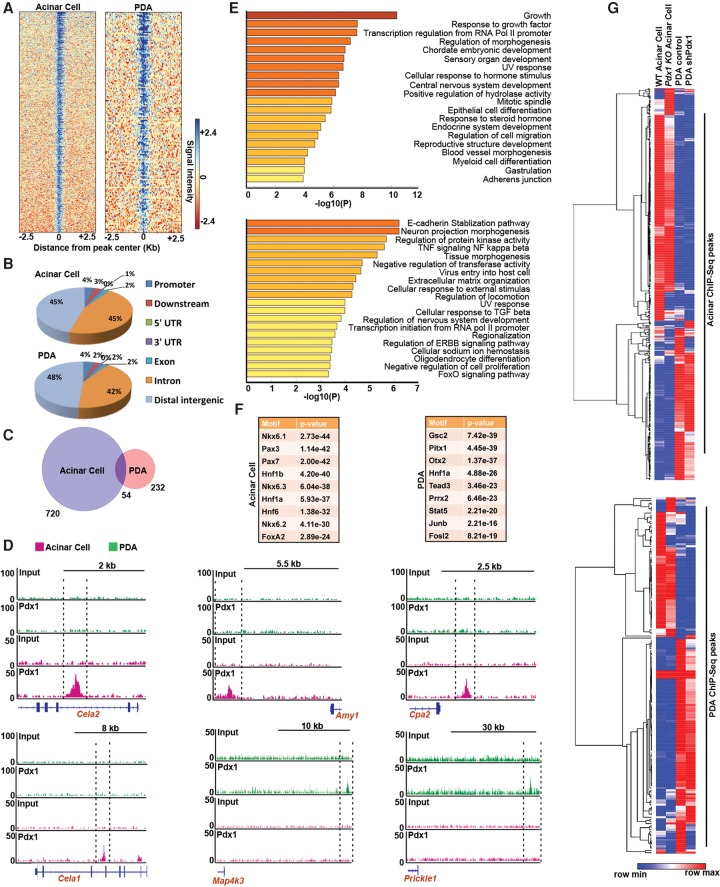
Our data demonstrate that, during the earliest transitional stages from acinar cells toward neoplasia, Pdx1 restrains neoplastic transformation. In contrast, Pdx1 maintains the tumorigenic properties of transformed PDA cells (Supplemental Fig. S4C), a shift that is accompanied by robust changes in gene expression profiles. To determine how Pdx1 performs these discordant functions in normal and transformed cells, we performed PDX1 ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) profiling on primary mouse acinar cells and mouse PDA-derived cell lines (Fig. 5A). Region gene association studies revealed that PDX1 binds mostly to intergenic regulatory regions (Fig. 5B; Supplemental Fig. S6A). Notably, there was minimal overlap between PDX1-bound regions in acinar and PDA cells, indicating that PDX1 regulates distinct gene sets in primary and transformed cells (Fig. 5C). Pathway enrichment analysis of PDX1-bound regions revealed that, in acinar cells, PDX1 binds to genes associated with endocrine development and differentiation, embryonic development, and epithelial cell differentiation (Fig. 5E, top). Examples of the latter include Nr5a2, FoxA2, and Onecut1, transcription factors driving pancreatic development. In addition, PDX1 binding was enriched on Cela1, Cela2, Cpa2, and Amy1, genes that define the acinar differentiation state (Fig. 5D). In tumor cells, PDX1 bound to genes in important networks regulating tumorigenesis, including the E-cadherin stabilization pathway (associated with epithelial-to-mesenchymal transition [EMT]), TNF-NFκβ, cellular response to TGFβ, and regulation of ERBB signaling (Fig. 5E; bottom). For instance, PDX1 was uniquely bound to the MAPK pathway gene Map4k3 and the Wnt/β-catenin pathway component Prickle1 in PDA cells but not acinar cells (Fig. 5D). PDX1 enrichment in these regions correlates with active transcription of associated genes. Acinar-specific genes (such as Amy1 and Cela1) display higher H3K4me3 and H3K27ac enrichment in acinar cells compared with PDA cells (Supplemental Fig. S6C,E,F). Conversely, PDA-specific genes (Map3k1, Map4k3, Mapkapk2, and Cdkn2b) exhibit an increased repressive mark (H3K27me3) in acinar cells compared with PDA cells (Supplemental Fig. S6D). These data highlight an important regulatory shift in PDX1 function from maintenance of an acinar-specific differentiation program to promotion of oncogenic signaling in PDA. Motif analysis revealed that PDX1-bound regions in acinar cells were also enriched for transcription factors defining the pancreas differentiation state, such as Hnf4a, Nkx6.1, Hnf1a, and Hnf1β (Fig. 5F; Supplemental Fig. S6B). In contrast, in PDA, motifs were enriched for some pancreas differentiation factors but also oncogenic transcription factors such as Gsc2, Prrx2, Stat5, c-Jun, and c-Fos.
Figure 5.
