Abstract
The moss Physcomitrella patens performs efficient homologous recombination, which allows for the study of individual gene function by generating gene disruptions. Yet, if the gene of study is essential, gene disruptions cannot be isolated in the predominantly haploid P. patens. Additionally, disruption of a gene does not always generate observable phenotypes due to redundant functions from related genes. However, RNA interference (RNAi) can provide mutants for both of these situations. We show that RNAi disrupts gene expression in P. patens, adding a significant tool for the study of plant gene function. To assay for RNAi in moss, we constructed a line (NLS-4) expressing a nuclearly localized green fluorescent protein (GFP):β-glucuronidase (GUS) fusion reporter protein. We targeted the reporter protein with two RNAi constructs, GUS-RNAi and GFP-RNAi, expressed transiently by particle bombardment. Transformed protonemal cells are marked by cobombardment with dsRed2, which diffuses between the nucleus and cytoplasm. Cells transformed with control constructs have nuclear/cytoplasmic red fluorescence and nuclear green fluorescence. In cells transformed with GUS-RNAi or GFP-RNAi constructs, the nuclear green fluorescence was reduced on average 9-fold as soon as 48 h after transformation. Moreover, isolated lines of NLS-4 stably transformed with GUS-RNAi construct have silenced nuclear GFP, indicating that RNAi is propagated stably. Thus, RNAi adds a powerful tool for functional analysis of plant genes in moss.
The moss Physcomitrella patens has a relatively simple developmental pattern resembling the basic organization of most land plants. The two predominant tissue types, protonemal and gametophyte, are composed of either filaments or sheets of single cells, respectively. Thus, cell lineage analysis and cell biological studies can be performed easily. Like other land plants, P. patens metabolism and development are controlled by similar environmental signals and growth substances such as auxin (Imaizumi et al., 2002), cytokinin (Schulz et al., 2001), and abscisic acid (Knight et al., 1995). Additionally, P. patens has a relatively small genome, only three times larger than the Arabidopsis genome. An extensive expressed sequence tag (EST) database, of which at least 60,000 are publicly available, shows that the P. patens proteome contains many of the same genes present in higher plants (Rensing et al., 2002).
Unlike other land plants, P. patens has the unique ability to perform efficient homologous recombination (Schaefer and Zrÿd, 1997; Schaefer, 2001, 2002). Because P. patens is predominately haploid, gene targeting can be used to rapidly generate gene disruptions to allow the study of basic functions of individual genes. However, this approach is experimentally difficult when the gene of interest belongs to a large family and shares function with other family members. Due to redundant function, single-gene disruptions may not result in any observable phenotype. Moreover, if a gene is essential, its disruption can never be isolated in a haploid organism.
RNA interference (RNAi) has been successfully used as a tool in many organisms to silence individual genes and multiple members of a gene family (Sharp, 2001; Hannon, 2002; Tijsterman et al., 2002). In particular, RNAi in plants is systemic (Palauqui et al., 1997; Voinnet and Baulcombe, 1997). Additionally, in plants, RNAi transgenes can be inherited in subsequent generations (Chuang and Meyerowitz, 2000), creating a population of transgenic plants with reduced expression of the gene of interest. In ferns, RNAi in regenerating spores provides a simple single-cell system to study gene function (Klink and Wolniak, 2000; Stout et al., 2003). Here, we show that P. patens also uses RNAi for gene silencing. In a transient assay, RNAi silences the targeted gene product for at least 8 d after transformation. In addition, we are able to produce transgenic moss expressing the RNAi construct resulting in constitutive silencing. Because RNAi-induced silencing reduces the level of a gene product rather than removing it completely, this technique can be used to target essential genes that are inaccessible by gene replacement. RNAi can also provide moss mutants for genes in large gene families by silencing multiple members simultaneously. Together with targeted gene replacement and the ease of single-cell study due to the simple moss body plan, RNAi adds a powerful tool for this model organism to understand gene function in plants.
RESULTS
We searched the P. patens EST database for genes involved in RNAi-induced silencing and identified a sequence with similarity to an RNA-dependent RNA polymerase, which is implicated in at least one step of the RNAi mechanism in various organisms. To test whether RNAi functions in moss, we constructed a transgenic moss line expressing a reporter gene, which we used to assay for RNAi-induced silencing. We transformed protoplasts with a plasmid containing a kanamycin resistance marker and a green fluorescent protein (GFP):β-glucuronidase (GUS) fusion with an engineered nuclear localization sequence. The GFP:GUS fusion was expressed from the cauliflower mosaic virus 35S promoter. We isolated a kanamycin-resistant line expressing GFP uniformly in every nucleus (NLS-4). NLS-4 growth is comparable with wild type and has no observable phenotype, indicating that the plasmid DNA integrated into a nonessential locus and that expression of GFP:GUS does not alter the morphology and growth of P. patens.
