Abstract
Listeria monocytogenes, one of most problematic foodborne pathogens, is responsible for listeriosis in both humans and animals and mainly transmitted through the food chain. In this report, we propose a simple, rapid, and nearly instrument-free molecular technique using multiple cross displacement amplification (MCDA) label-based gold nanoparticles lateral flow biosensor (LFB) for specific, sensitive, and visual detection of L. monocytogenes. The MCDA-LFB method was carried out at a constant temperature (61°C) for only 20 min during the reaction stage, and then the amplification mixtures were directly detected by using LFB, eliminating the use of an electrophoresis instrument, special reagents, or amplicon analysis equipment. The whole procedure, from sample processing to result indicating, was finished within 1 h. The analytical specificity of MCDA-LFB method was successfully determined by distinguishing the target bacterium from other pathogens. The analytical sensitivity of the MCDA-LFB assay was 10 fg of genomic templates per reaction in pure culture, which was in complete accordance with MCDA by gel electrophoresis, real-time turbidity, and colorimetric indicator. The assay was also successfully applied to detecting L. monocytogenes in pork samples. Therefore, the rapidity, simplicity, and nearly equipment-free platform of the MCDA-LFB technique make it possible for food control, clinical diagnosis, and more. The proof-of-concept assay can be reconfigured to detect various target sequences by redesigning the specific MCDA primers.
Keywords: Listeria monocytogenes, gold nanoparticles, MCDA, MCDA-LFB
Introduction
Listeria monocytogenes, a Gram-positive, facultative intracellular, foodborne bacteria pathogen, is the causative agent of animals and human listeriosis.1 The opportunistic organism is widespread in the natural environment, thus can easily contaminate various raw and processed foods, such as meat, poultry, fish, vegetables, fermented sausages, milk, and dairy products.2 L. monocytogenes has the ability to survive and grow even under high concentrations of salt or bile, low temperature and pH, carbon starvation, oxidative stress, and other adverse conditions.3 The occurrence of the target pathogen is considered as a serious public threat because of its severity of the illness manifested by gastroenteritis, septicemia, meningitis, spontaneous abortion, pneumonia, central nervous system infections, etc.4
Human foodborne listeriosis was mainly linked to the consumption of L. monocytogenes-contaminated food products and particularly associated with ready-to-eat foods.5 The groups at high risk are pregnant women, newborn infants, older adults, and immunocompromised individuals.6 Despite its low incidence, human listeriosis reminds of great public health concern due to its high case fatality rate (20%–30%).7 Over the past two decades, numerous listeriosis outbreaks were reported all over the world, such as Japan, New Zealand, USA, France, England, Germany, and other European countries.8 Hence, it is crucial to devise a rapid, highly sensitive, and specific assay for L. monocytogenes detection.
Conventional techniques for the L. monocytogenes detection are culture-based methods, which required trained personnel and consist of multiple steps. Moreover, the slow growth rate of the pathogenic bacteria is also a challenge for detection by culture-based methods as the time to results requires up to 7 days to produce reliable results.9,10 The absence of high avidity antibodies has restricted the use of antibody-based detection of the target pathogen, and as a result, much effort has been devoted to nucleic acid-based assays for efficient L. monocytogenes detection.11,12 In this regard, polymerase chain reaction (PCR)-based identification methods have been established for detecting the target pathogens in enrichment cultures.2 However, these assays required advanced and expertise laboratory system, were expensive and not feasible in resource-challenged regions of the world.
The growing use of molecular detection techniques has emphasized simplicity and speed as key criteria for adoption in field detection, “on-site” diagnosis, point-of-care testing and more, and the isothermal amplification methodologies were well-suited for these applications. Multiple cross displacement amplification (MCDA) (Chinese IP Office Patent Application CN201510280765.X) was a novel isothermal amplification technique devised by our group and successfully applied to diagnostic detection of pathogens in food and clinical samples.13,14 MCDA displayed unique advantages of rapidity, simplicity, repeatability, sensitivity, and specificity, generating amplicons from as few as three bacterial cells. The MCDA products were detected by visual inspection of color changes, by use of spectrophotometric apparatus to measure turbidity, or by agarose gel electrophoresis. However, all of the monitoring techniques are not specific to template sequences, thus the nonspecific reaction products can cause the false positive results. To overcome the technical shortcomings, further speedup and simplify the process of MCDA technology, a novel lateral flow biosensor (LFB) was successfully devised and applied to detecting MCDA amplicons in this study. Then, the MCDA assay combined with the novel LFB (MCDA-LFB) was developed for simple, visual, and rapid detection of L. monocytogenes. The analytical sensitivity and specificity of L. monocytogenes-MCDA-LFB assay were evaluated by pure cultures and the practical application was also successfully verified by detecting the target pathogen in pork samples.
Materials and methods
Reagents and apparatus
The sheep anti-digoxigenin antibody (anti-Dig), the streptavidin-immobilized 30 nm gold nanoparticles (SA-G), and biotinylated bovine serum albumin (biotin-BSA) were purchased from the Resenbio Co., Ltd. (Xi’an, People’s Republic of China). The backing card, absorbent pad, nitrocellulose (NC) membrane, conjugate pad, and sample pad were purchased from the Jie Yi Biotechnology Co., Ltd. (Shanghai, People’s Republic of China). The Loopamp kits and Loopamp™ Fluorescent Detection Reagent (FD) were purchased from Eiken Chemical (Beijing, People’s Republic of China). The DNA extraction kits (QIAamp DNA minikits; Qiagen, Hilden, Germany) were purchased from Qiagen (Beijing, People’s Republic of China).
Construction of gold nanoparticle-based dipstick biosensor
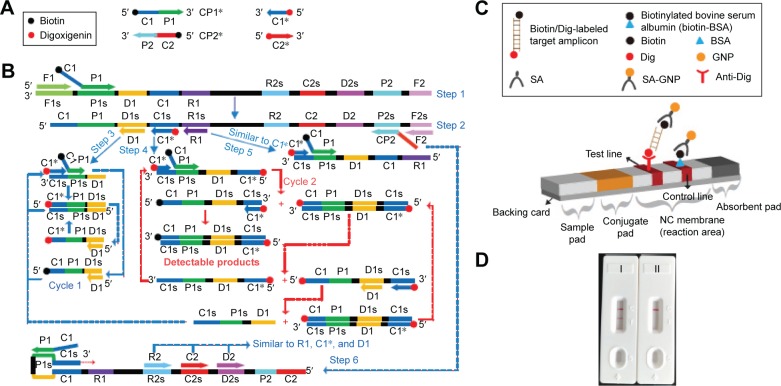
The LFB (4×6 mm) consists of an immersion pad, a conjugate pad, an NC membrane, and an absorbent pad and assembled on a plastic adhesive backing card orderly by overlapping 2 mm among them. A schematic diagram of the biosensor was shown in Figure 1. The SA-G in 0.01 M phosphate-buffered saline (PBS, pH 7.4) was deposited on the conjugate pad. Anti-Dig (0.2 mg/mL) and biotin-BSA (2.5 mg/mL) in 0.01 M PBS (pH 7.4) were immobilized at the test line (TL) and control line (CL) with each line separated by 5 mm, respectively. The assembled cards were subsequently cut at 4 mm widths. All the biosensors were packaged in a plastic bag containing a desiccant gel and dryly stored at the room temperature until use.
