Davoli et al. identified ribosomal protein translation and proteasomal protein degradation as critical nononcogene dependencies for PI3K-driven tumors. Their results suggest that disruption of protein turnover homeostasis via ribosome or proteasome inhibition may be a novel treatment strategy for PI3K mutant human tumors.
Keywords: PI3K, genetics screen, synthetic lethality
Abstract
Activating mutations in the phosphoinositide 3-kinase (PI3K) signaling pathway are frequently identified in cancer. To identify pathways that support PI3K oncogenesis, we performed a genome-wide RNAi screen in isogenic cell lines harboring wild-type or mutant PIK3CA to search for PI3K synthetic-lethal (SL) genes. A combined analysis of these results with a meta-analysis of two other large-scale RNAi screening data sets in PI3K mutant cancer cell lines converged on ribosomal protein translation and proteasomal protein degradation as critical nononcogene dependencies for PI3K-driven tumors. Genetic or pharmacologic inhibition of either pathway alone, but not together, selectively killed PI3K mutant tumor cells in an mTOR-dependent manner. The expression of ribosomal and proteasomal components was significantly up-regulated in primary human colorectal tumors harboring PI3K pathway activation. Importantly, a PI3K SL gene signature containing the top hits of the SL genes identified in our meta-analysis robustly predicted overall patient survival in colorectal cancer, especially among patients with tumors with an activated PI3K pathway. These results suggest that disruption of protein turnover homeostasis via ribosome or proteasome inhibition may be a novel treatment strategy for PI3K mutant human tumors.
Devising strategies to selectively kill cancer cells while sparing their wild-type counterparts has been a persistent challenge in cancer drug development (Engelman 2009). Selectivity can be achieved by targeting the oncogenic “driver” mutations upon which cancer cells become dependent for growth and survival (Jonkers and Berns 2004; Weinstein and Joe 2006, 2008). For example, tyrosine kinase (TK) inhibitors are initially efficacious in treating tumors harboring oncogenic receptor TK (RTK) mutations (Paez et al. 2004); however, many tumors ultimately become resistant to these drugs (Linardou et al. 2009). An alternative approach is to search for genes that exhibit synthetic lethality with oncogenic mutations (Fece de la Cruz et al. 2015). This approach is based on the concept of nononcogene addiction, which postulates that tumorigenic mutations confer certain critical cellular dependences on the activity of specific genes that are themselves not oncogenes but are required to a greater extent for cell growth and survival in tumor cells relative to wild-type cells (Solimini et al. 2007; Luo et al. 2009). This concept broadens the list of potential therapeutic targets beyond the oncogene itself. RNAi screens have been used extensively for the identification of such nononcogenic addictions, although the utility of this approach is often hindered by the inefficiency and off-target effects of reagents (Adamson et al. 2012; Mohr et al. 2014).
Phosphoinositide 3-kinase (PI3K) is an intracellular lipid kinase that phosphorylates the 3′-hydroxyl position on the inositol ring of phosphotidylinositols (PtdIns), leading to activation of AKT1 kinase, a central regulator of cell growth (Whitman et al. 1988). A key downstream transducer of the PI3K–AKT pathway is the serine–threonine kinase mTOR, whose activation promotes protein synthesis and other anabolic processes (Yuan and Cantley 2008). mTOR stimulates the translation of specific mRNAs containing highly structured 5′ untranslated regions (UTRs) and 5′ oligopyrimidine tract mRNAs, encoding cell cycle regulators (such as Cyclin D1 and Myc) and translation factors. It accomplishes this primarily through the phosphorylation of S6K kinase, which enhances translation, and phosphorylation-dependent inhibition of EIF4EBP1/2, a negative regulator of translation. Furthermore, by promoting S6K-dependent phosphorylation of the transcription factor UBF, mTOR increases the rate of rDNA synthesis (Hannan et al. 2003). More recently, mTOR has been implicated in the promotion of NFE2L1-dependent transcription of proteasomal genes (Zhang et al. 2014).
PIK3CA encodes the catalytic subunit p110α of PI3K. Activating “hot spot” mutations in PIK3CA, such as E545K (in the helical domain) and H1047R (in the kinase domain) (Chalhoub and Baker 2009), are found in ∼30% of colon, prostate, and breast cancers, among others (Samuels and Velculescu 2004; Chalhoub and Baker 2009). Constitutive PI3K activation in cancer can also occur secondary to PTEN inactivation or RTK-activating mutation or amplification (Yuan and Cantley 2008). In colorectal cancer, PI3K mutations likely occur after the transformation of colon polyps to malignant lesions and are associated with poor clinical outcomes (Engelman 2009; He et al. 2009).
Given the prevalence and importance of PI3K mutation in human cancer, a number of isoform-specific PI3K inhibitors, including pan-PI3K inhibitors and dual PI3K–mTOR inhibitors have been developed, and >15 of these drugs are in various phases of clinical trials (Wong et al. 2010; Klempner et al. 2013). However, in contrast to the effects of targeted inhibitors in other examples of oncogene addiction such as BRC-ABL or EGFR mutation, single-agent PI3K pathway inhibitors thus far have not had comparable success (Luo et al. 2003; Engelman 2009; Klempner et al. 2013). In some preclinical studies, many of these drugs led to tumor stasis rather than cell death in vivo, and substantial tumor shrinkage was not observed (Fan et al. 2007; Raynaud et al. 2007; Serra et al. 2008). Interestingly, the combination of MEK inhibition with PI3K/mTOR dual inhibition has a synergistic effect in lung adenocarcinoma (Engelman et al. 2008).
PI3K activation during normal development is likely to be part of a highly coordinated process of cell proliferation and growth control. Mutational activation of PI3K outside of this normal regulatory environment is likely to have profound and possibly unbalanced effects on cellular physiology that could produce novel vulnerabilities in tumors. To identify specific vulnerabilities of PI3K mutant tumors, we investigated the genetic dependencies of human cancer cells bearing oncogenic mutations in PIK3CA. Through a meta-analysis of our genome-wide RNAi synthetic lethality screen in isogenic colon cancer cells and large data sets of RNAi-based screens in other cancer cell lines with oncogenic PI3K mutations, we uncovered a particular sensitivity of human cancer cells with oncogenic PI3K hyperactivation to inhibition of ribosomal protein translation and proteasomal protein degradation. By using a gene expression data set from cancer cell lines and human primary tumors, we demonstrate that multiple molecular components of the translation pathway and the proteasome are up-regulated in tumors bearing PI3K mutations. Importantly, a gene expression signature derived from the meta-analysis of our PI3K synthetic-lethal (SL) interactions significantly predicts overall survival in colorectal cancer patients. These results suggest that the disruption of protein turnover homeostasis via ribosome or proteasome inhibition may be an effective treatment strategy for PI3K mutant human tumors.