We used two constructs to test for silencing of GFP:GUS expression. GUS-RNAi construct targets the GUS region, and GFP-RNAi construct targets the GFP region of the GFP:GUS reporter mRNA. Both GFP-RNAi and GUS-RNAi constructs are expressed from the maize (Zea mays) ubiquitin promoter and are designed to produce a double-stranded RNA hairpin in the cell. As a control, we used an RNAi construct (LUC-RNAi) designed to silence luciferase, an exogenous gene not present in NLS-4. We expressed these RNAi constructs both transiently by particle bombardment and constitutively by protoplast transformation followed by selection of transgenic moss lines.
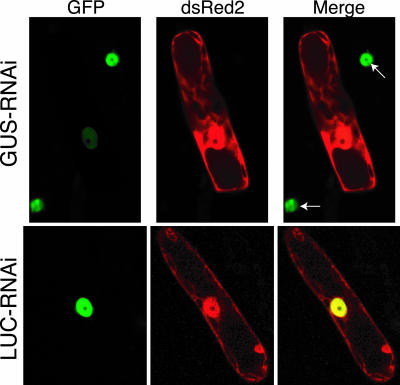
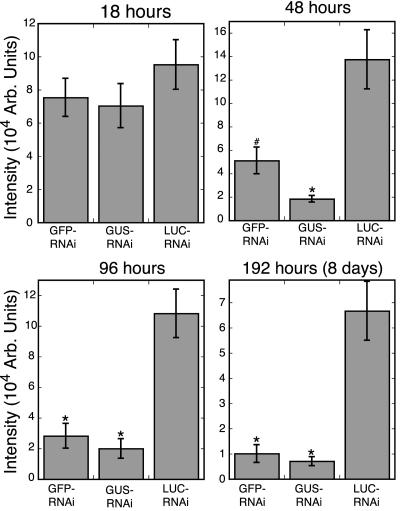
For transient expression of RNAi constructs, we marked transformed protonemal cells by cobombarding a plasmid expressing dsRed2 fluorescent protein. We identified dsRed2-expressing cells by confocal microscopy and then measured the intensity of the GFP signal in the nucleus. Figure 1 shows a representative cell cotransformed with GUS-RNAi (top row) or LUC-RNAi (bottom row) and dsRed2. In moss protonemal cells, dsRed2 is cytoplasmic and freely diffuses into the nucleus. In cells expressing GUS-RNAi, we observed a markedly reduced GFP signal in the nucleus as compared with neighboring non-transformed cells (Fig. 1, arrows). Reduction of nuclear GFP fluorescence is specific to the GUS-RNAi construct, because the nuclear GFP signal in cells transformed with the control LUC-RNAi construct was not affected (Fig. 1, bottom row). Specific reduction of nuclear GFP signal was similarly observed in cells transformed with GFP-RNAi construct (data not shown; Fig. 2). Significant reduction of the nuclear GFP was evident after 48 h and was stable for at least 8 d after transformation with either GUS-RNAi or GFP-RNAi constructs (Fig. 2).
Figure 1.
Transient RNAi in P. patens. NLS-4 (a transgenic moss line expressing nuclearly localized GFP) was cobombarded with dsRed2 and either GUS-RNAi or LUC-RNAi constructs. Transformed cells are marked with dsRed2 fluorescent protein. The nuclear GFP signal is reduced when the cell is transformed with GUS-RNAi (top row) and unaffected in neighboring untransformed cells (arrows). Transformation with LUC-RNAi has no effect on the nuclear GFP signal (bottom row). Images were collected with the Zeiss confocal microscope.
Figure 2.
Time course of transient RNAi. The intensity of nuclear GFP signal in cells transformed with each RNAi construct is shown for the indicated time points after particle bombardment. At least 15 cells were measured for each construct. Error bars indicate se. Statistical analysis was carried out with the Student's t test for unpaired data with unequal variance. The asterisk shows data sets are statistically different from LUC-RNAi with a t-probability of <0.001. #, A t-probability of 0.0056. Images were collected with the Leica confocal microscope.