Figure 1.
The outline of MCDA combined with LFB.
Notes: (A) Schematic depiction of the new cross primer (CP1*) and amplification primer (D1*). (B) Outline of MCDA with CP1* and D1*. (C) Schematic illustration of the principle of LFB for visualization of MCDA amplicons. (D) Interpretation of the results: I, positive (two red lines at the readout area) and II, negative (only the control line zone showed a red band). C1*, 5′-labeled with Dig when used in MCDA-LFB assay; CP1*, 5′-labeled with biotin when used in MCDA-LFB assay.
Abbreviations: BSA, bovine serum albumin; Dig, digoxigenin; GNPs, gold nanoparticles; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification; NC, nitrocellulose; SA, streptavidin.
Visual detection of MCDA products using the biosensor
A 0.3 μL aliquot of MCDA products was deposited to the sample application region of the biosensor. Then, a 100 μL aliquot of running buffer (10 mM PBS, pH 7.4 with 1% Tween 20) was also deposited to the sample application region, and the biosensor allowed to absorb the whole running buffer. After 2 min, the MCDA product detection was visualized in the form of red lines on the NC membrane.
Primer design for L. monocytogenes-MCDA-LFB assay
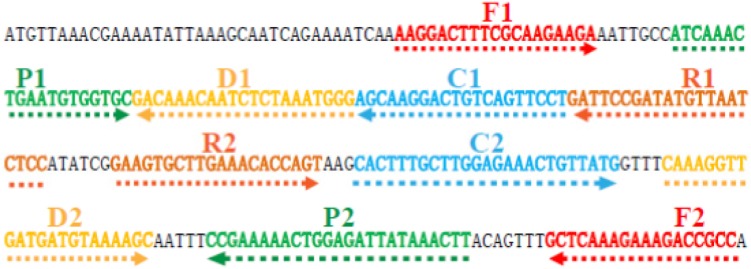
To design L. monocytogenes-specific MCDA primers, a set of MCDA primers was designed by PrimerExplorer V4 (Eiken Chemical, Japan) and primer software PRIMER PREMIER 5.0 according to the specific lmo0733 gene (GenBank accession no 985919).15 Blast analysis confirmed that the MCDA primer set was specific for L. monocytogenes strains. The details of primer design, primers sequences, locations, and modifications of MCDA primers were shown in Table 1 and Figure 2. All of the oligomers were synthesized and purified by TsingKe Biological Technology (Beijing, People’s Republic of China) at high-performance liquid chromatography purification grade.
Table 1.
The primers used in this study
| Primers | Sequences and modifications (5′–3′) | Length |
|---|---|---|
| F1 | AAGGACTTTCGCAAGAAGA | 19 nt |
| CP1 | AGGAACTGACAGTCCTTGCTATCAAACTGAATGTGGTGC | 39 mer |
| CP1* | Biotin-AGGAACTGACAGTCCTTGCTATCAAACTGAATGTGGTGC | 39 mer |
| C1 | AGGAACTGACAGTCCTTGCT | 20 nt |
| C1* | Dig-AGGAACTGACAGTCCTTGCT | 20 nt |
| D1 | CCCATTTAGAGATTGTTTGTC | 21 nt |
| R1 | GGAGATTAACATATCGGAATC | 21 nt |
| R2 | GAAGTGCTTGAAACACCAGT | 20 nt |
| D2 | CAAAGGTTGATGATGTAAAAGC | 22 nt |
| C2 | CACTTTGCTTGGAGAAACTGTTATG | 25 nt |
| CP2 | CACTTTGCTTGGAGAAACTGTTATGAAGTTTATAATCTCCAGTTTTTCGG | 50 mer |
| F2 | GGCGGTCTTTCTTTGAGC | 18 nt |
Notes: C1*, 5′-labeled with Dig when used in MCDA-LFB assay; CP1*, 5′-labeled with biotin when used in MCDA-LFB assay.
Abbreviations: LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification; mer, monomeric; nt, nucleotide.
Figure 2.
Location and sequence of lmo0733 gene (Listeria monocytogenes-specific gene) used to design multiple cross displacement amplification primers.
Notes: The nucleotide sequence of the sense strand of lmo0733 is shown. Right arrows and left arrows indicate sense and complementary sequences that were used.
Bacterial strains and genomic template preparation
A total of 95 bacterial strains, including 50 strains of L. monocytogenes, 23 strains of other Listeria species and 22 strains of non-Listeria bacteria, were used in the current study (Table 2). All strains were stored in 15% (w/v) glycerol broth at −70°C and then were refreshed three times on nutrient agar plate at 37°C. According to the manufacture’s protocol, the genomic templates were extracted from all culture strains using DNA extraction kits and were determined with ultraviolet spectrophotometer (Nano drop ND-1000, Calibre, Beijing, People’s Republic of China) at A260/280. The DNA templates were stored under at −20°C before the templates were used. The strains of L. monocytogenes serovar 1/2a (EGD-e) were used for confirmation performance, optimal temperature, and sensitivity analysis conducted in the study. The genomic templates of L. monocytogenes serovar 1/2a (EGD-e) were serially diluted (10 ng, 10 pg, 10 fg, 1 fg, and 0.1 fg per μL) and a 1 μL aliquot of each dilution was applied for sensitivity examination of MCDA-LFB detection.
Table 2.