Results
A genome-wide screen for genes exhibiting synthetic lethality with oncogenic PI3K
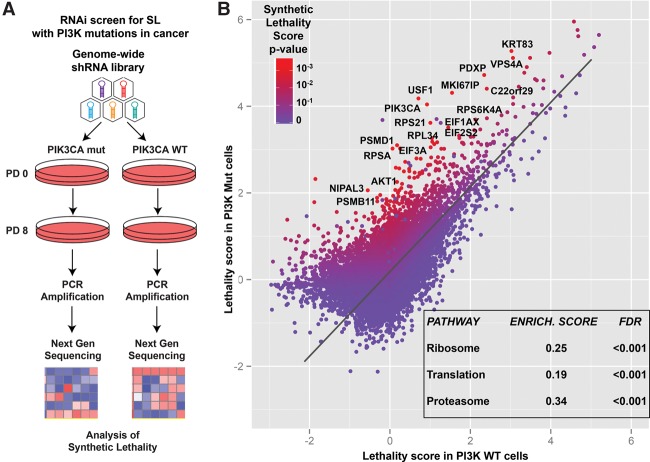
A genome-wide shRNA screen was performed in a pair of isogenic HCT116 colon cancer cell lines harboring either PIK3CA oncogenic mutation H1047R (PI3K Mut) or not (PI3K wild-type) (Samuels et al. 2005). These lines were derived from the parental HCT116 line after inactivation of either the wild-type or the Mut allele (Samuels et al. 2005). PI3K Mut and PI3K wild-type cells were transduced with a retroviral shRNA library targeting 18,300 human genes (four shRNA per genes), and, after passaging for eight population doublings, the relative distribution of each shRNA in the cell population was determined by next-generation sequencing (NGS) of PCR-amplified half-hairpins as described previously (Fig. 1A).
Figure 1.
A genome-wide shRNA screen to identify genes synthetically lethal with oncogenic PIK3CA mutations. (A) Schematic diagram depicting the experimental procedure used for the genome-wide shRNA screen in the isogenic PI3K Mut and wild-type HCT116 cell line pair (see the text for details). (B) Graphical distribution where each dot represents a single gene present in the genome-wide library. For each gene, the plot contains its corresponding lethality score (average among the shRNAs targeting the gene) in the PI3K Mut and PI3K wild-type cells. As in the heat map legend, each dot is also colored according to its P-value for synthetic lethality with PI3K Mut versus wild-type cells. The inset within the graph contains the three top enriched nonredundant pathways found among the list of PI3K SL genes ranked by the SL P-value (see also Supplemental Table S1).
To determine the vulnerabilities associated with PI3K mutation, we first used EdgeR (Robinson et al. 2010) to estimate a lethality score; i.e., a quantification of the decrease in fitness for each shRNA separately in the PI3K wild-type and Mut cell line (Supplemental Fig. S1). The EdgeR-based method for NGS-based genetic screenings is superior to the simple use of log2 fold change (in shRNA abundance comparing end point versus start point) for quantifying the fitness change determined by each reagent in the genetic library (see the Materials and Methods). Finally, RIGER (RNAi gene enrichment ranking) (Luo et al. 2008) KS metrics were applied to the Δ lethality score (difference between lethality scores) between the PI3K Mut and wild-type cells for each shRNA to determine a synthetic lethality score for each gene represented in the library (Fig. 1B; see the Materials and Methods; Supplemental Fig. S1; Supplemental Table S1).
A significant SL interaction score was found for multiple genes in the PI3K pathway, such as PIK3CA itself (P < 0.001), AKT1 (P = 0.04), and RPS6 (P = 0.004, Fig. 2A). Additionally, a gene set derived from the Reactome database that contained genes essential for PI3K pathway activation (i.e., “PI3K.AKT.MTOR.Activation.Pathway”) was enriched ∼80% over expected (P < 0.05, Fisher's exact test) (Supplemental Table S2). Using the gene list ranked by the synthetic lethality score, we performed a gene set enrichment analysis (GSEA) (Subramanian et al. 2005), which showed that the proteasome pathway (Kyoto Encyclopedia of Genes and Genomes [KEGG]), the ribosome pathway (KEGG), and the related translation pathway (Reactome) were significantly enriched in the list of genes ranked by their PI3K synthetic lethality score (Fig. 2B,C; Supplemental Table S3). The pathway of splicing was also enriched (P = 0.01; false discovery rate [FDR] = 0.12).
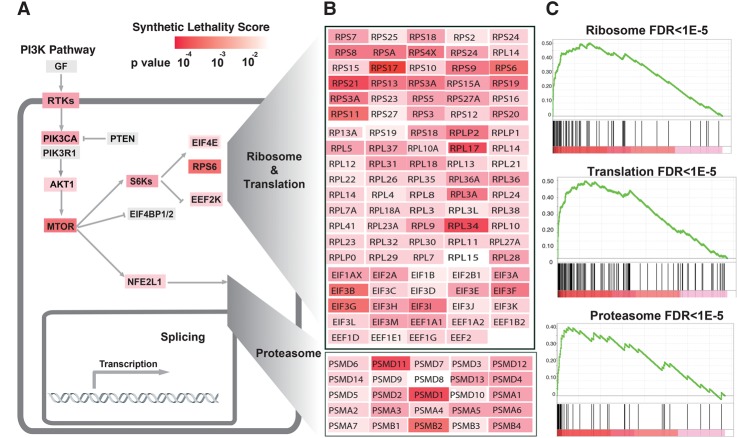
Figure 2.
Meta-analysis of the shRNA PI3K synthetic lethality screen in isogenic cell lines and the Achilles and COLT-Cancer RNAi data sets. (A) The major dependencies in the PI3K pathway are represented, and the combined P-value of each gene for synthetic lethality with the PI3K pathway is indicated with the heat map (see the text for details on the combined P-value; Supplemental Tables S2, 8). (B) The ribosome/translation pathway is represented with all of the genes present in the library and color-coded according to their combined P-value and heat map as in A (Supplemental Table S8). (C) Enrichment plots for the indicated pathways derived from GSEA on the list of genes ranked by the combined P-value for synthetic lethality with PI3K mutation (Supplemental Table S8).
Meta-analysis of three data sets for synthetic lethality with activation of PI3K
Genetic screens with isogenic cancer cell lines allow the examination of the phenotypic effect of a specific genetic alteration in an otherwise identical genetic background. An alternative approach is to perform RNAi screens across a number of different cancer cell lines, where contextual lethalities can be inferred by comparing groups of cancer cell lines classified according to their genetic alterations (Cowley et al. 2014). Although unsuitable for analysis of the effect of one genetic determinant at a time, the latter method represents a complementary approach to the use of isogenic cell lines and allows the testing of the reproducibility of genotype-specific vulnerabilities within different genetic contexts (Cowley et al. 2014). We therefore performed an integrated analysis of the data from our PI3K SL screen in isogenic HCT116 cells with two other data sets from RNAi screens performed by other institutions on a panel of different cancer cell lines: the Project Achilles data set of RNAi screens performed in 102 cancer cell lines (Cheung et al. 2011) and the COLT-Cancer data set of RNAi screens in 72 cancer cell lines (Supplemental Fig. S2; Koh et al. 2012; Marcotte et al. 2012).