PDX1 dynamically switches its function between acinar cells and PDA. (A) PDX1 ChIP-seq was performed on acinar cells isolated from wild-type mice and the mouse PDA cell line NB-490. The heat map shows enrichment of PDX1 binding in acinar and PDA cells. (B) PDX1 ChIP-peak distribution across different genomic regions in primary acinar cells or PDA cells (NB490). (C) Overlap in PDX1-bound regions between acinar and PDA cells. (D) ChIP-seq signal tracks of PDX1 on the indicated genes in acinar and PDA cells. (E) Functional categories of PDX1 putative target genes by using pathway enrichment analysis. The list shows the top 20 pathways enriched in PDX1 ChIP-seq targets in acinar (top) and PDA (bottom) cells (F) Sequence motifs identified by motif analysis of PDX1-bound regions in acinar and PDA cells. (G) Heat map showing relative expression of genes bound by PDX1 in either acinar cells (top) or PDA cells (bottom) in four different cells types: wild-type acinar cells, Pdx1-null acinar cells, wild-type mouse PDA cells, and PDA cells expressing shRNA against Pdx1.
To show direct correspondence between PDX1 binding and effects on gene expression, we next integrated RNA-seq and ChIP-seq data (Fig. 5G). We examined the expression of genes bound by PDX1 in either acinar cells (Fig. 5G, top) or PDA cells (Fig. 5G, bottom) in four different cell types: wild-type and Pdx1-null acinar cells and wild-type and Pdx1 knockdown murine PDA cells. It is evident from the analysis that the expression of genes bound by PDX1 in either cell type is significantly different between wild-type acinar and PDA cells. Also, genes bound by PDX1 in acinar cells are differentially expressed largely between wild-type and Pdx1-null acinar cells. On the other hand, expression of genes bound by PDX1 in PDA cells is mostly affected by Pdx1 depletion in PDA cells, with only a fraction affected in acinar cells. The dynamic changes in PDX1 binding to acinar-specifying genes to oncogenic signaling pathways reveal an important underlying mechanism of the context-dependent roles of PDX1.
Distinct PDX1 expression levels in PDA subtypes
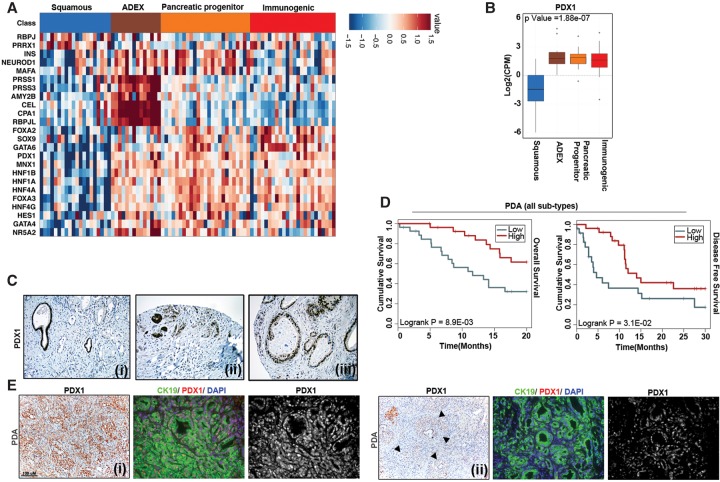
Recent next-generation sequencing analysis has revealed that PDA is a conglomeration of four different subtypes with distinct histopathological characteristics and disparate survival prognoses (Collisson et al. 2011; Bailey et al. 2016). Interestingly, in the squamous subtype that carries the worst prognosis, the PDX1 promoter is highly methylated, suggesting epigenetic silencing of PDX1. This seemingly contradictory finding led us to analyze the expression pattern of pancreas lineage-specifying factors, including PDX1, in different PDA subtypes (Fig. 6A). Unsupervised expression clustering for these factors revealed PDX1 to be highly expressed in the pancreatic progenitor ADEX and immunogenic PDA subtypes but observed at low levels in the squamous subtype (Fig. 6B). We further confirmed the heterogeneous PDX1 expression pattern among PDA patients by immunohistochemistry (Fig. 6C). Furthermore, in two different cohorts that encompassed all PDA subtypes, low PDX1 expression was significantly associated with lower overall and disease-free survival (Fig. 6D; Supplemental Fig. S7A). However, PDX1 expression did not have any statistically significant correlation with disease outcome in the pancreatic progenitor subclass, where patients display high PDX1 (Supplemental Fig. S7B).
Figure 6.
Low PDX1 expression predicts poor PDA prognosis. Expression of transcription factors regulating pancreatic organogenesis (A) and PDX1 (B) in four different pancreatic cancer subtypes based on the study by Bailey et al. (2016). (C) Representative PDX1 staining in human PDA with low (panel i), intermediate (panel ii), and high (panel iii) PDX1 expression in infiltrating tumors. Normal ducts in panel i and islets in panel ii stained positive for PDX1. (D) Kaplan-Meier survival curve of PDA patients with low or high PDX1 RNA expression. n = 183. (E) Representative staining for PDX1/cytokeratin 19 in Ptf1aCre/+;KrasLSL-G12D/+;Trp53f/+ mouse pancreata showing variable expression (high [panel i] and low [panel ii]) of PDX1 in mouse tumor tissues. Solid arrowheads indicate regions that have reduced PDX1 expression.
We also observed that genetically engineered mouse models faithfully recapitulate heterogeneity in PDX1 expression (Fig. 6E). In some areas of PDA, expression was very high (Fig. 6E, panel i), whereas others displayed a complete lack of PDX1 expression (Fig. 6E, panel ii). Thus, not only does PDX1 have distinct functions in acinar versus cancer cells, but reduced PDX1 levels correlate with the most aggressive squamous tumor type.