Bacterial strains used in this study
| Bacteria | Serovar | Strain no (source of strain) | No. of strains |
|---|---|---|---|
| Listeria monocytogenes | 1/2a | EGD-e | 1 |
| ATCC51772 | 1 | ||
| Isolated strains (ICDC) | 5 | ||
| 3a | ATCC51782 | 1 | |
| Isolated strains (ICDC) | 5 | ||
| 1/2c | Slcc2372 | 1 | |
| Isolated strains (ICDC) | 5 | ||
| 3c | Slcc2479 | 1 | |
| 1/2b | Slcc2755 | 1 | |
| Isolated strains (ICDC) | 5 | ||
| 3b | Slcc2450 | 1 | |
| 7 | Slcc2482 | 1 | |
| NCTC10890 | 1 | ||
| 4a | ATCC19114 | 1 | |
| Isolated strains (ICDC) | 5 | ||
| 4c | ATCC19116 | 1 | |
| Isolated strains (ICDC) | 3 | ||
| 4b | ATCC19115 | 1 | |
| Isolated strains (ICDC) | 5 | ||
| 4d | ATCC19117 | 1 | |
| Isolated strains (ICDC) | 5 | ||
| 4e | ATCC19118 | 1 | |
| Isolated strains (ICDC) | 1 | ||
| Listeria ivanovii | U | ATCCBAA-678 | 1 |
| U | Isolated strains (ICDC) | 5 | |
| Listeria innocua | U | ATCCBAA-680 | 1 |
| U | Isolated strains (ICDC) | 2 | |
| Listeria grayi | U | ATCC25402 | 1 |
| U | Isolated strains (ICDC) | 5 | |
| Listeria seeligeri | U | ATCC35967 | 1 |
| U | Isolated strains (ICDC) | 1 | |
| Listeria welshimeri | U | ATCC35897 | 1 |
| U | Isolated strains (ICDC) | 5 | |
| Bacillus cereus | U | Isolated strains (ICDC) | 2 |
| Enteropathogenic Escherichia coli | U | Isolated strains (ICDC) | 1 |
| Enterotoxigenic E. coli | U | Isolated strains (ICDC) | 1 |
| Enteroaggregative E. coli | U | Isolated strains (ICDC) | 1 |
| Enteroinvasive E. coli | U | Isolated strains (ICDC) | 1 |
| Enterohemorrhagic E. coli | U | EDL933 | 1 |
| Plesiomonas shigelloides | U | ATCC51903 | 1 |
| Shigella boydii | U | Isolated strains (ICDC) | 1 |
| Shigella sonneri | U | Isolated strains (ICDC) | 1 |
| Shigella flexneri | U | Isolated strains (ICDC) | 1 |
| Enterobacter cloacae | U | Isolated strains (ICDC) | 1 |
| Enterococcus faecalis | U | ATCC35667 | 1 |
| Yersinia enterocolitica | U | ATCC23715 | 1 |
| Streptococcus pneumoniae | U | Isolated strains (ICDC) | 1 |
| Aeromonas hydrophila | U | ATCC7966 | 1 |
| Vibrio vulnificus | U | Isolated strains (ICDC) | 1 |
| Proteus vulgaris | U | Isolated strains (ICDC) | 1 |
| Vibrio fluvialis | U | Isolated strains (ICDC) | 1 |
| Streptococcus bovis | U | Isolated strains (ICDC) | 1 |
| Vibrio parahaemolyticus | U | ATCC17802 | 1 |
| Klebsiella pneumoniae | U | ATCC700603 | 1 |
| Salmonella enteric | U | ICDC-NPSa001 | 1 |
Abbreviations: ATCC, American Type Culture Collection; ICDC, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention; NCTC, National Collection of Type Culture; Slcc, Seeliger’s Special Listeria Culture Collection; U, unidentified serotype.
The standard MCDA assay
To determine the suitability of lmo0733-MCDA primers, the MCDA reaction was performed as the standard MCDA condition, which has been described in previous studies.13 Briefly, the MCDA reaction was conducted in 25 μL amplification mixtures containing the following components: 0.8 μM cross primers CP1*, 1.6 μM cross primer CP1, 2.4 μM cross primer CP2, 0.8 μM each of amplification primers C1* and C2, 1.2 μM each of amplification primers R1, R2, D1, and D2, 0.4 μM each of displacement primers F1 and F2, 12.5 μL 2× reaction mix (Loopamp kits), 1.25 μL of Bst DNA polymerase (10 U), and 1 μL DNA template. The monitoring techniques, including colorimetric indicator (FD), turbidimeters (LA-320C), gel electrophoresis, and LFB detection, were applied to confirming the MCDA products.
Then, we evaluated the optimal amplification temperature of lmo0733-MCDA primers. The MCDA amplification mixtures were carried out at a constant temperature ranging from 60°C to 67°C for 1 h and then incubated at 85°C for 5 min to terminate the MCDA reactions. Mixtures with 1 μL genomic template of Listeria ivanovii strain (L. ivanovii, ATCCBAA-678) and Salmonella strain (ICDC-NPsa001) were selected as negative controls, and mixtures with 1 μL double distilled water were used as a blank control.
The analytical sensitivity of the L. monocytogenes-MCDA by four monitoring techniques
The genomic templates of L. monocytogenes serovar 1/2a (EGD-e) were serially diluted to verify the limit of detection (LoD). The LoD was defined by genomic DNA amount of the template. The analytical sensitivity of MCDA-LFB was tested as described above, and was further confirmed by colorimetric indicator (FD reagent), 2% agarose gel electrophoresis, and real-time turbidity analysis. Three replicates of each dilution were determined to verify the analytical sensitivity.
The analytical specificity of the L. monocytogenes-MCDA-LFB assay
To evaluate the analytical specificity of L. monocytogenes-MCDA-LFB assay, the MCDA reactions were conducted under the conditions described above with purely genomic templates from 50 L. monocytogenes, 23 strains of other Listeria species, and 22 strains of non-Listeria bacteria (Table 2). The MCDA products were analyzed using LFB detection. The evaluations were performed twice independently.