We first applied ATARIS (analytic technique for assessment of RNAi by similarity) on the Achilles and COLT-Cancer data sets to identify shRNAs that, based on their phenotypic pattern across multiple cell lines, are more likely to affect their intended target genes relative to their off-target effects (Shao et al. 2013). Next, by usng the ATARIS solutions as quantification of the phenotypic effect of the inhibition of each gene across the two RNAi data sets, we performed a two-class comparison between PI3K Mut and PI3K wild-type cell lines in both data sets (Wilcoxon test) (see the Materials and Methods; Supplemental Tables S4, 5). PI3K Mut cells were defined as those cancer cell lines bearing an oncogenic activating mutation in PIK3CA, while PI3K wild-type cells were defined as those cell lines harboring a wild-type PIK3CA gene that also did not have PI3K-activating mutations or amplifications in KRAS, HRAS, NRAS, BRAF, AKT1, PTEN, EGFR, or ERBB2 oncogenes (Supplemental Tables S4, S5). As a confirmation of our analysis to identify genes synthetically lethal with PI3K hyperactvation, we found that the PI3K.AKT.mTOR.Activation.Pathway was strongly enriched in both RNAi data sets (Achilles data set: 46% enrichment; COLT-Cancer data set: 84% enrichment) (Supplemental Table S2). As a negative control, we compared cancer cell lines harboring or not harboring oncogenic KRAS mutations in the Achilles data set. In this comparison, depletion of KRAS itself scored as highly significant SL, as expected, while there was no enrichment of the PI3K.AKT.mTOR.Activation.Pathway among the SL genes (Supplemental Tables S2, S6).
Using the Fisher method, we obtained a combined P-value, derived from all three PI3K data sets (Supplemental Table S7). This meta-analysis resulted in an enrichment of the PI3K.AKT.mTOR.Activation.Pathway relative to the three individual analyses; i.e., the analysis of the screen in isogenic lines, the Achilles data set analysis, and the COLT-Cancer data set analysis. In fact, while the three individual analyses resulted in, at most, ∼80% enrichment (observed versus expected) of the PI3K.AKT.MTOR.Activation.Pathway, the combined analyses resulted in a 102% enrichment (P = 0.03, Fisher exact test) (Fig. 2; Supplemental Table S2). This result highlights the utility of performing a meta-analysis that includes screens performed on isogenic cell lines and parallel screens performed on multiple cell lines.
A GSEA on the gene list, ranked by the synthetic lethality combined P-value, showed a high enrichment for genes implicated in the ribosome/translation (enrichment in the top 10% = 445%), proteasome (enrichment in the top 10% = 497%), and splicing pathways (enrichment in the top 10% = 308%; FDR < 1 × 10−4 for all three pathways) (Supplemental Table S8). Importantly, the enrichment score (GSEA ES) of the genes in these three pathways was more prominent in the combined analysis compared with the analysis of the screening on isogenic cell lines by itself (cf. Supplemental Table S3 and Supplemental Table S8). The common enrichment from multiple orthogonal screens and databases of these three pathways is very strong evidence for their involvement as nononcogene dependencies for tumors with activating PI3K mutations.
Based on this combined analysis, we designed a focused shRNA library (10 shRNAs per gene) to perform a secondary screen in the same cell lines. Analysis of this secondary screen, performed in a fashion similar to the genome-wide screen, showed that 312 of 1045 genes (29%) targeted by the library had significant SL scores in PI3K Mut cells compared with wild-type cells (Supplemental Table S1). Although the smaller size of this library compared with the one used for the genome-wide screen rendered pathway analysis more difficult, the translation, proteasome, and ribosome pathways were all significantly enriched (FDR < 0.3), corroborating our previous findings.
Inhibition of protein synthesis is synthetically lethal with PI3K oncogenic mutations
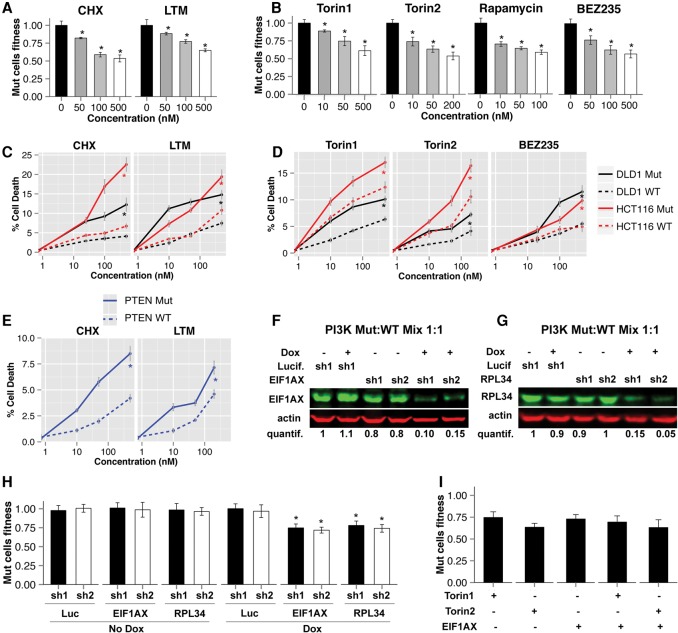
To further examine the synthetic lethality of the identified candidate pathways, we performed a multicolor competition assay (MCA) (Smogorzewska et al. 2007) in which GFP-expressing HCT116 PI3K wild-type cells and dsTomato-expressing HCT116 PI3K Mut cells were combined and cocultured for at least six population doublings (5–7 d) in the absence or presence of a drug or after the transduction with an shRNA. Treatment with the translation elongation inhibitors cycloheximide (CHX) or lactimidomycin (LTM) resulted in an ∼30%–40% decrease in the relative survival of Mut versus wild-type cells, which is similar to that observed for the mTOR inhibitors Torin1, Torin2, and rapamycin or the dual PI3K/mTOR inhibitor NVP-BEZ235 (Fig. 3A,B; Supplemental Fig. S3E). Cell death assays with 7-amino-actinomycinD (7-AAD) staining (see the Materials and Methods) revealed that CHX or LTM induced significantly more cell death in PI3K Mut versus wild-type cells (Fig. 3C,D). The increase in cell death with translation inhibitors in PI3K Mut cells versus wild-type cells was higher than for Torin1/2 or NVP-BEZ235.
Figure 3.
Synthetic lethality of oncogenic PI3K hyperactivation with ribosome and translation inhibition. (A,B) MCAs were performed using the indicated drugs and concentrations. Mixtures of HCT116 PI3K Mut and wild-type cells (expressing different fluorescent proteins) were passaged in the presence of the drug for 5–7 d. At the end of the assay, the relative percentages of Mut and wild-type cells were quantified by FACS. “Mut cell fitness” represents the ratio between the percentage of Mut cells in the sample treated with the drug and the percentage of Mut cells in the sample treated with DMSO (drug concentration of 0 nM). (BEZ235) NVP-BEZ235. The presence of an asterisk indicates a statistical significance for the corresponding measurement and the relative control (see the Materials and Methods; Supplemental Fig. S3E). (C–E) Results of cell death assays performed in HCT116 and DLD1 cells of the indicated genotypes. After the treatment of cells with the indicated drug and incubation for 48–72 h, FACS analysis was performed to assess the fraction of cell death after staining with 7-AAD. The fraction of cell death at the different concentrations is reported. (BEZ235) NVP-BEZ235. A drug concentration of 0 nM corresponds to DMSO control. (F,G) A mixture of HCT116 PI3K Mut and wild-type cells transduced with pHAGE-Ind10-mirE encoding the indicated shRNAs was treated with or without 1 µg/mL doxycycline to induce expression of the shRNA. Western blot was performed and visualized using the Licor system with the indicated antibodies after 72 h of treatment. Quantification of the amount of protein was performed using Image Studio Analysis software and is reported below the Western blots (see also Supplemental Fig. S3D). (H) Cells infected with the indicated shRNAs as in F were selected and cultured for an additional 5–7 d in the presence or absence of 1 µg/mL doxycycline before FACS analysis was performed to determine the fitness of Mut cells (relative to wild-type), as in A and B. Analysis was performed as described in A. (I) MCAs were performed as in A by treating the cells with the different reagents (shRNAs or drugs) individually or in combination. Both Torin1 and Torin2 were used at a concentration of 50 nM. The difference between the combination of Torin1 + EIF1AX-shRNA and the two reagents used individually and between Torin2 + EIF1AX-shRNA and the two reagents used individually was not statistically significant.