Because the dsRed2 emission spectrum slightly overlaps with the GFP emission spectrum, we used two different detectors to collect data to ensure that the detected GFP signal has no contribution from dsRed2. First, we collected data on a confocal microscope (Zeiss, Thornwood, NY) using a spectral imager detector, which uses linear unmixing to resolve multiple chromofore signals. We measured the dsRed2 emission spectrum in a cell only expressing dsRed2 and the GFP emission spectrum in a cell only expressing GFP. We used an excitation wavelength of 488 nm and collected the emission spectra with a 494- to 579-nm bandwidth. The collected spectra were then used for linear unmixing, removing the dsRed2 signal from cells expressing both dsRed2 and GFP. Second, we collected data on a confocal microscope (Leica Microsystems, Heidelberg) using a photomultiplier tube (PMT) as the detector with an extremely narrow wavelength collection (490-502 nm). We compared the data collected with these two methods and found that the reduction of the nuclear GFP signal is 8.8-fold using the spectral imager (GUS-RNAi construct, 19 cells; LUC-RNAi construct, 18 cells) and 8.7-fold using the narrow bandwidth on the PMT (GUS-RNAi construct, 93 cells; LUC-RNAi construct, 75 cells). Thus, the observed nuclear GFP signal is not affected by expression of dsRed2 in the nucleus.
Transient silencing is rapid. However, to determine the effect of silencing a particular gene on the overall growth and morphology of P. patens, systemic silencing is required, as would be the case in a transgenic moss line constitutively expressing an RNAi construct. To test whether silencing can be propagated in a transgenic line, we transformed NLS-4 protoplasts with GUS-RNAi and LUC-RNAi, both modified by the addition of a hygromycin resistance cassette. After transformation and regeneration of protoplasts, stable moss lines are isolated by a first selection in antibiotic, a relaxation of the selection, and then a second selection in antibiotic. After the first selection, more than 90% of the resistant colonies transformed with GUS-RNAi showed a significant reduction of nuclear GFP fluorescence (data not shown). Resistant colonies transformed with LUC-RNAi had no reduction of GFP expression in the nucleus (data not shown). After relaxation of the selection, we observed that many of the GUS-RNAi colonies began to express GFP. A possible explanation is that resistant colonies that contain the RNAi construct episomally (Ashton et al., 2000) but have not integrated the RNAi construct begin to lose the construct in the absence of selection and thus lose silencing. Another possibility is that the GFP transgene may be reactivated by other mechanisms.
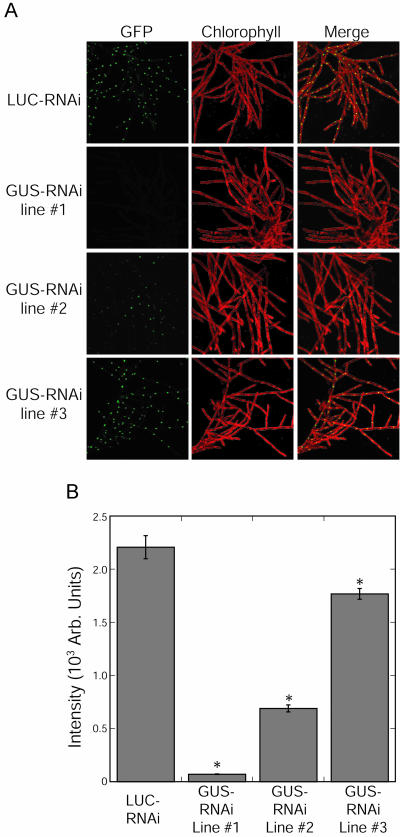
From four transformations, we isolated three stable lines containing GUS-RNAi and at least three lines containing LUC-RNAi. This number of stable lines per transformation is expected for untargeted transformation of P. patens (Schaefer, 2001). The LUC-RNAi lines all had uniform GFP expression in the nucleus. In Figure 3A (top row), we show a representative line expressing LUC-RNAi. The three stable GUS-RNAi lines are also shown in Figure 3A. Line 1 has very little detectable nuclear GFP, and lines 2 and 3 have increasing amounts of nuclear GFP expression. We measured the intensity of the GFP signal and show the results in Figure 3B. As has been observed in other systems, the level of RNAi-induced silencing in the generated P. patens stable lines ranges from strong (line 1) to weak (line 3). Additionally, we observed silencing of the nuclear GFP in protonemal and gametophyte tissues (data not shown). Taken together, these results demonstrate that transgenic moss can be produced with long-term silencing of the desired transgene.
Figure 3.
Stable RNAi in P. patens. A, P. patens lines are shown after selection for stable insertion of the indicated constructs. Top row is a line transformed with LUC-RNAi showing no reduction of GFP expression in the nucleus. All other rows show lines transformed with GUS-RNAi with increasing levels of GFP expression in the nucleus. B, The intensity of nuclear GFP in the transformed stable lines. At least 206 cells were quantified for each line. The asterisk shows data sets are statistically different from LUC-RNAi with a t-probability of <0.001.