Applicability of the L. monocytogenes-MCDA-LFB assay
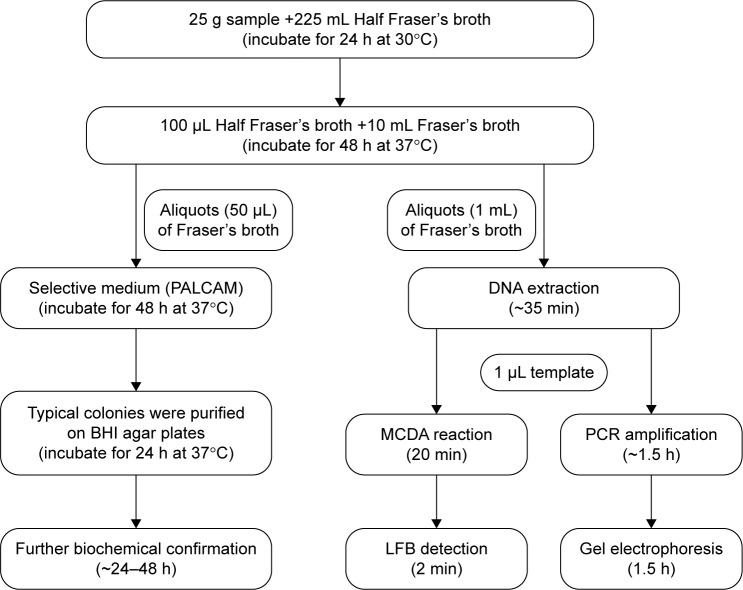
To test the availability of the MCDA-LFB assay for detection of L. monocytogenes, the analysis results of MCDA-LFB assay for 61 pork samples (ground meat samples) obtained from one meat wholesale market in Beijing were compared with conventional PCR and culture-biotechnical detection results for the same samples (Figure 3). The culture-biotechnical detection of pork samples was conducted according to the ISO 11290-1 standard method. In brief, 25 g of each pork sample was added into 225 mL Half Fraser’s broth (Oxoid, Hampshire, UK) and homogenized, followed by incubating the Listeria enrichment broth at 30°C for 24 h. Then, aliquots (100 μL) of Half Fraser’s broth were transferred to 10 mL of Fraser’s broth and homogenized, followed by incubating the broth at 37°C with shaking (250 rpm) for 48 h. A portion (50 μL) of positive Fraser’s broth cultures was streaked onto Polymyxin-acriflavine-LiCl-ceftazidime-aesculin-mannitol Agar (PALCAM) agar plates (Oxoid), which were incubated for 48 h at 37°C. The identification of L. monocytogenes strains was confirmed by gram staining, characteristic colony morphology, and various tests (motility, catalase, oxidase CAMP test, indole, urease, and aesculin hydrolysis). A portion (1 mL) of the Fraser’s broth was subjected to extract genomic DNA, which is used as a template in the MCDA-LFB and PCR assay. A L. monocytogenes-PCR assay has been developed in a report, which amplified a 113 bp DNA fragment of lmo0733 and was employed to verify the presence of target pathogens in pork samples.16
Figure 3.
Schematic representation of MCDA-LFB method compared with PCR and standard culture methods.
Abbreviations: BHI, brain heart infusion; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification; PCR, polymerase chain reaction.
Results
Development of the MCDA-LFB assay
A schematic illustration of the principle of MCDA-LFB technique is presented in Figure 1. In the MCDA system, the cross primers (CP1 or CP2) are modified at the 5′ end with biotin and the amplification primers (C1 or C2) are labeled at the 5′ end with Dig (Figure 1A). The new CP1, CP2, C1, and C2 primers are termed as CP1*, CP2*, C1*, and C2*, respectively. For clarity, the CP2* and C2* primers are not shown in outline of MCDA reaction (Figure 1B). During the reaction stage, the CP1* primer initiates MCDA reaction at the P1s sequence of the target and this new strand is displaced by upstream synthesis from the displacement primer F1 (step 1). Then, five primers (D1, C1*, R1, CP2, and F2) anneal to the newly synthesized strand and the polymerase (Bst) extended in tandem generating four distinct products (step 2). The D1 product serves as the template for subsequent elongation and cycling steps (step 3, Cycle 1). The C1* product is also used as the template by C1* and CP1* primers, and enters a cyclic process (step 4, Cycle 2). In the cycle, a larger amounts of double-labeled detectable products, which contain Dig-labeled C1* primer and a biotin-labeled CP1* primer, are successfully constructed. The details of the amplification process for R1 and CP2 products (steps 5 and 6) have been described in previous report.13 Furthermore, a double-labeled detectable product (CP2*/C2* product), which is similar to the detectable CP1*/C1* product, can be formed when the C2 primer is modified with a Dig at the 5′ end and CP2 primer for biotin.
A schematic illustration of the principle of LFB for visualization of MCDA amplicons is shown in Figure 1C. The biosensor detects MCDA amplicons through specific recognition of the Dig labels at the end of products, which are produced by using Dig labeled primers (C1* primer). The amplicons labeled with biotin bind streptavidin-conjugated gold nanoparticles for visualization at the other end. The MCDA products are deposited on the sample application region of the biosensor, and then the running buffer is also deposited on the sample application area. The running buffer is used for rehydrating and releasing the dried detector reagents (SA-G) from the conjugate region to the detection zone by capillary migration. At the detection zone, the target amplicons are specifically captured by the immobilized anti-Dig at the TL, which provides a visual response to the presence/absence of the target amplicons. The detector reagents rapidly accumulate in the reaction zone of the strip through biotin/streptavidin interaction, resulting in a visual red colored band at the TL. The proper functioning of the strip is demonstrated by the CL formation, which contains biotin-BSA that captured excess detector reagent. Therefore, the appearance of two red lines in the readout region suggests a positive test and only one red line in the CL suggests a negative test (Figure 1D).
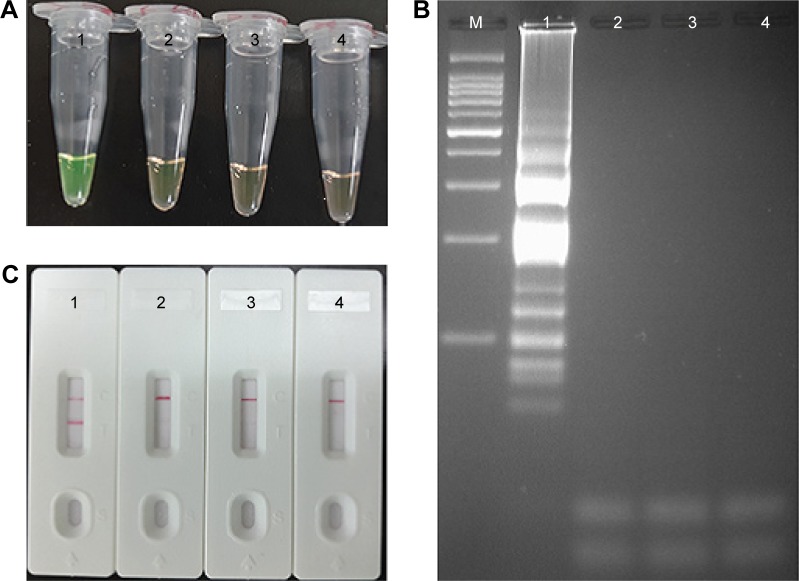
Validation and confirmation of L. monocytogenes-MCDA-LFB products
To validate the feasibility of L. monocytogenes-MCDA primers, the MCDA reactions were conducted in the presence or absence of genomic DNA templates within 1 h at a constant temperature (61°C). The positive L. monocytogenes-MCDA tube was visualized by naked eye as green, whereas the negative and blank controls remained light gray (Figure 4A). Agarose gel electrophoresis of the MCDA products exhibited the typical ladder-like patterns in the positive MCDA amplification but not in the negative and blank controls (Figure 4B). It was also displayed that two visible red bands (TL and CL) were observed in the positive amplification and only the CLs were seen in negative and blank controls (Figure 4C). These results indicated that the L. monocytogenes-MCDA primer set was a good candidate for development of the MCDA-LFB method for L. monocytogenes detection.