Importantly, we confirmed the selective induction of cell death in PI3K Mut versus wild-type cells with CHX and LTM by using a different pair of isogenic colon cancer cell lines: DLD1 (carrying a different oncogenic mutation in PIK3CA and E545K) (Fig. 3C,D, black lines; Samuels et al. 2005). Moreover, we found a similar effect in PTEN−/− cells, which exhibit PI3K pathway activation, compared with their wild-type counterparts (Fig. 3E; Supplemental Fig. S3A). In contrast, KRAS Mut versus wild-type cell lines did not show a significant difference in the induction of cell death from protein synthesis inhibition as well as mTOR signaling inhibition (Supplemental Fig. S3B,C).
We next studied the effects of genetic inhibition of the protein translation process by transducing cells with lentiviral vectors (pHAGE-pInd10-mirE) expressing doxycycline-inducible shRNAs that target translation factors that scored in the top hits of SL genes in the meta-analysis of the three data sets (Supplemental Table S7). In MCAs, depletion of the elongation factor EIF1AX (combined P-value = 9 × 10−5) to ∼10%–15% of its endogenous level by two independent shRNAs resulted in a significant selective inhibition of HCT116 PI3K Mut versus wild-type cells compared with two negative control shRNAs targeting Luciferase (Fig. 3F,H; Supplemental Fig. S3D). Depletion of the ribosomal protein RPL34 (P = 1 × 10−16) had a similar selective decrease in viability for HCT116 PI3K Mut compared with wild-type cells. Additionally, in MCAs, the combination of individual shRNAs targeting EIF1AX with Torin1 and Torin2 did not further increase the selective growth inhibition of PI3K Mut cells versus wild-type compared with treatment with the drugs or EIF1AX shRNAs individually (Fig. 3I). These results collectively indicate that PI3K pathway activation due to oncogenic PIK3CA mutation or PTEN inactivation confers a specific susceptibility to ribosome and protein translation that is likely dependent on mTOR.
Proteasome inhibition shows synthetic lethality with PI3K oncogenic mutations
In addition to the ribosome/translation pathway, the proteasome machinery was also significantly enriched among the list of PI3K SL hits (Fig. 2). We investigated whether the expression of components of the proteasome pathway is up-regulated in response to oncogenic PI3K activation by analyzing a gene expression profiling data set from PTEN−/− and PTEN+/+ isogenic HCT116 cancer cell lines (data set Gene Expression Omnibus [GEO] GDS2446) (Kim et al. 2007). This data set contains the expression profile of parental HCT116, one PTEN+/+ control clone, and three PTEN−/− clones. Limma-based analysis (Ritchie et al. 2015) followed by GSEA showed that multiple components of the proteasome were up-regulated in PTEN−/− cells compared with PTEN+/+ (FDR = 0.003) (Fig. 4A; Supplemental Table S9). Indeed, 10 out of 48 proteasomal genes are contained in the top 10% of the genes ranked by differential overexpression in PTEN−/− versus PTEN+/+ cells, resulting in an enrichment of 111% over expected (Supplemental Table S9). PSMA1 and PSMB5, two of the most significantly overexpressed genes in PTEN−/− versus wild-type cells (GEO GDS2446), were also up-regulated (greater than twofold) in PI3K Mut versus wild-type HCT116 and DLD1 cells (Fig. 4B; Supplemental Fig. S4A,B). This increase was mTOR-dependent, since the expression of both genes in PI3K Mut cells was reduced by Torin2 and rapamycin treatment (Fig. 4B; Supplemental Fig. S4A,B; data not shown).
Figure 4.
Synthetic lethality of oncogenic PI3K activation with inhibition of the proteasome in vitro and in vivo. (A) GSEA enrichment plot for the proteasome pathway among genes ranked by the P-value of overexpression in HCT116 PTEN−/− versus PTEN+/+ cells (from data set GEO GDS2446). (B) Quantitative PCR (qPCR) for the indicated mRNAs in cells with the indicated genotype treated with DMSO, 20 nM Torin2, or 20 nM rapamycin for 48 h prior to harvesting the cells. (C) PI3K Mut and wild-type cells were treated with 50 µM CHX, and cells were harvested and analyzed at the indicated times. Western blots were performed for total ubiquitin conjugates as well as B-actin (a long-lived protein that serves as a loading control). (D) The quantification of the ubiquitin conjugates in each sample in C relative to β-actin is plotted (average values). The asterisk indicates a significant difference between Mut and wild type after Wilcoxon test at 4 and 6 h (three replicates). (E) Quantification of proteasome activity measured with proteasome-glow chymotrypsin-like cell-based assay in cells of the specified genotype treated for 48 h with 20 nM Torin2 or 20 nM rapamycin. (F) MCAs were performed by using the indicated drug and concentration as in Figure 3A. The percentage of Mut over wild-type cells at the end of the assay relative to the treatment with DMSO is shown. (CLL) Clasto-lactacystin lalactone. (G) MCAs were performed as in Figure 3A by treating the cells with the different drugs as single agents or in combination. All drugs were used at 50 nM concentration. Asterisks refer to a significant difference between samples treated with CHX versus CHX + MG132 and LTM versus LTM + MG132. (H) DLD1-PI3K wild-type and Mut cells were injected in the flanks of nu/nu nude mice. After tumor formation, mice were treated with vehicle or bortezomib, and tumor volume was monitored. The change of tumor volume over time for PI3K wild-type and PI3K Mut tumors treated with vehicle or bortezomib is shown. P-value (P = 0.004) refers to the comparison between the change in tumor volume (of bortezomib-treated vs. vehicle-treated tumors) between PI3K Mut and PI3K wild-type tumors (Wilcoxon test). The experiment was repeated twice, and both times each cohort included at least seven mice.
Up-regulation of proteasome subunits suggests that the activity of the proteasome is enhanced in PI3K mutants. We therefore analyzed proteasomeal activity in PI3K Mut versus wild-type cells. We found that the kinetics of ubiquitin conjugate degradation after protein synthesis inhibition by CHX was significantly faster in PI3K Mut than wild-type cells, suggesting an enhanced rate of proteasome-mediated protein degradation in PI3K Mut compared with wild-type cells (Fig. 4C,D). In addition, we measured the chymotrypsin-like activity with a luciferase-based reporter assay, which confirmed a significant increase in proteasome activity in PI3K Mut versus wild-type cells (Fig. 4E).