PDX1 activity regulates EMT
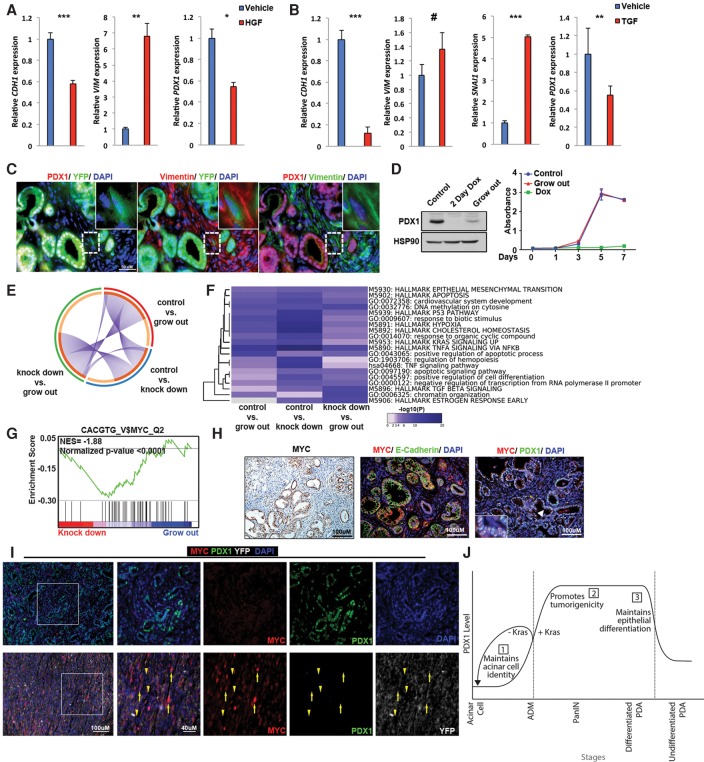
In contrast to our cell culture data, which suggest that reducing PDX1 expression in established tumors would be beneficial, patient data revealed that low-PDX1 patients show worse survival. Metastasis to distant organs, rather than primary tumor burden, is frequently the cause of death in PDA patients (Yachida et al. 2010). Intriguingly, the squamous subtype, which loses PDX1 expression, displays molecular characteristics of increased metastatic potential. We reasoned that, independent of its effects on proliferation and survival, PDX1 down-regulation might enhance tumor aggressiveness. EMT, where epithelial cells lose epithelial marks in order to disseminate into a vascular or lymphatic system, enhances metastasis (Kalluri and Weinberg 2009). To test whether PDX1 down-regulation is part of the EMT program, we examined two models: HGF (hepatocyte growth factor) treatment of HPAF-II cells and TGFβ treatment of Panc.1 cells (Song et al. 2010). As shown in Figure 7, both HGF and TGFβ treatment induced EMT, accompanied by reduced PDX1 expression (Fig. 7A,B). To further confirm the potential role of Pdx1 in EMT in vivo, we used Ptf1aCre/+;KrasLSL-G12D/+;Trp53f/+;R26YFP/+ animals. In these animals, cells expressing Cre recombinase were irreversibly marked with YFP, thus allowing for detection of epithelial-derived cancer cells that have undergone EMT and now express mesenchymal markers; e.g., YFP/Vimentin-double-positive. Similar to our in vitro observations, epithelial cells that have converted to a mesenchymal state exhibited low PDX1, supporting the notion that, in order to undergo EMT, epithelial cells need to reduce PDX1 expression (Fig. 7C). Likewise, in matched tissue sections from mouse PDA, invasive tumor cells displayed lower PDX1 expression compared with adjacent PanIN lesions, suggesting that PDX1 loss may confer a metastatic advantage (Supplemental Fig. S8).
Figure 7.
PDX1 loss is associated with EMT. (A) Quantitative PCR analysis of E-cadherin, Vimentin, and PDX1 expression in HPAF-II cells treated with either vehicle or HGF. (B) Quantitative PCR analysis of E-cadherin, Vimentin, Snail1, and PDX1 expression in Panc.1 cells treated with either vehicle or TGFβ. (C) Fluorescent images of lineage-labeled cells derived from pancreatic epithelium of Ptf1aCre/+;KrasLSL-G12D/+;Trp53f/+;Rosa26LSL-YFP animals. Staining shows that epithelial cells that have undergone EMT (YFP-positive/Vimentin-positive) have lost PDX1 expression. (D) Mouse PDA cells (NB490 sh1) were cultured and passaged with continual dox treatment for 3 wk to select for cells resistant to Pdx1 knockdown (“grow out”). Western blot analysis of PDX1 and HSP90 from “grow out” cells treated with dox and NB490 sh1 cells left untreated (control) or treated with dox for 48 h (2-d dox). Growth curves of NB 490 sh1 cells left untreated (control) or treated with dox and “grow out” cells treated with dox. (E) Overlap among differentially expressed genes (1.5-fold up or down) in control, knockdown, and “grow out” cells. Purple curves link identical genes. The inner circle represents gene lists, where hits are arranged along the arc. Genes shared by multiple lists are colored dark orange, and genes unique to a list are shown in light orange. (F) Heat map of enriched terms across differentially expressed genes (1.5-fold up or down) in the indicated groups, colored by P-value. (G) MYC-binding signature enrichment plots in knockdown versus “grow out” cells. The plot shows the enrichment of MYC-binding targets in “grow out” cells. (H) Immunofluorescence staining from a Ptf1aCre/+;KrasLSL-G12D/+;Trp53f/+ mouse showing colocalization of MYC and PDX1 in PanIN. (I) Immunofluorescence staining of a well-differentiated tumor from an 11-wk-old PDX1-Cre;KrasLSL-G12D/+;Trp53R172H/+ mouse (top panels) and a poorly differentiated invasive tumor from a 28-wk-old PDX1-Cre;KrasLSL-G12D/+;Trp53R172H/+;Rosa26LSL-YFP mouse (bottom panels). Yellow arrows highlight YFP-positive tumor cells, while yellow arrowheads highlight YFP-negative stromal cells. (J) Model depicting PDX1 functions at different stages of pancreatic cancer tumorigenesis. At early stages, PDX1 maintains acinar identity and inhibits Kras-driven acinar dedifferentiation. In the later malignant stage, PDX1 maintains tumorigenicity of cells by supporting cell proliferation and inhibiting apoptosis. During metastasis, malignant cells lose PDX1 expression to undergo EMT.