Figure 4.
Verification and detection of Listeria monocytogenes-MCDA products.
Notes: (A) Reaction products of L. monocytogenes-MCDA assay were visually detected by observation of the color change. (B) Agarose gel electrophoresis of L. monocytogenes-MCDA products was shown. (C) LFB applied for visual detection of L. monocytogenes-MCDA products. Tube 1/lane 1/strip 1, positive amplification of L. monocytogenes strain (EGD-e); tube 2/lane 2/strip 2, negative control of Listeria ivanovii strain (ATCCBAA-678); tube 3/lane 3/strip 3, negative control of Salmonella strain (ICDC-NPSa001); tube 4/lane 4/strip 4, blank control (DW). Lane M, DNA maker DL 100.
Abbreviations: DW, double distilled water; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
The temperature optimization of the MCDA-LFB assay during the reaction stage
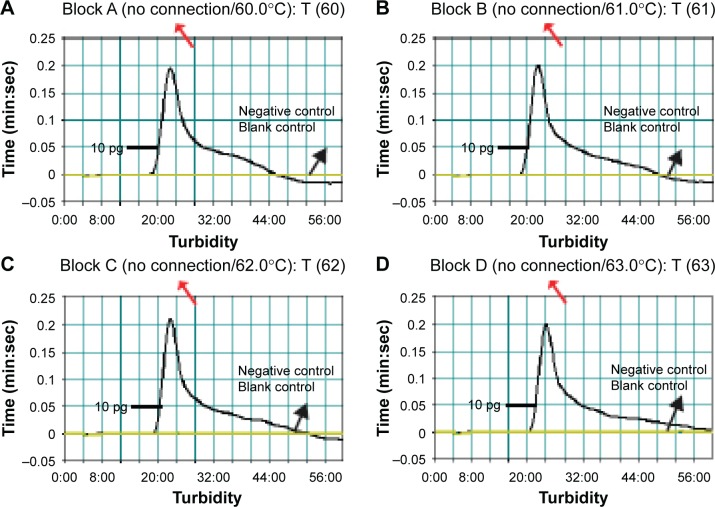
According to the standard MCDA conditions, the MCDA was carried out using the EGD-e DNA as the template at the level of 10 pg genomic templates per reaction to examine the optimal assay temperature during the amplification stage. Various amplification temperatures ranging from 60°C to 67°C at 1°C intervals were compared for optimal assay temperature. The real-time measurement of turbidity was used for analyzing the MCDA amplifications and the typical kinetics graphs corresponding to eight temperatures were produced. As shown in Figure 5, the optimal results were yielded from assay temperature of 60°C–63°C (Figure 5). The reaction temperature of 61°C was selected for the rest of MCDA-LFB tests conducted in this report.
Figure 5.
Optimal reaction temperature for Listeria monocytogenes-MCDA primer sets.
Notes: The standard MCDA reactions for detection of L. monocytogenes were monitored by real-time measurement of turbidity and the corresponding curves of concentrations of DNA were marked in the figures. The threshold value was 0.1 and the turbidity of >0.1 was considered to be positive. Eight kinetic graphs (A–H) were generated at various temperatures (60°C–67°C, 1°C intervals) with target pathogens DNA at the level of 10 pg per reaction. The graphs from (A) to (D) show robust amplification.
Abbreviation: MCDA, multiple cross displacement amplification.
Analytical sensitivity of MCDA-LFB technique in pure culture
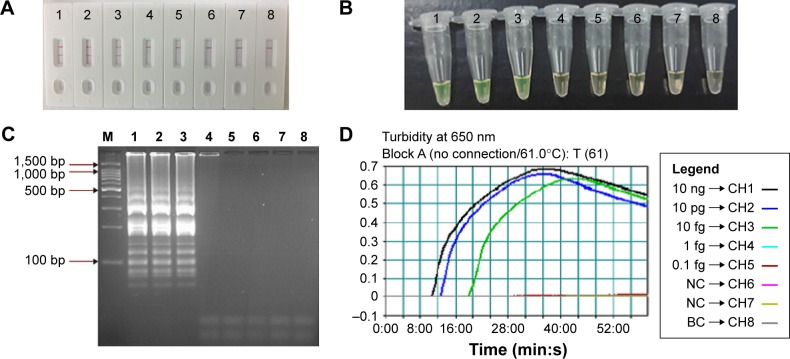
To test the LoD of L. monocytogenes-MCDA-LFB assay, the assay was evaluated using serial dilutions (from 10 ng to 0.1 fg per μL) of pure L. monocytogenes (EGD-e) DNA. As shown in Figure 6A, the MCDA products were analyzed by LFB detection, and the LoD of MCDA-LFB assay for detecting lmo0733 gene was 10 fg of genomic templates per reaction. As expected, the biosensor displayed clear visible red lines for both TL and CL when the products came from positive MCDA reactions, and only the CL were obtained from negative MCDA amplifications, negative control, and blank control.
Figure 6.
Analytical sensitivity of MCDA-LFB assay using serially diluted genomic DNA with Listeria monocytogenes strain EGD-e.
Notes: A total of 4 monitoring methods, including LFB (A), colorimetric indicator (FD) (B), gel electrophoresis (C), and real-time turbidity (D) were applied for analyzing the amplification products. The serial dilutions (10 ng, 10 pg, 10 fg, 1 fg, and 0.1 fg) of target templates were subjected to standard MCDA reactions. Strips (A)/tubes (B)/lanes (C)/turbidity signals (D) 1–8 represented the DNA levels of 10 ng, 10 pg, 10 fg, 1 fg, and 0.1 fg per reaction, negative control (10 pg of Listeria ivanovii genomic DNA), negative control (10 pg of Salmonella genomic DNA) and blank control (DW). The genomic DNA levels of 10 ng, 10 pg, and 10 fg per reaction produced the positive reactions.