We further assessed the synthetic lethality between proteasome inhibition and PI3K mutations using pharmacological inhibition. Treatment of the cells with different inhibitors of the proteasome (MG-132, clasto-lactacystin lalactone and bortezomib) resulted in selective inhibition of cell growth in PI3K Mut versus wild-type cells (Fig. 4F) accompanied by increased cell death (Supplemental Fig. S4C). Similar selective inhibition was found in PTEN−/− compared with PTEN+/+ cells (Supplemental Fig. S4D) but not in KRAS Mut versus KRAS wild-type cells (Supplemental Fig. S4E). In contrast, the autophagy inhibitor bafilomycin A1 did not exhibit a selective effect on PI3K Mut cells (Supplemental Fig. S4F). These data indicate that PI3K pathway hyperactivation results in increased proteasome activity that renders tumor cells more sensitive to proteasome inhibition relative to wild-type cells.
PI3K pathway hyperactivation leads to an increase in protein turnover by increasing both protein synthesis and protein degradation. We reasoned that PI3K Mut (or PTEN−/−) cells are more sensitive to translation or proteasome inhibition because antagonism of either pathway would result in an “uncoupling” of protein synthesis from protein degradation, thereby impairing cell survival via effects on the homeostasis of protein turnover. In support of this is the fact that an MCA that used simultaneous translation and proteasome inhibition did not result in a synergistic effect on cell survival (Fig. 4G). In fact, the combined inhibition of translation and the proteasome significantly diminished the inhibition of either pathway alone in PI3K Mut versus wild-type cells (Fig. 4G).
We also tested the effect of bortezomib on tumor growth in mice. We subcutaneously injected DLD1-PI3K wild-type and Mut cells into the flanks of athymic nude mice and waited for tumor formation (see the Materials and Methods). Mice were then treated with either vehicle (PBS) or 0.5 mg/kg bortezomib by intraperitoneal injection once daily for 28 d and monitored for tumor growth. We found that, although both PI3K wild-type and PI3K Mut tumors exhibited a response to the drug, the PI3K Mut cell tumors showed a significantly stronger response to bortezomib than PI3K wild-type cell tumors (Fig. 4H; P = 0.004). These results suggest that oncogenic PI3K mutations in tumors promote sensitivity to bortezomib in vivo.
A PI3K SL signature predicts survival in colorectal cancer patients
We next analyzed the expression signatures of genes in pathways that we found to be synthetically lethal with mutant PI3K activation in human primary tumors. We chose to analyze The Cancer Genome Atlas (TCGA) gene expression data set along with the clinical data of patients with colorectal carcinoma for several reasons: (1) Our screen was performed in isogenic human colorectal cancer cell lines. (2) The majority of cell lines used for the two-class comparison of PI3K Mut and wild-type lines in the Achilles data set was also derived from colorectal cancer. (3) Oncogenic PI3K mutations correlate with a higher rate of recurrence and worse outcome in nonmetastatic colorectal cancer (He et al. 2009; Ogino et al. 2009).
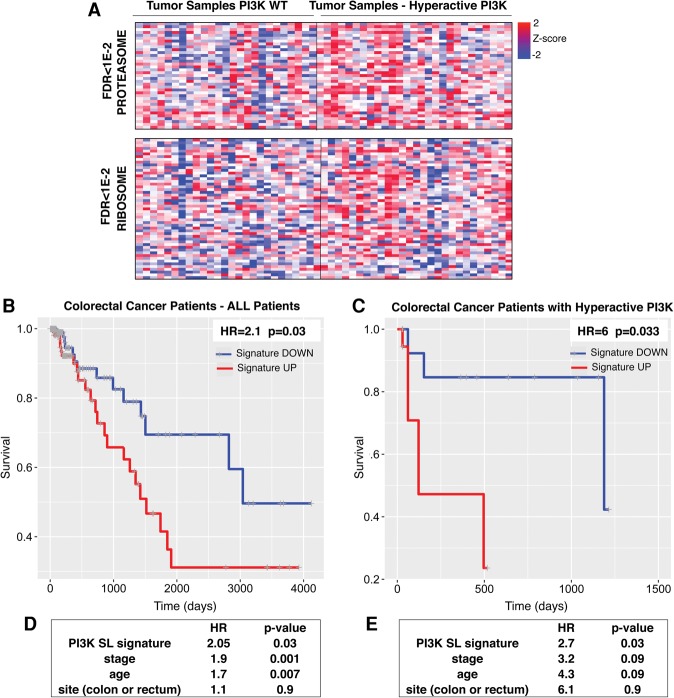
In order to compare tumors with or without PI3K pathway activation, samples were first classified according to the presence or absence of an oncogenic mutation in PIK3CA and/or the presence or absence of an inactivating mutation or deletion in PTEN. Additionally, given the role of RTK mutation/amplification in promoting PI3K pathway hyperactivation, tumors that had mutations or amplifications in EGFR or ERBB2 were classified among the tumors with PI3K pathway activation (see the Materials and Methods). Limma analysis of gene expression followed by GSEA showed that components of both the ribosome and the proteasome pathway were significantly enriched in tumors with PI3K pathway activation (FDR < 1 × 10−2, Fig. 5A).
Figure 5.
Analysis of a transcriptome data set of primary colon adenocarcinoma and prediction of survival based on PI3K SL signature. (A) Gene expression profiles of primary colorectal carcinomas with the normal or hyperactive PI3K pathway are presented showing the enrichment (based on GSEA) of ribosome and proteasome genes in tumors with a hyperactive PI3K pathway (TCGA data). (B,C) A PI3K SL gene signature was derived by considering the top hits of the SL genes identified in the meta-analysis (P < 0.02; 350 genes) (Supplemental Table S7). Survival analysis (Kaplan-Meier curve) comparing colorectal cancer patients (TCGA data) with a high or low PI3K SL signature score (top and bottom half) in all colorectal tumors (Supplemental Tables S10) or in the subset of tumors with a hyperactive PI3K pathway (Supplemental Tables S11). HR and P-value (Wald test) are shown. (D,E) The tables show the HR and P-value of the multivariable Cox proportional hazard model, including the PI3K SL signature and other clinical covariates for all colorectal cancer patients (D) or the subset with the hyperactive PI3K pathway (E).
Since oncogenic PI3K mutations correlate with a worse outcome in nonmetastatic colorectal cancer (He et al. 2009), we asked whether a gene expression signature derived from the analysis of our identified PI3K SL genes could predict survival in patients. In order to derive a PI3K SL signature for colon cancer, we considered the meta-analysis described above and selected the genes with a combined P-value of <0.02 (350 genes). We derived an average expression score for this PI3K SL gene signature for each tumor sample in the data set of colorectal cancer patients. By comparing tumors with higher or lower PI3K SL gene signatures, PI3K SL gene signature significantly predicts survival among all colorectal cancer patients (HR = 2.1; P = 0.03) (Fig. 5B; Supplemental Table S10). Furthermore, when we analyzed only patients classified as having PI3K hyperactivation (as described above), our PI3K SL gene signature showed an even greater ability to predict overall survival (HR = 6; P = 0.033) (Fig. 5C; Supplemental Table S11). The PI3K SL gene signature remained a significant parameter in the prediction of survival after taking into consideration the stage, age, and site of the disease (Fig. 5D,E).