The observed functions of PDX1 in PDA maintenance and metastasis appear contradictory. Primary tumor cells require sustained PDX1 expression for survival but attenuate PDX1 expression as part of EMT and metastasis. To reconcile these functions, we hypothesized that tumor cells use mechanisms to compensate for PDX1 loss. To identify such compensatory mechanisms, we generated cell lines that are resistant to Pdx1 knockdown. By serially passaging PDA cells expressing dox-inducible Pdx1 shRNA grown continuously in dox, Pdx1 knockdown cells grew out within weeks (Fig. 7D) and were as proliferative as the parental line, indicating that they have acquired traits to resist Pdx1 loss.
To identify the mechanism by which these cells tolerate the absence of Pdx1, we performed BrU-seq (bromouridine [BrU] sequencing) to identify new transcripts synthesized after acquiring resistance to reduced Pdx1 (Paulsen et al. 2014). Cells were treated with BrU to label nascent RNA. Labeled RNA was precipitated with an anti-BrdU antibody and sequenced. With the cutoff set at 1.5-fold change in expression, we found 348 genes differentially expressed between control and Pdx1 knockdown, 569 genes differentially expressed between knockdown and “grow out,” and 385 genes differentially expressed between control and “grow out” cells. When compared with each other, there was little overlap between each data set, suggesting that “grow out” cells initiate a unique program to become resistant to Pdx1 depletion (Fig. 7E). Pathway analysis revealed several pathways that were enriched in “grow out” cells: “TGFβ signaling,” “chromatin organization,” and “estrogen response pathway” (Fig. 7F). Among them, “chromatin organization” was particularly interesting, as chromatin remodeling is a frequent mechanism of chemoresistance. Two-thirds of the genes comprising the “chromatin organization” pathway were found to be up-regulated in “grow out” cells. Among them, we detected an overrepresentation of histones, suggesting transcriptional amplification (Supplemental Fig. S9A). Interestingly, when genes differentially expressed between “knockdown” and “grow out” cells were used for GSEA for motif enrichment, the MYC-binding motif was the only significantly enriched motif (Fig. 7G), which was associated with enrichment of the Myc regulatory network (Supplemental Fig. S9B). MYC has been shown to induce transcriptional amplification in tumor cells, which contributes to tumor maintenance (Lin et al. 2012). Thus, we conclude that, at the metastasis stage, tumor cells reduce Pdx1 to undergo EMT, temporarily sacrificing proliferative potential, and then use MYC to revive cell growth. In KPC animals, uniform MYC and PDX1 colocalization was observed consistently in PanIN lesions (Fig. 7H). Interestingly, when PanINs had progressed to PDA, there was a divergence of this coexpression pattern. We found two groups of tumors in KPC animals: well differentiated and undifferentiated. Well-differentiated tumors were PDX1-high and MYC-low, whereas undifferentiated invasive tumors were MYC-high and PDX1-low (Fig. 7I). This is consistent with human PDA, where metastasis-prone squamous subtype cancers display MYC pathway activation, are PDX1-low, and have mutations in chromatin modifier genes (Bailey et al. 2016).
Discussion
PDX1 function in the adult pancreas is historically considered to be restricted to β cells (Ahlgren et al. 1998; Gannon et al. 2008; Gao et al. 2014). Despite this common perception, prior studies have shown Pdx1 expression in normal adult exocrine pancreata and increased expression in PDA, observations that imply possible regulatory roles of PDX1 in neoplastic development and progression (Roy and Hebrok 2015). Our analysis indicates temporally distinct and even seemingly antagonistic functions of PDX1 during maintenance of mature acinar differentiation, injury-induced acinar regeneration, PanIN, PDA, and metastasis. At the onset of oncogenic Kras-driven transformation, PDX1 restrains acinar cells from transdifferentiating into ductal cells, thus acting as a “tumorigenesis-suppressor.” In contrast, PDX1 assumes a tumor-promoting role when ADM lesions progress toward neoplasia and into the malignant state. In cancer cells, PDX1 supports the malignant phenotype by controlling a distinct group of genes compared with normal acinar cells, suggesting that not only the level of expression but redirected binding to specific regulatory elements are critical aspects of its diverse activities. Finally, in a subset of epithelial tumor cells, PDX1 loss promotes EMT and metastasis to distant organs (Fig. 7J).