Abbreviations: BC, blank control; DW, double distilled water; FD, fluorescent detection reagent; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
Using the MCDA amplification by self-trail, the LoD of the approaches was further confirmed by direct visual inspection of the MCDA products with FD reagent and electrophoresis in 2% agarose gels stained with ethidium bromide. The positive amplifications by FD reagent were indicated by a color change from light gray to green and the positive results by electrophoresis were observed the typical ladder-like patterns after 2% agarose gels electrophoresis when compared with negative amplification (negative reactions, negative control, and blank control; Figure 6B and C). Moreover, the MCDA reactions were analyzed by real-time turbidity detection and the decreasing concentrations of pure L. monocytogenes DNA were displayed from left to right. The analytical sensitivity of the two assays and turbidity analysis was also 10 fg per reaction (Figure 6D). These results indicated that the analytical sensitivity by FD reagent, real-time turbidity, and agarose gel electrophoresis detection for L. monocytogenes-MCDA amplifications was conformity with biosensor analysis.
The time optimization of the MCDA-LFB assay during the reaction stage
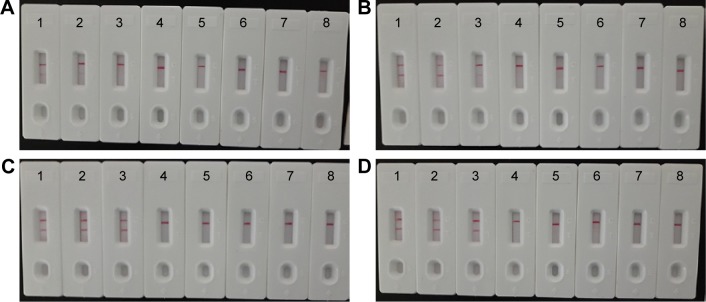
In this report, we then examined the optimal duration of time required for the MCDA-LFB assay during the reaction stage. Various reaction times ranging from 10 to 25 min at 5 min intervals were compared for optimal assay time according to the standard MCDA conditions (Figure 7A, 7B, 7C and 7D). The lowest genomic DNA level (10 fg of L. monocytogenes templates per tube) exhibited two red lines when the reaction only lasted for 20 min at 61°C (Figure 7C). A reaction time of 20 min was selected as the optimal time for the MCDA-LFB assay during the reaction stage (Figure 7C). Therefore, the whole procedure, including specimen (such as food sample) processing (35 min), isothermal reaction (20 min), and result reporting (5 min), could be finished within 60 min.
Figure 7.
The optimal duration of time required for MCDA-LFB assay.
Notes: Four different reaction times (A) 10 min, (B) 15 min, (C) 20 min, and (D) 25 min were tested and compared at 61°C. Strips 1–8 represent DNA levels of 10 ng of Listeria monocytogenes templates, 10 pg of L. monocytogenes templates, 10 fg of L. monocytogenes templates, 1 fg of L. monocytogenes templates, 0.1 fg L. monocytogenes templates per tube, negative control (Listeria ivanovii, 10 pg per reaciton), negative control (Salmonella, 10 pg per reaction), and blank control (DW). The best sensitivity was seen when the amplification lasted for 20 min (C).
Abbreviations: DW, double distilled water; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
The analytical specificity of MCDA-LFB assay
The analytical specificity of the MCDA-LFB assay targeting lmo0733 gene was evaluated with genomic DNA extracted from 50 L. monocytogenes strains to 45 non-L. monocytogenes strains (roughly 10 ng of genomic templates for each pathogen; Table 2). The positive results were seen in L. monocytogenes strains within a 20 min incubation period at 61°C. As shown in Figure 8, two red bands, including TL and CL, were produced in the LFB, and the sample was considered as positive. By contrast, the detection was negative for the non-L. monocytogenes strains and blank control after a 20 min incubation period. When only a band was observed (CL) in the LFB, the sample was defined as negative. The results demonstrated that the MCDA-LFB assay has a 100% analytical specificity for L. monocytogenes detection.
Figure 8.
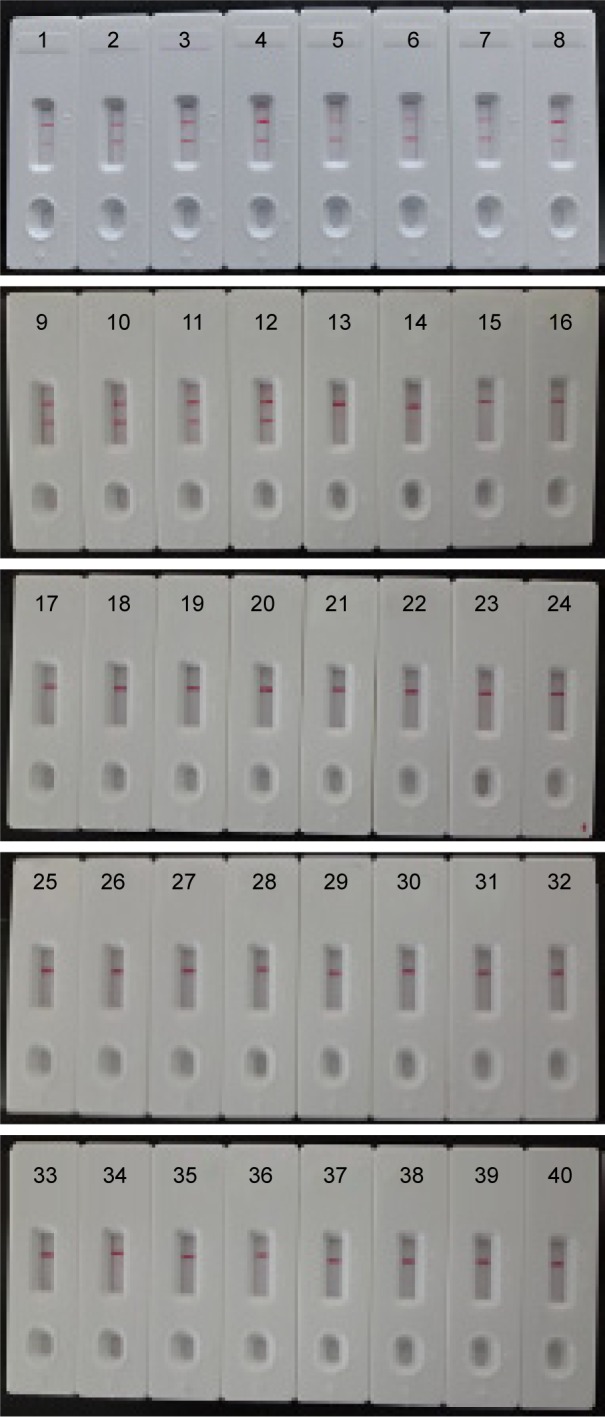
The specificity of MCDA-LFB assay for different strains.