Discussion
The discovery of synthetic lethalities through high-throughput RNAi screens is often hindered by the low efficiency of reagents, off-targets effects, and heterogeneity in RNAi among cancer cell lines (Hart et al. 2014). Here, we show that a functional genomics approach that uses a meta-analysis performed on three independent screening data sets, performed with different strategies (one in a pair of isogenic cell lines, the other two on several different cancer cell lines) and different libraries, is superior to the analysis of each of the individual data sets in detecting positive control genes, which, in our case, were the known critical players of the PI3K pathway. This suggests the other SL genes identified by this meta-analysis are likely to be of higher confidence and greater potency than those identified by the individual screens.
Our analysis identified novel vulnerabilities for cancer cells harboring PI3K pathway activation. The two major pathways that our combined analysis identified as SL with oncogenic PI3K pathway activation were the ribosome/translation and the proteasome pathways. Protein synthesis is enhanced and modulated by PI3K activation of the mTOR pathway (Hay and Sonenberg 2004). Our data suggest that PI3K mutation also enhances protein degradation by the proteasome. These findings are consistent with recent studies that suggest that mTOR activation and a consequent increase in protein synthesis can lead to a concomitant increase in protein degradation that promotes the maintenance of cellular amino acid homeostasis (Zhang et al. 2014). If this is true, it would predict that the uncoupling of ribosomal protein synthesis from proteasomal protein degradation via the inhibition of either pathway, but not both pathways simultaneously, would compromise cell viability—particularly in the context of mTOR activation. This hypothesis is in agreement with our data showing that simultaneous proteasome and translation inhibition not only did not have a synergistic effect but actually diminished the PI3K-selective vulnerability of inhibiting either pathway alone.
Despite increasing interest in exploring protein synthesis inhibition as a therapeutic strategy in cancer, this approach has not yet been used in the clinic, mainly due to pharmacokinetic barriers and toxicity of existing drugs (Hsieh et al. 2011; Malina et al. 2012). Interestingly, 4E1RCat, which inhibits CAP-dependent translation by blocking the binding of EIF4G to EIF4E-bound nascent mRNA transcripts, improved the response to chemotherapy in a murine model of EuMyc lymphoma (Cencic et al. 2011). In contrast, targeting the proteasome has already emerged as a viable strategy for some types of malignancies (Adams 2003). Bortezomib (Velcade) has been approved for the treatment of patients with multiple myeloma and mantle cell lymphoma (http://www.fda.gov). In solid tumors, a number of clinical trials performed in patients with advanced (usually metastatic) tumors did not show a significant benefit of treatment with bortezomib as a single agent (Shah et al. 2004; Yang et al. 2006). Among the many potential reasons for inefficacy in these trials are the dose used in the trial, the advanced stage of the disease, and the treatment with bortezomib as a single agent. Furthermore, selection of the patients for the clinical trial, based on tumor molecular markers that may predict the response to a specific agent, is a crucial aspect of new clinical trials for targeted therapy. Our data indicate that the presence of PI3K oncogenic mutations might confer a particular susceptibility to proteasome inhibitors. Additionally, our data suggest that the expression levels of some proteasomal factors might also represent useful molecular biomarkers to identify patients who may benefit from treatment with proteasome inhibitors.
The relevance of our analysis of SL for tumors with a hyperactive PI3K pathway in cancer patients is highlighted by the survival analysis in colorectal cancer patients. Among patients with hyperactive PI3K, a gene expression signature derived from PI3K SL hits in colon cancer cell lines significantly correlates with survival. In other words, patients with hyperactive PI3K who show a lower expression of the genes in this defined signature have better survival. Importantly, this expression signature is a significant predictor of survival also in multivariate analysis taking into account the stage of the disease. This result highlights the fact that the PI3K SL genes identified by our analysis also likely represent bona fide SL factors in human tumors. Our findings may also be relevant for wild-type PI3K tumors that harbor other genetic or epigenetic alterations that result in AKT and mTOR pathway activation. Thus, these PI3K SL factors may represent not only biomarkers that can predict patient outcome but also potential therapeutic targets.
Materials and methods
shRNA genome-wide library and shRNA constructs
The shRNA library targeting 18,300 human genes (four shRNA per gene) was cloned into the retroviral MSCV vector. PI3K Mut and PI3K wild-type HCT116 cells were infected at an MOI (multiplicity of infection) of 0.5, selected in puromycin, and passaged for eight population doublings. Cells were harvested right after selection (start population) and after eight population doublings (end population). The relative distribution of each shRNA in the cell population was determined by NGS of PCR-amplified half-hairpins (Schlabach et al. 2008). Briefly, for NGS, DNA was harvested from the cells by standard procedures, and half-hairpins were amplified by PCR. Indexing primers were finally added by PCR amplification before Illumina sequencing. The screen was performed in triplicate. For transduction of individual shRNA targeting a specific gene and for the secondary shRNA screen, shRNAs were cloned into pHAGE-Ind-10-mirE-inducible lentiviral vector.
Cell culture
Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin–strepomycin, and 2 mM glutamine. For virus production, 293T cells were transfected using Trans-IT Minus, and the supernatant was harvested after 36 h and stored at −80°C. For the infections, cells were exposed to the appropriate amount of virus for 24 h in the presence of 8 µg/mL polybrene. For the shRNA screens, the cells were infected with an MOI of 0.5.
Western blotting and quantitative PCR (qPCR)
Protein gels for Western blotting were run using standard procedures. The proteins were detected using the LI-COR system (LI-COR Biosciences). After transfer, the nitrocellulose membranes were blocked for 30 min with Odyssey blocking buffer containing PBS with 0.2% Tween, incubated overnight in the same buffer containing PBS with the specific primary antibody, washed in PBS 0.2% Tween, and incubated with Alexa fluor-conjugated secondary antibodies (Invitrogen) before a second wash in PBS 0.2% Tween. Signals were detected with the LI-COR system and quantified using Image Studio software (LI-COR Biosciences).
For qPCR, total RNA was isolated using the RNAeasy minikit (Qiagen), and cDNA was synthesized using the High Capacity RNA-to-cDNA kit (Invitrogen) according to the manufacturer's instructions. RT-qPCR was performed in triplicate using the gene expression assay (Applied Biosystems) on an Applied Biosystems Fast 7500 machine. GAPDH and ACTB were used as internal controls for normalization and relative quantification.