CONCLUSIONS
We show that RNAi functions in moss to silence expression of a reporter gene. This is significant because it provides a new tool for studying gene function in P. patens. Transient RNAi is stable for at least 8 d, allowing for rapid determination of potential phenotypes, because numerous physiological assays may be performed in this time frame. For example, induction of bud formation by cytokinin requires 48 to 72 h (Schulz et al., 2001). Auxin treatment causes transformation of chloronemal tip cells into caulonemal cells (Imaizumi et al., 2002) and transformation of budding cells back into caulonemal cells within 48 to 72 h (P. Perroud, personal communication). The response to polarized light and to directional light can be observed within 8 d as well (Cove and Knight, 1987).
We also show that RNAi can be propagated by production of transgenic moss lines. P. patens is predominately haploid, and cultures are routinely propagated in the haploid state. Thus once a transgenic line is isolated, there is no need to study silencing in subsequent generations. In addition, multiple independent transgenic lines can be easily generated to verify that the observed phenotype is due to introduction of the RNAi construct.
Because RNAi silences gene expression, RNAi provides an opportunity to study essential genes in P. patens. We can also probe the function of large gene families because using a single-construct RNAi may silence multiple related genes with sequence similarity. Additionally, large-scale analysis of plant gene function may be possible using RNAi in moss. With the cDNA sequence information available from extensive P. patens EST databases (such as at http://www.moss.leeds.ac.uk), a large number of moss genes can be silenced to determine their function. This large-scale approach is not possible with gene disruptions, because cDNA sequence information is usually not sufficient for efficient gene targeting. Thus, RNAi provides a new and significant tool for studying the function of plant genes in moss.
MATERIALS AND METHODS
DNA Constructs
The GFP:GUS fusion construct was a gift from D.W. Galbraith (Grebenok et al., 1997). We subcloned GFP:GUS as a SacI/XhoI fragment into SacI/XhoI sites in pMBL5 (J. Machuka The Physcomitrella EST Program at the University of Leeds [UK] and Washington University [St. Louis]) for transformation into moss protoplasts. GUS-RNAi and LUC-RNAi plasmids were gifts from R. Zentella (Zentella et al., 2002). The maize ubiquitin promoter controlled expression of GUS- and LUC-inverted repeats. To allow for selection of transgenic moss, we subcloned a PvuII fragment from pGL2 (Schaefer and Zrÿd, 1997) containing the hygromycin marker controlled by the cauliflower mosaic virus 35S promoter into a blunted EcoRI site in GUS-RNAi and a blunted HindIII site in LUC-RNAi plasmid. We replaced the GUS-inverted repeats in GUS-RNAi by subcloning GFP-inverted repeats from pCGT1824 (gift from C. Taylor) as an EcoRV fragment into blunted SacI and BamHI sites in GUS-RNAi, creating the GFP-RNAi plasmid.
Moss Transformation
Transformation of moss protoplasts and isolation of transgenic moss lines was carried out as described (Schaefer et al., 1991). To verify that the transformed lines were stable, colonies were blended and plated onto two plates containing cellophanes and non-selection media. After at least 1 week of growth, one cellophane was transferred to media containing hygromycin. The line was considered stable if both the plate without selection and the plate with selection grew equally well.
Transient transformation by particle bombardment was carried out using a PDS-1000 He System particle gun (Bio-Rad Laboratories, Hercules, CA). In brief, 2.5 μg of DNA was added to 25 μL of 1-μm gold in 50% (v/v) glycerol. For cobombardment, we used 1.25 μg of dsRed2 plus 1.25 μg of RNAi construct or 0.8 μg of dsRed2 plus 1.7 μg of RNAi construct. Both the 1:1 and 1:2 ratios of dsRed2:RNAi construct yielded similar silencing results (data not shown). While vortexing, 25 μL of 2.5 m CaCl2 and 10 μL of 0.1 m spermidine were added. The gold and DNA mixture was vortexed for an additional minute, incubated for 1 min at room temperature, and spun for 30 s at maximum speed in a microcentrifuge. The DNA/gold was washed with 70 μL of 70% (v/v) ethanol and 70 μL of 100% (v/v) ethanol without disturbing the pellet. The DNA/gold was resuspended in 24 μL of 100% (v/v) ethanol. A 6-μL drop was spotted onto a Macro carrier, which had been presoaked in 100% (v/v) ethanol and dried. We used 4- to 10-d-old moss protonemal tissue for particle bombardment. We used a rupture pressure of 900 psi and a vacuum of 27 in Hg.