The roles of PDX1 during embryogenesis have been studied extensively and show that it is expressed in all pancreatic epithelial cells at the onset of organogenesis (Jonsson et al. 1994; Offield et al. 1996). While PDX1 is required to establish the acinar cell lineage, only low-level expression is detected in adult enzyme-producing cells (Hale et al. 2005; Kodama et al. 2016). Thus, it was surprising that Pdx1 depletion in adult mice led to perturbations in exocrine function (Holland et al. 2002). However, ectopic Pdx1 expression in progenitors induces abundant ADM (Kawaguchi et al. 2002; Miyatsuka et al. 2006), and Elastase-Cre-driven Pdx1 overexpression in exocrine precursors triggers marked dysmorphogenesis in the exocrine pancreas (Heller et al. 2001). These studies indicate that Pdx1 expression needs to be tightly controlled to properly maintain adult acinar tissue functionality. Supporting this notion, our data show that Pdx1 deletion in mature acinar cells in the presence of oncogenic/inflammatory signals produces a phenotype comparable with ectopic overexpression in embryonic progenitor cells. Of note, our analysis further suggests that once acinar identity is eroded, Pdx1 reactivation supports tumor progression by controlling a distinct transcriptional program capable of promoting neoplasia, the first indication that PDX1 fulfills different roles within the same cells but in the different cellular contexts of homeostasis and disease.
The notion that a defined network of transcriptional factors, such as Ptf1a, Mist1, and Nr5a2, maintains acinar integrity and impairs mutant Kras-driven ADM has been described before (Shi et al. 2013; Flandez et al. 2014; von Figura et al. 2014b; Krah et al. 2015). Our results indicate that Pdx1 joins this list of “acinar differentiation” regulators. Consistent with this idea, our RNA-seq analysis suggests that PDX1 maintains acinar identity by direct binding to regulatory elements of acinar differentiation genes such as Amy1, Cela2, Cela1, and Cpa2. PDX1 exerts its transcriptional activity through interaction with distinct sets of coregulators, such as PBX1b and MEIS2, in acinar cells (Swift et al. 1998). In future work, it will be interesting to examine changes in the composition of coregulators during ADM, PanIN, and PDA development and whether this alters PDX1 binding to a diverse set of target genes.
Our data indicate that a subset of human PDA, centro-acinar, and ductal cells expresses PDX1, an observation analogous to the results obtained in mice. Intriguingly, in human PDA samples, PDX1 levels vary widely. Earlier studies described either no correlation or high PDX1 to be associated with poor prognosis (Koizumi et al. 2003; Park et al. 2011). In contrast, using two different patient cohorts, we show conclusively that low, not high, PDX1 expression is an independent predictor of poor disease-free survival. Also, considering the heterogeneity of PDX1 expression in KPC mouse models and PDA subtypes, it is possible that not all subtypes were included in the smaller patient cohort size (n = 35 in Koizumi et al. 2003; n = 67 in Park et al. 2011). Our analysis of a larger cohort, totaling 325 patients and representing all subgroups, may explain the newly evident correlation between survival and PDX1 expression.
Brunicardi and colleagues (Yu et al. 2016) had shown previously that PDX1 knockdown reduces cell survival in human PDA lines. We observed a similar effect, but our examination of multiple PDA lines suggests that all tumor cells are not equally vulnerable to PDX1 deficiency. Based on our observation of disparate PDX1 levels in different PDA patients harboring tumors of differing subtypes, it is possible that the gene network intrinsic to these various PDA subtypes likely predicts the response to PDX1 knockdown.
The exact role of PDX1 in metastasis is hard to pinpoint. Prior work on a mouse model of PDA has revealed that a subpopulation of PDX1+ cells has an inherent capacity to metastasize (Ischenko et al. 2014). However, other data together with our findings point to distinct PDX1 activities in resident and metastatic PDA cells. One recent study showed PDX1 to be down-regulated during EMT (David et al. 2016). In vivo, metastatic lesions also exhibit loss of nuclear PDX1 expression, further suggesting a possible regulatory role of PDX1 in EMT (Morton et al. 2008). Similarly, we found that EMT in PDA attenuates PDX1 expression both in vitro and in vivo. PDX1 is known to activate expression of the epithelial marker E-cadherin (Marty-Santos and Cleaver 2016), supporting the suggestion that its loss might be necessary for EMT. Consistent with this notion, in the context of oncogenic Kras, our preliminary analysis suggests that Pdx1-null animals display more frequent delamination events and higher numbers of circulating tumor cells compared with their wild-type counterpart (data not shown). However, PDX1 overexpression is not sufficient to reverse EMT (Ischenko et al. 2014), suggesting that PDX1 loss is associated with, and may initiate, a broadly remodeled cancer phenotype.
Evidence for this hypothesis comes from selecting for cells capable of expanding in the absence of Pdx1. In these cells, we observed up-regulation of a Myc regulatory network, which is negatively regulated by PDX1 and confers metastatic properties to PDA cells (Chen et al. 2007; Bailey et al. 2016). Our data support the possibility of dual-targeting PDX1 and MYC in PDX1-dependent tumors as a viable therapeutic opportunity for the PDA patients.