Data analysis of shRNA screen in HCT116 cell lines
Sequence read alignment was performed using Bowtie software (Langmead et al. 2009). EdgeR was used to estimate the lethality score in PI3K Mut and wild-type cell lines (Robinson et al. 2010). The shRNAs with a read count >10 in at least one replicate were considered. The negative binomial distribution (EdgeR classic method) was used to determine the significance in the relative representation of each shRNA between the end and start populations. The lethality score for each shRNA was defined as the log10 of the P-value multiplied by the sign corresponding to the direction of the fold change (+1 for genes increasing between start and end population and −1 for the opposite behavior). Thus, a positive lethality score corresponds to an shRNA that undergoes a significant drop between the start and the end populations and vice versa for a negative lethality score (Fig. 1B; Supplemental Fig. S1). Afterward, RIGER KS metrics (Luo et al. 2008) were applied to the Δ lethality score between the PI3K Mut and wild-type cells for each shRNA to determine a synthetic lethality score for each gene in the library. The Δ lethality score represented the difference between the lethality score of the PI3K Mut versus wild-type cells.
Meta-analysis and combined P-value
We applied ATARIS on the Achilles and COLT-Cancer data sets (Marcotte et al. 2012). Next, by using the ATARIS solutions as quantification of the phenotypic effect of the inhibition of each gene across the two RNAi data sets, we performed a two-class comparison between PI3K (or KRAS) Mut and PI3K (or KRAS) wild-type cell lines in both data sets (Wilcoxon test). For PI3K synthetic lethality, PI3K Mut cells were defined as those cancer cell lines bearing an oncogenic activating PIK3CA mutation, while PI3K wild-type cells were defined as those cell lines having a wild-type PIK3CA gene that also did not have mutations or amplifications in KRAS, HRAS, NRAS, BRAF, AKT1, PTEN, EGFR, or ERBB2 oncogenes. Similarly, for KRAS synthetic lethality, KRAS mutant cells were defined as those cancer cell lines bearing an oncogenic activating KRAS mutation, while PI3K wild-type cells were defined as those cell lines having a wild-type KRAS gene that also did not have mutations or amplifications in KRAS, HRAS, NRAS, BRAF, EGFR, or ERBB2 oncogenes. The list of the cell lines used for these comparisons is in Supplemental Tables S4 and S5. The Fisher method was used to obtain a combined P-value between the P-values derived from the individual analyses of the screen on the isogenic HCT116 cell lines, the Achilles’ data set, and the COLT-Cancer data set. Q-values were determined using the Benjamini-Hochberg method (see also Supplemental Table S2).
Cell death assay
7-AAD is a fluorescent DNA intercalator commonly used to stain dead cells. Briefly, 1 × 104 or 2 × 104 cells were plated in 100 µL in each well of a 96-well plate. The next day, the cells were treated with the drug to be tested or solvent only. After 48–72 h, the cells were washed twice by PBS and trypsinized. Cells in each well were then resuspended in 100 µL of PBS containing 2 µL of 7-AAD staining solution (1 g/mL; BD Bosciences) and incubated for 5–15 min at 4°C in the dark prior to FACS analysis. The presence of an asterisk in the figure panels relative to cell death assays indicates a statistical significance for the difference in the cell death rate between PI3K Mut and wild-type at the corresponding experimental conditions.
Gene expression analysis
For the comparison between HCT116 PTEN−/− and PTEN+/+, we used the GEO data set GDS2446 (Kim et al. 2007). For each gene, we calculated the P-value for overexpression in HCT116 PTEN−/− cells versus PTEN+/+ using a one-sided t-test (Supplemental Table S9). GSEA was performed through the stand-alone software available at http://www.broadinstitute.org/gsea/index.jsp, using the classic method (Subramanian et al. 2005).
Primary tumor sample analysis and survival analysis
We used TCGA data sets of gene expression data (Agilent microarray data set and/or RNA sequencing [RNA-seq] data), gene mutation, and copy number variation of nonmetastatic colorectal tumor samples (http://gdac.broadinstitute.org). Tumor samples having a functionally relevant mutation in PIK3CA, PTEN, or ERBB2; a deletion in PTEN; or amplification or overexpression in ERBB2 or EGFR were classified as having a hyperactive PI3K pathway. Tumors that were normal for all of these parameters were considered wild type for PI3K pathway. A Limma-based analysis (Ritchie et al. 2015) was performed to compare the PI3K hyperactive versus the PI3K wild-type tumors.
The PI3K SL gene expression signature was derived from the PI3K SL gene list (combined analysis of the three data sets) (Supplemental Table S7), including the genes with a combined P-value of <0.02 (350 genes). For the survival analysis, a univariate or multivariate Cox proportional hazard model was used to determine the impact of each parameter on the overall survival (Wald test P-value is reported).
Drugs and antibodies
The following chemicals were used: Torin1 (Cayman Chemical), Torin2 (Cayman Chemical), LTM (Millipore), CHX (Sigma-Aldrich), rapamycin (Sigma-Aldrich), NVP-BEZ235 (Cayman Chemical), MG-132 (EMD Millipore), clasto-lactacystin lalactone (Sigma), epoxomicin (Sigma), bortezomib (Selleckchem), and doxycycline (Fisher). The following antibodies were used: ubiquitin antibody clone FK-2 (Enzo Life Sciences), PSMA1 antibody (Sigma), EIF1AX antibody (Novus Biologicals), RPL34 antibody (Abcam), and actin antibody (Santa Cruz Biotechnology).
Drug-based MCA
The competition assay used to test candidate genes/drugs (PI3K signaling pathway inhibitors) from the screen was modified from a previous protocol (Luo et al. 2007; Smogorzewska et al. 2007) and was carried out in 96-well plates in independent triplicates. For this assay, 20,000 GFP-labeled PI3K Mut and ds-Tomato-labeled wild-type cells were mixed and seeded at a ratio of 1:3 to 1:1 in each well. As an alternative to the GFP- and ds-Tomato-labeled cells, GFP-labeled PI3K Mut and unlabeled wild-type cells (or vice versa) were used. Cells were treated with drugs or DMSO (control) for 5–7 d prior to FACS analysis. The fitness of Mut cells (relative to wild type) was determined at the end of the assay as the ratio between the percentage of Mut cells in the sample treated with the drug (or shRNA) (see below) and the percentage of Mut cells in the sample treated with DMSO (or control shRNA). In the figures (e.g., Fig. 3A), a concentration of 0 nM of the drug corresponds to the DMSO control treatment. The presence of an asterisk in the figure panels relative to MCA indicates a statistical significance for the difference in the fitness of Mut cells in the treatment samples (drug or shRNA) as compared with the control samples (DMSO or control shRNA) and was derived using the Wilcoxon test.
shRNA-based MCA
mir30-based shRNAs were designed and synthesized as clonable PCR products and cloned into pHAGE-pInd-10-mirE, an inducible lentiviral vector (Supplemental Table S10). Cells were transduced with the vectors for the expression of the shRNAs, selected, and cultured in the presence or absence of 1 µg/mL doxycycline for an additional 5–7 d. FACS analysis was then performed using a BD FACSAria II (BD Biosciences). The MCA was performed as described above, comparing the samples treated with shRNA specific for the gene of interest with the samples treated with the shRNAs specific for Luciferase.
Assay for proteasome activity
The measurement of proteasome activity was carried out by using a Promega Proteasome-Glo chymotrypsin-like cell-based assay. Proteasome-Glo cell-based assays are homogeneous, luminescent assays that individually measure the chymotrypsin-like, trypsin-like, or caspase-like protease activity associated with the proteasome complex in cultured cells. Briefly, appropriate substrate and inhibitors were added to Proteasome-Glo cell-based reagent and incubated at room temperature for 30 min. After equilibration, equal volumes of this reagent were added to 48-h drug-treated cells in a 96-well plate and then shaken. The luminescence was read after 10–30 min of incubation.