Microscopy and Image Analysis
Protonemal tissue was placed in water on a glass slide and covered with a coverslip. We used two systems for collecting images: a Zeiss LSM 510 Meta NLO and a Leica Confocal System TCS2. Both systems imaged GFP and dsRed2 simultaneously using an argon laser (488-nm excitation). For detection, the Zeiss system used a spectral imager, and the Leica system used two PMTs. The spectral imager collected emission at a bandwidth of 494 to 579 nm. We collected dsRed2 and GFP emission spectra in cells expressing the individual chromophores. We used the software provided with the Zeiss microscope system to perform linear unmixing of data in which both chromophores were expressed. The images shown in Figure 1 have been unmixed. For the Leica microscope system, PMT1 detected emission from 490 to 502 nm for the GFP signal, and PMT2 detected emission from 615 to 650 nm for the dsRed2 signal. The gain for PMT1 was identical for all images. The GFP intensity in the nucleus was measured using ImageJ 1.29X (http://rsb.info.nih.gov/ij/).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Acknowledgments
We thank R. Zentella, C. Taylor, and D. Galbraith for providing us with plasmids. We thank H. Berg for technical assistance using the Zeiss confocal microscope system at the Donald Danforth Plant Science Center Integrated Microscopy Facility in St. Louis. We are grateful to Y. Sakata and W.L. Lee for careful reading of the manuscript.
This work was supported by a post-doctoral research fellowship from the Helen Hay Whitney Foundation to M.B. and by the National Science Foundation (grant nos. IBN 0112461 to R.S.Q. and DBI 0216150 to Howard Berg from the Donald Danforth Plant Science Center Integrated Microscopy Facility, St. Louis).
References
- Ashton NW, Champagne CEM, Weiler T, Verkoczy LK (2000) The bryophyte Physcomitrella patens replicates extrachromosomal transgenic elements. New Phytol 146: 391-402 [Google Scholar]
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985-4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove DJ, Knight CD (1987) Gravitropism and phototropism in the moss, Physcomitrella patens. In H Thomas, D Grierson, eds, Developmental Mutants of Higher Plants. Cambridge University Press, London
- Grebenok RJ, Pierson E, Lambert GM, Gong FC, Afonso CL, Haldeman-Cahill R, Carrington JC, Galbraith DW (1997) Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J 11: 573-586 [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244-251 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kadota A, Hasebe M, Wada M (2002) Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14: 373-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Wolniak SM (2000) The efficacy of RNAi in the study of the plant cytoskeleton. J Plant Growth Regul 19: 371-384 [DOI] [PubMed] [Google Scholar]
- Knight CD, Sehgal A, Atwal K, Wallace JC, Cove DJ, Coates D, Quatrano RS, Bahadur S, Stockley PG, Cuming AC (1995) Molecular responses to abscisic acid and stress are conserved between moss and cereals. Plant Cell 7: 499-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16: 4738-4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Rombauts S, Van de Peer Y, Reski R (2002) Moss transcriptome and beyond. Trends Plant Sci 7: 535-538 [DOI] [PubMed] [Google Scholar]
- Schaefer DG (2001) Gene targeting in Physcomitrella patens. Curr Opin Plant Biol 4: 143-150 [DOI] [PubMed] [Google Scholar]
- Schaefer DG (2002) A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53: 477-501 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd J-P (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11: 1195-1206 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd J-P, Knight CD (1991) Stable transformation of the moss Physcomitrella patens. Mol Gen Genet 226: 418-424 [DOI] [PubMed] [Google Scholar]
- Schulz PA, Hofmann AH, Russo VE, Hartmann E, Laloue M, von Schwartzenberg K (2001) Cytokinin overproducing ove mutants of Physcomitrella patens show increased riboside to base conversion. Plant Physiol 126: 1224-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA (2001) RNA interference: 2001. Genes Dev 15: 485-490 [DOI] [PubMed] [Google Scholar]
- Stout SC, Clark GB, Archer-Evans S, Roux SJ (2003) Rapid and efficient suppression of gene expression in a single-cell model system, Ceratopteris richardii. Plant Physiol 131: 1165-1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Ketting RF, Plasterk RH (2002) The genetics of RNA silencing. Annu Rev Genet 36: 489-519 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC (1997) Systemic signalling in gene silencing. Nature 389: 553. [DOI] [PubMed] [Google Scholar]
- Zentella R, Yamauchi D, Ho TH (2002) Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14: 2289-2301 [DOI] [PMC free article] [PubMed] [Google Scholar]