In summary, our findings point to PDX1 as a context-dependent mediator of PDA initiation and progression. Based on our comprehensive analysis, it is evident that to define PDX1 as an entirely pro- or anti-cancer factor is naïve, and, without further understanding its diverse functions, treating it as a therapeutic target, as has been proposed (Yu et al. 2016), is premature. Future studies are needed to identify how PDX1 might interact with divergent sets of coregulators to modulate critical aspects of cancer formation. Unraveling the temporal activities of PDX1 should provide important insights into subtype-specific disease vulnerabilities that can be targeted for personalized therapeutic intervention.
Materials and methods
Mouse lines
The following mice strains were used: Ptf1aCre, Ptf1aCreERTM, and Pdx1f/f (Gannon et al. 2008; Roy and Hebrok 2015); KrasLSL-G12D, PDX1-Cre, and Trp53R172H (gifts of David Tuveson, Cold Spring Harbor Laboratory); and Trp53f/f, R26RLSL-YFP, and NOD scidγ (purchased from the Jackson Laboratory). Mice were crossed on a mixed background. The University of California at San Francisco and University of Michigan Institutional Care and Use of Animals Committees (IACUCs) approved all of the mouse experiments.
RNA-seq for acinar cells
RNA was harvested using the RNEasy Plus kit (Qiagen, 74136) and assessed for quality using the TapeStation (Agilent). Samples with an RNA integrity number of ≥8 were rRNA-depleted using Ribo Minus (Invitrogen, K1550-04). rRNA-depleted samples were prepared using the TruSeq mRNA sample preparation version 2 kit (Illumina, RS-122-2001 and RS-122-2002). The entire fraction of 0.1–3 µg of rRNA-depleted total RNA was fragmented and copied into first strand cDNA using reverse transcriptase and random primers. cDNA 3′ ends were adenylated, and adapters were ligated. One of the adapters had a 6-nucleotide barcode that was unique for each sample, allowing us to multiplex in a HiSeq flow cell (Illumina). Products were purified and enriched by PCR to create the final cDNA library. Libraries were checked for quality and quantity by TapeStation (Agilent) and quantitative PCR using a library quantification kit for Illumina platforms (Kapa Biosystems, KK4835). They were clustered on the cBot (Illumina) and sequenced 24 samples per lane on six lanes of a 50-cycle single-end HiSeq 4000. HiSeq control software version 3.3.52 was used according to manufacturer's protocols. Demultiplexing and FastQ file generation were done using bcl2fastq version 2.17.1.14.
ChIP-seq
Chromatin was prepared from isolated acinar cells or cell lines using the truChIP chromatin-shearing reagent kit (Covaris, 520154). Chromatin was sheared in 1-mL millitubes (Covaris, 520135) using a Covaris S200 (5% duty cycle, intensity of 4, 200 cycles per burst, for 8 min) to obtain chromatin fragments 100–700 base pairs in length. Fifty micrograms of fragmented chromatin was incubated overnight with 2.5 μL of rabbit anti-PDX1 antibody (Millipore, 07-696). Complexes were isolated using 50 μL of Dynabeads (Invitrogen, 10004D) previously blocked with 1 mg/mL BSA. Two nanograms of immunoprecipitated DNA was prepared as a standard Illumina library using the ThruPLEX DNA-seq kit (Rubicon) according to the manufacturer's protocol. Samples were PCR-amplified and pooled. Final libraries were checked for quality and quantity by TapeStation (Agilent) and quantitative PCR using Kapa's library quantification kit for Illumina sequencing platforms (Kapa Biosystems, KK4835). They were clustered on the cBot (Illumina) and sequenced three samples per lane on a 50-cycle single-end HiSeq 2000 in high-output mode using version 3 reagents. The raw reads were aligned to mm10 assembly by Bowtie1 using unique mapping mode, meaning the reads that could be mapped to more than one location were discarded. MACS2 was used for transcription factor peak calling, and then blacklist islands were removed from the peaks. Peaks were annotated by HOMER for their locations relative to nearby genes.
BrU-seq
BrU-seq on “grow out” cells treated with dox and NB490 sh1 cells left untreated or treated with dox for 48 h was performed as described previously (Paulsen et al. 2014). Briefly, BrU (Aldrich) was added to the culture medium of cells to a final concentration of 2 mM, and cells were then incubated for 30 min at 37°C. Cells were lysed in TRIzol reagent (Invitrogen), and cell lysates were scraped off and collected. Total RNA was isolated, and BrU-labeled RNA was captured by incubation of isolated total RNA with anti-BrdU antibodies (BD Biosciences) conjugated to magnetic Dynabeads (Invitrogen) under gentle agitation for 1 h at room temperature. cDNA libraries were prepared from isolated Bru-labeled RNA using the Illumina TruSeq library kit and sequenced using Illumina HiSeq sequencers at the University of Michigan DNA Sequencing Core. Sequencing and read mapping were carried out as described previously (Paulsen et al. 2014).