Effect of bortezomib on tumor growth in mice
nu/nu mice were purchased from Charles River Laboratories International, Inc. DLD1-PI3K wild-type and Mut cell lines were detected as pathogen-free at Charles River Laboratories International, Inc., and cultured in RPMI-1640 with 10% FBS and 1% penicillin–streptomycin. Cells were resuspended in serum-free medium mixed with an equal amount of Matrigel (BD Biosciences). Mice were injected with 2 million cells per shot at two locations per mouse in the flanks. The mice were randomly grouped, and treatment was started when tumor size reached 100–200 mm3. The experiment was repeated twice, and, every time, each cohort included at least seven mice. Tumor sizes were monitored weekly, and volumes were calculated with the following formula: mm3 = length × width × width × 0.5.
Supplementary Material
Acknowledgments
We are grateful to the members of the Elledge laboratory for helpful comments and ideas on the project. We apologize to our colleagues whose work we could not cite due to space limitations. This work was funded by National Institutes of Health grant 1U01CA199252-01, a Department of Defense grant to S.J.E., and a National Institutes of Health Pathway to Independence Award (K99/R00) to T.D. S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.290122.116.
References
- Adams J. 2003. The proteasome: structure, function, and role in the cell. Cancer Treat Rev 29: 3–9. [DOI] [PubMed] [Google Scholar]
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. 2012. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 14: 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, Dingledine R, Fu H, Vajda S, Talbot PJ, et al. 2011. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J Virol 85: 6381–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ. 2009. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 4: 127–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. 2011. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci 108: 12372–12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, East-Seletsky A, Ali LD, Gerath WF, Pantel SE, et al. 2014. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data 1: 140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA. 2009. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 9: 550–562. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. 2008. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Cheng CK, Nicolaides TP, Hackett CS, Knight ZA, Shokat KM, Weiss WA. 2007. A dual phosphoinositide-3-kinase α/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res 67: 7960–7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fece de la Cruz F, Gapp BV, Nijman SM. 2015. Synthetic lethal vulnerabilities of cancer. Annu Rev Pharmacol Toxicol 55: 513–531. [DOI] [PubMed] [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, et al. 2003. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23: 8862–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. 2014. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol 10: 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev 18: 1926–1945. [DOI] [PubMed] [Google Scholar]
- He Y, Van't Veer LJ, Mikolajewska-Hanclich I, van Velthuysen ML, Zeestraten EC, Nagtegaal ID, van de Velde CJ, Marijnen CA. 2009. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res 15: 6956–6962. [DOI] [PubMed] [Google Scholar]
- Hsieh AC, Truitt ML, Ruggero D. 2011. Oncogenic AKTivation of translation as a therapeutic target. Br J Cancer 105: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Berns A. 2004. Oncogene addiction: sometimes a temporary slavery. Cancer Cell 6: 535–538. [DOI] [PubMed] [Google Scholar]
- Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. 2007. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol Cell Biol 27: 662–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempner SJ, Myers AP, Cantley LC. 2013. What a tangled web we weave: emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov 3: 1345–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh JL, Brown KR, Sayad A, Kasimer D, Ketela T, Moffat J. 2012. COLT-Cancer: functional genetic screening resource for essential genes in human cancer cell lines. Nucleic Acids Res 40: D957–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardou H, Dahabreh IJ, Bafaloukos D, Kosmidis P, Murray S. 2009. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol 6: 352–366. [DOI] [PubMed] [Google Scholar]
- Luo J, Manning BD, Cantley LC. 2003. Targeting the PI3K–Akt pathway in human cancer: rationale and promise. Cancer Cell 4: 257–262. [DOI] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. 2009. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JM, Cen LP, Zhang XM, Chiang SW, Huang Y, Lin D, Fan YM, van Rooijen N, Lam DS, Pang CP, et al. 2007. PI3K/akt, JAK/STAT and MEK/ERK pathway inhibition protects retinal ganglion cells via different mechanisms after optic nerve injury. Eur J Neurosci 26: 828–842. [DOI] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. 2008. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci 105: 20380–20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina A, Mills JR, Pelletier J. 2012. Emerging therapeutics targeting mRNA translation. Cold Spring Harb Perspect Biol 4: a012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, Sircoulomb F, Medrano M, Fedyshyn Y, Koh JL, et al. 2012. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov 2: 172–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr SE, Smith JA, Shamu CE, Neumuller RA, Perrimon N. 2014. RNAi screening comes of age: improved techniques and complementary approaches. Nat Rev Mol Cell Biol 15: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, Chan AT, Engelman JA, Kraft P, Cantley LC, et al. 2009. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol 27: 1477–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- Raynaud FI, Eccles S, Clarke PA, Hayes A, Nutley B, Alix S, Henley A, Di-Stefano F, Ahmad Z, Guillard S, et al. 2007. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res 67: 5840–5850. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Velculescu VE. 2004. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle 3: 1221–1224. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Schmidt-Kittler O, Cummins JM, DeLong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. 2005. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7: 561–573. [DOI] [PubMed] [Google Scholar]
- Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, et al. 2008. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res 68: 8022–8030. [DOI] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. 2008. Cancer proliferation gene discovery through functional genomics. Science 319: 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MH, Young D, Kindler HL, Webb I, Kleiber B, Wright J, Grever M. 2004. Phase II study of the proteasome inhibitor bortezomib. (PS-341) in patients with metastatic neuroendocrine tumors. Clin Cancer Res 10: 6111–6118. [DOI] [PubMed] [Google Scholar]
- Shao DD, Tsherniak A, Gopal S, Weir BA, Tamayo P, Stransky N, Schumacher SE, Zack TI, Beroukhim R, Garraway LA, et al. 2013. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res 23: 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129: 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solimini NL, Luo J, Elledge SJ. 2007. Non-oncogene addiction and the stress phenotype of cancer cells. Cell 130: 986–988. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe AK. 2006. Mechanisms of disease: oncogene addiction - A rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol 3: 448–457. [DOI] [PubMed] [Google Scholar]
- Weinstein IB, Joe A. 2008. Oncogene addiction. Cancer Res 68: 3077–3080. [DOI] [PubMed] [Google Scholar]
- Whitman M, Downes CP, Keeler M, Keller T, Cantley L. 1988. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature 332: 644–646. [DOI] [PubMed] [Google Scholar]
- Wong KK, Engelman JA, Cantley LC. 2010. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev 20: 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Gonzalez-Angulo AM, Reuben JM, Booser DJ, Pusztai L, Krishnamurthy S, Esseltine D, Stec J, Broglio KR, Islam R, et al. 2006. Bortezomib (Velcade) in metastatic breast cancer: pharmacodynamics, biological effects, and prediction of clinical benefits. Ann Oncol 17: 813–817. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. 2008. PI3K pathway alterations in cancer: variations on a theme. Oncogene 27: 5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD. 2014. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 513: 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.