Notes: The MCDA reactions were carried out using different genomic DNA templates and were monitored by means of visual format. Biosensors 1–12, Listeria monocytogenes strains of serovar 1/2a (ATCC51772), 3a (ATCC51782), 1/2c (Slcc2372), 3c (Slcc2479), 1/2b (Slcc2755), 3b (Slcc2540), 7 (NCTC10890), 4a (ATCC19114), 4c (ATCC19116), 4b (ATCC19115), 4d (ATCC19117), and 4e (ATCC19118); biosensor 13–17, Listeria ivanovii (ATCCBAA-678), Listeria innocua (ATCCBAA-680), Listeria grayi (ATCC25402), Listeria seeligeri (ATCC35897), and Listeria welshimeri (ATCC35897); biosensors 18–39, Bacillus cereus, Enteropathogenic Escherichia coli, Enterotoxigenic E. coli, Enteroaggregative E. coli, Enteroinvasive E. coli, Enterohemorrhagic E. coli, Plesiomonas shigelloides, Shigella boydii, Shigella sonneri, Shigella flexneri, Enterobacter cloacae, Enterococcus faecalis, Yersinia enterocolitica, Streptococcus pneumonia, Aeromonas hydrophil, Vibrio vulnificus, Vibrio fluvialis, Vibrio parahaemolyticus, Proteus vulgaris, Streptococcus bovis, Klebsiella pneumonia, and Salmonella enteric; and biosensor 40, blank control (DW).
Abbreviations: ATCC, American Type Culture Collection; DW, double distilled water; LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
Evaluation of the L. monocytogenes-MCDA-LFB assay using pork samples
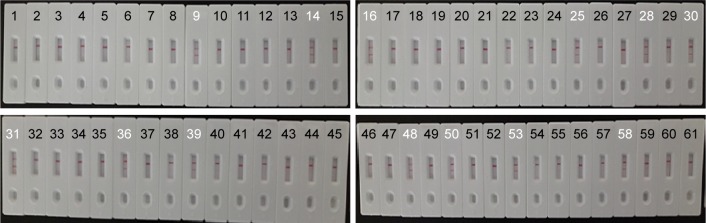
To evaluate the feasibility of L. monocytogenes-MCDA-LFB approach as a reliable surveillance tool for target pathogen in food samples, 61 pork samples were analyzed by the culture-biotechnical, PCR, and MCDA-LFB assays. The results were shown in Figure 9 and summarized in Table 3. In case of practical samples, 10 (16.4%) and 13 (21.3%) pork samples were L. monocytogenes positive by PCR and MCDA-LFB methods, respectively (Figure 9). The MCDA-LFB diagnostic accuracy obtained was 100% when compared to the conventional culture-based assay (21.3%; Table 3). These results suggested that the L. monocytogenes-MCDA-LFB method developed here has higher detection ability when compared to the traditional PCR approach.
Figure 9.
MCDA-LFB assay for the detection of Listeria monocytogenes in raw meat samples.
Notes: A total of 61 pork samples were analyzed using L. monocytogenes-MCDA-LFB assay, and 13 samples (samples 9, 14, 16, 25, 28, 30, 31, 36, 39, 48, 50, 53, and 58) were L. monocytogenes positive.
Abbreviations: LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification.
Table 3.
Comparison of culture-biotechnical, MCDA-LFB, conventional PCR, for the detection of Listeria monocytogenes in raw meat samples
| Detection methods | Pork samples (n=61)
|
|
|---|---|---|
| Positive | Negative | |
| Culture | 13 | 48 |
| MCDA-LFB | 13 | 48 |
| PCR | 10 | 51 |
Abbreviations: LFB, lateral flow biosensor; MCDA, multiple cross displacement amplification; PCR, polymerase chain reaction.
Discussion
L. monocytogenes, recognized as an important foodborne pathogen worldwide, is mainly transmitted through the food chain and accounts for listeriosis – a severe illness of human and animals with a high case fatality rate.17 The recent food recalls and well-publicized outbreaks due to this bacterium have increased the demand for more rapid, simple, specific, and sensitive techniques for detection of L. monocytogenes in various samples. Here, we proposed an approach using MCDA combined with gold nanoparticles-based LFB (MCDA-LFB) to offer a simple, rapid, highly sensitive, and accurate method for detection of the target pathogen. In the MCDA-LFB assay, gold nanoparticles were used as reporter particles, because they are stable in both dried and liquid forms and their colors do not fade after staining on membranes.18 Gold nanoparticles also display distinct physical and chemical attributes including high surface-to-volume ratio, unique optoelectronic properties (surface plasmon resonance), and good biocompatibility with DNA, RNA, and proteins, and thus offer a well-suited platform for colorimetric sensing of target analytes.19 Moreover, the proof-of-concept strategy (MCDA-LFB) may be reconfigured to detect a wide variety of nucleic acid sequences by redesigning the specific MCDA primers.
To develop specific assays for L. monocytogenes detection, a large number of approaches, including PCR- and loop-mediated isothermal amplification-based approaches, were established for species-specific identification of this bacterium, and the primer sequences were mainly derived from two gene sequences: iap and hlyA gene.3 However, the two target genes were not specific to L. monocytogenes, iap gene was present in all six Listeria species and hlyA gene for Listeria seeligeri, L. ivanovii, and L. monocytogenes.15 In this study, the MCDA primer sequences were based on the species-specific gene (lmo0733), which was unique to L. monocytogenes.15 The analytical specificity of L. monocytogenes-MCDA-LFB assay was successfully verified with the genomic DNA extracted from 50 L. monocytogenes to 45 non-L. monocytogenes strains, and the positive results were yielded in the assay of L. monocytogenes strains but not for non-L. monocytogenes strains and blank control (Figure 8). Thus, the proposed assay was highly selective to target pathogen over other bacteria and could be as a potential tool for screening L. monocytogenes with high specificity.