Acinar cell explants
Pancreata were harvested, washed twice in Hank's buffered salt solution (HBSS), minced, and incubated with 0.2 mg/mL Collagenase P (Roche, 11249002001) for 15 min at 37°C. Tissue was washed three times in HBSS containing 5% FBS and filtered through 500-μm and 105-μm polypropylene mesh (Spectrum Laboratories, 106436 and 106418). Filtrate was centrifuged through 30% FBS in HBSS and resuspended in complete Waymouth medium (1× Waymouth MB 752/1 medium, 50 mg/mL gentamycin, 0.4 mg/mL soybean trypsin inhibitor, 1 mg/mL dexamethasone). For RNA-seq experiments, suspension was plated in nonadherent plates, treated with 50 ng/mL TGFα, and maintained at 37°C in a 5% CO2 atmosphere. Acinar cells used in transdifferentiation assays were plated in 2.5 mg/mL bovine collagen I (Trevigen, 3442-050-01). Collagen/cell mixture was overlaid with complete Waymouth medium supplemented with 1.9 pmol/L cerulein, changed the next day and every other day thereafter.
Human subjects
The PDAC tissue microarray has been described previously (Nguyen Kovochich et al. 2013) and included three separate 1.0-mm tumor cores from each of 145 treatments: naïve or AJCC stage I or II PDACs resected at University of California at Los Angeles Medical Center from 1990 to 2005. All work was performed with prior institutional review board approval. Human subjects used for RNA-seq-based analysis have been described previously (Bailey et al. 2016).
Supplementary Material
Acknowledgments
We thank Dr. David Tuveson for the PDX1-Cre, Trp53R172H, and KrasLSL-G12D mice. We thank Dr. Nabeel Bardeesy for the NB508 and NB490 mouse PDA cell lines. We also thank Debbie Ngow for tissue processing, Michelle T. Paulsen and Karina E. Villanueva for technical help, and all of the Hebrok/Crawford laboratory members for helpful discussions. This work was supported by National Institutes of Health/National Cancer Institute grants P30CA046592 to the University of Michigan Comprehensive Cancer Center, R01CA172045 and R01CA112537 to M.H., and R01 CA159222 to H.C.C.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.291021.116.
References
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. 2003. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 17: 3112–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. 1998. β-Cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes Dev 12: 1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, Miller DK, Christ AN, Bruxner TJC, Quinn MC, et al. 2016. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531: 47–52. [DOI] [PubMed] [Google Scholar]
- Chen L, Yan H-X, Chen J, Yang W, Liu Q, Zhai B, Cao H-F, Liu S-Q, Wu M-C, Wang H-Y. 2007. Negative regulation of c-Myc transcription by pancreas duodenum homeobox-1. Endocrinology 148: 2168–2180. [DOI] [PubMed] [Google Scholar]
- Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, et al. 2011. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 17: 500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Huang Y-H, Chen M, Su J, Zou Y, Bardeesy N, Iacobuzio-Donahue CA, Massagué J. 2016. TGF-β tumor suppression through a lethal EMT. Cell 164: 1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La O J-P, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. 2008. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci 105: 18907–18912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandez M, Cendrowski J, Cañamero M, Salas A, del Pozo N, Schoonjans K, Real FX. 2014. Nr5a2 heterozygosity sensitises to, and cooperates with, inflammation in KRas(G12V)-driven pancreatic tumourigenesis. Gut 63: 647–655. [DOI] [PubMed] [Google Scholar]
- Gannon M, Ables ET, Crawford L, Lowe D, Offield MF, Magnuson MA, Wright CVE. 2008. pdx-1 function is specifically required in embryonic β cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev Biol 314: 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, McKenna B, Li C, Reichert M, Nguyen J, Singh T, Yang C, Pannikar A, Doliba N, Zhang T, et al. 2014. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metab 19: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G. 1995. Expression of murine STF-1, a putative insulin gene transcription factor, in β cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development 121: 11–18. [DOI] [PubMed] [Google Scholar]
- Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD, et al. 2008. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci 105: 18913–18918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale MA, Kagami H, Shi L, Holland AM, Elsässer H-P, Hammer RE, MacDonald RJ. 2005. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev Biol 286: 225–237. [DOI] [PubMed] [Google Scholar]
- Heller RS, Stoffers DA, Bock T, Svenstrup K, Jensen J, Horn T, Miller CP, Habener JF, Madsen OD, Serup P. 2001. Improved glucose tolerance and acinar dysmorphogenesis by targeted expression of transcription factor PDX-1 to the exocrine pancreas. Diabetes 50: 1553–1561. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4: 437–450. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7: 469–483. [DOI] [PubMed] [Google Scholar]
- Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. 2002. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci 99: 12236–12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruban RH, Wilentz RE, Kern SE. 2000. Genetic progression in the pancreatic ducts. Am J Pathol 156: 1821–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischenko I, Petrenko O, Hayman MJ. 2014. Analysis of the tumor-initiating and metastatic capacity of PDX1-positive cells from the adult pancreas. Proc Natl Acad Sci 111: 3466–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371: 606–609. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. 2009. The basics of epithelial-mesenchymal transition. J Clin Invest 119: 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CVE. 2002. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 32: 128–134. [DOI] [PubMed] [Google Scholar]
- Kodama S, Nakano Y, Hirata K, Furuyama K, Horiguchi M, Kuhara T, Masui T, Kawaguchi M, Gannon M, Wright CVE, et al. 2016. Diabetes caused by Elastase-Cre-mediated Pdx1 inactivation in mice. Sci Rep 6: 21211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M, Doi R, Toyoda E, Masui T, Tulachan SS, Kawaguchi Y, Fujimoto K, Gittes GK, Imamura M. 2003. Increased PDX-1 expression is associated with outcome in patients with pancreatic cancer. Surgery 134: 260–266. [DOI] [PubMed] [Google Scholar]
- Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JP, Pan FC, Akiyama H, Wright CVE, Jensen K, et al. 2012. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah NM, De La O J-P, Swift GH, Hoang CQ, Willet SG, Chen Pan F, Cash GM, Bronner MP, Wright CV, MacDonald RJ, et al. 2015. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. Elife 4: e07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ballian N, Belaguli NS, Patel S, Li M, Templeton NS, Gingras M-C, Gibbs R, Fisher W, Brunicardi FC. 2008. PDX-1 acts as a potential molecular target for treatment of human pancreatic cancer. Pancreas 37: 210–220. [DOI] [PubMed] [Google Scholar]
- Marty-Santos L, Cleaver O. 2016. Pdx1 regulates pancreas tubulogenesis and E-cadherin expression. Development 143: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, et al. 2003. Notch mediates TGFα-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 3: 565–576. [DOI] [PubMed] [Google Scholar]
- Miyatsuka T, Kaneto H, Shiraiwa T, Matsuoka TA, Yamamoto K, Kato K, Nakamura Y, Akira S, Takeda K, Kajimoto Y, et al. 2006. Persistent expression of PDX-1 in the pancreas causes acinar-to-ductal metaplasia through Stat3 activation. Genes Dev 20: 1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JP IV, Cano DA, Sekine S, Wang SC, Hebrok M. 2010. β-Catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest 120: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JP, Klimstra DS, Mongeau ME, Lewis BC. 2008. Trp53 deletion stimulates the formation of metastatic pancreatic tumors. Am J Pathol 172: 1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Kovochich A, Arensman M, Lay AR, Rao NP, Donahue T, Li X, French SW, Dawson DW. 2013. HOXB7 promotes invasion and predicts survival in pancreatic adenocarcinoma. Cancer 119: 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122: 983–995. [DOI] [PubMed] [Google Scholar]
- Ohlsson H, Karlsson K, Edlund T. 1993. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J 12: 4251–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Bankaitis ED, Boyer D, Xu X, Van de Casteele M, Magnuson MA, Heimberg H, Wright CVE. 2013. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development 140: 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Hong S-M, Klimstra DS, Goggins MG, Maitra A, Hruban RH. 2011. Pdx1 expression in pancreatic precursor lesions and neoplasms. Appl Immunohistochem Mol Morphol 19: 444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Magnuson B, Wilson TE, Ljungman M. 2014. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods 67: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S, Folias AE, Hebrok M. 2014. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell 16: 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. 2014. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74: 2913–2921. [DOI] [PubMed] [Google Scholar]
- Roy N, Hebrok M. 2015. Regulation of cellular identity in cancer. Dev Cell 35: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, DiRenzo D, Qu C, Barney D, Miley D, Konieczny SF. 2013. Maintenance of acinar cell organization is critical to preventing Kras-induced acinar–ductal metaplasia. Oncogene 32: 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2016. Cancer statistics, 2016. CA Cancer J Clin 66: 7–30. [DOI] [PubMed] [Google Scholar]
- Song SY, Gannon M, Washington MK, Scoggins CR, Meszoely IM, Goldenring JR, Marino CR, Sandgren EP, Coffey RJ, Wright CV, et al. 1999. Expansion of Pdx1-expressing pancreatic epithelium and islet neogenesis in transgenic mice overexpressing transforming growth factor α. Gastroenterology 117: 1416–1426. [DOI] [PubMed] [Google Scholar]
- Song Y, Washington MK, Crawford HC. 2010. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res 70: 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift GH, Liu Y, Rose SD, Bischof LJ, Steelman S, Buchberg AM, Wright CV, MacDonald RJ. 1998. An endocrine–exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2). Mol Cell Biol 18: 5109–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Figura G, Fukuda A, Roy N, Liku ME, Morris Iv JP, Kim GE, Russ HA, Firpo MA, Mulvihill SJ, Dawson DW, et al. 2014a. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 16: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Figura G, Morris JP, Wright CVE, Hebrok M. 2014b. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut 63: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Gannon M, Peshavaria M, Offield MF, Henderson E, Ray M, Marks A, Gamer LW, Wright CV, Stein R. 1997. Hepatocyte nuclear factor 3β is involved in pancreatic β-cell-specific transcription of the pdx-1 gene. Mol Cell Biol 17: 6002–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liu S, Yu J, Zhou G, Rao D, Jay CM, Kumar P, Sanchez R, Templeton N, Senzer N, et al. 2014. Vertically integrated translational studies of PDX1 as a therapeutic target for pancreatic cancer via a novel bifunctional RNAi platform. Cancer Gene Ther 21: 48–53. [DOI] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. 2010. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467: 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Liu S-H, Sanchez R, Nemunaitis J, Rozengurt E, Brunicardi FC. 2016. PDX1 associated therapy in translational medicine. Ann Transl Med 4: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.