Besides specificity, the analytical sensitivity was also an important factor of the newly developed method in practical application. Under the standard conditions, the L. monocytogenes-MCDA-LFB assay proposed here was capable of detecting as little as 10 fg of L. monocytogenes genomic DNA per tube in pure culture (Figure 6). The analytical sensitivity was further confirmed using gel electrophoresis, FD reagent, and reaction-turbidity as indicators, and the biosensor was as sensitive as gel electrophoresis, FD reagent, and turbidity detection (Figure 6). Due to the elimination of the requirement for electrophoresis, special reagents, and detection apparatus, the MCDA-LFB method was sometimes easy to determine, thus it was more suitable than other MCDA-based techniques for rapid, simple, and sensitive detection in various fields.
When the MCDA-LFB technique was applied to 61 pork samples and compared with PCR and standard culture assays. MCDA-LFB and culture-based methods could adequately detect the bacterial loads in the practical samples, and displayed higher detection sensitivity for target pathogen compared to PCR assay (Table 3). Three samples were confirmed to be positive by MCDA-LFB and culture-based approaches, but negative by PCR detection. However, as depicted in Figure 3, conventional culture-based assay was time-consuming, complicated, and labor-intensive, and the MCDA-LFB method was simpler and took only about 30 min after DNA extraction. Consequently, the whole procedure, including template preparation (35 min), isothermal reaction (20 min), and result indicating (2 min), was finished within 60 min (Figure 3). By comparison among three assays, the MCDA-LFB methodology is not only effective and accurate, but also simpler and more rapid.
The test result of L. monocytogenes-MCDA-LFB provided direct visualization by naked eyes, which was very easy to be interpreted. The interpretation of test results was based on the appearance of red lines on the reaction zone. The presence of two red lines (TL and CL) on the reaction region indicated a positive result for L. monocytogenes, whereas only a red line (CL) appeared in the reaction zone, indicating the negative results, negative controls, and blank control (Figures 1C, 4 and 6–9). In previous reports, MCDA reactions were analyzed using real-time turbidity, gel electrophoresis, and colorimetric indicator (such as FD reagent).13 First, the real-time turbidity detection suffered from background interference, and the use of gel electrophoresis to analyze MCDA products increased the risk of product degradation and contamination. Second, the assessment of color change with naked eye was potentially subjective, thus there was the possibility that a sample was somewhat ambiguous to the unaided eye when the concentration of target sequences was low. More importantly, MCDA generated a complex mixture of various amplicons due to use of 10 primers, thus these monitoring techniques could not differentiate the nonspecific and specific products.20 In the current study, we purposefully modified the MCDA assay by using two primers (CP1/C1 or CP2/C2) labeled with digoxigenin and biotin. This allowed labeled MCDA amplicons to be specifically analyzed using the biosensor housed inside of a plastic, leak-proof device, without the need for a turbidimeter, an electrophoresis instrument, or colorimetric indicator.
Comparing with PCR-based assays, MCDA-LFB method does not require accurate and rapid temperature control during the reaction stage, obviating the need for a sophisticated instrument for thermal management, and operates between temperatures ranging from 60°C to 67°C without losing amplification efficiency. Only a simple water bath or heater that maintains a constant temperature is sufficient, and this permits simplification of apparatus and reduction of costs. A wide variety of portable user-friendly instruments adapted for MCDA reaction exist, the dry block heater (HDT-100C, HengAo, Tianjing, People’s Republic of China) being one example. The portable (18×22 cm), battery-powered, $600 USD device supports 96 MCDA reactions per assay. Currently, the MCDA amplification can be performed using the commercial isothermal amplification kits (such as NEB Warmstart kits and Eiken Loopamp kits), and a MCDA reaction costs ~$3.5 USD. The cost of LFB devised in this study is estimated to be $2 USD per test. Combined with the elimination of labor costs because of the requirements for trained personnel in a certified laboratory, the MCDA-LFB assay becomes more cost-effective. Therefore, we deem that a MCDA-LFB assay would cost ~$6 USD per disposable and $600 USD or less for a dedicated apparatus.
Conclusion
In this study, we reported a rapid, simple, and nearly instrument-free molecular technique, which incorporated MCDA coupled with gold nanoparticle-based LFB for visual, specific, and sensitive detection of nucleic acid sequence. The proof-of-concept strategy may be reconfigured to detect various target sequences by redesigning the specific MCDA primers. The MCDA-LFB technology devised here obviated the thermal cycle steps, and incubating at a constant temperature using a simple block heater or water bath was sufficient to replicate target sequences to detectable levels within a short time (20 min for the amplification stage). Moreover, the use of the newly devised biosensor provided an objective and easily interpretable readout of approach’s results, alleviating the use of special reagents and expensive equipment. As a proof of concept, the pathogen (L. monocytogenes), which is responsible for severe listeriosis in both humans and animals, was detected by MCDA-LFB to demonstrate the capability of target identification. Our data showed that L. monocytogenes-MCDA-LFB assay was advantageous on quick results, easy operation, modest equipment requirements, high specificity and sensitivity, and cost and energy efficiency, thus could achieve the clinical diagnosis, infection control, and food hygiene inspection. Hence, these advantages made the MCDA-LFB system possible for point-of-care testing, genetic testing in resource-poor settings, and rapid testing of environment samples and food products.
Acknowledgments
We acknowledge the financial supports of the grants (Mega Project of Research on the Prevention and Control of HIV/AIDS, Viral Hepatitis Infectious Diseases 2013ZX10004-101 to Changyun Ye) from the Ministry of Science and Technology, People’s Republic of China, and grant (2015SKLID507 to Changyun Ye) from State Key Laboratory of Infectious Disease Prevention and Control, China CDC.
Footnotes
Author contributions
YW and CY conceived and designed the experiments; YW, HL, YW, HL, and LL performed the experiments; YW and HL analyzed the data; YW, HL, YW, HL, LL, JX, and CY contributed reagents/materials/analysis tools; YW, JX, and CY wrote the paper. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
Yi Wang and Changyun Ye have filed for a patent from the State Intellectual Property Office of the People’s Republic of China, which covers the novel method and sequences included in this manuscript (application number: CN201610892015.1). The other authors report no conflicts of interest in this work.
References
- 1.Vivant AL, Garmyn D, Piveteau P. Listeria monocytogenes, a down-to-earth pathogen. Front Cell Infect Microbiol. 2013;3:87. doi: 10.3389/fcimb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Law JW, Ab Mutalib NS, Chan KG, Lee LH. An insight into the isolation, enumeration, and molecular detection of Listeria monocytogenes in food. Front Microbiol. 2015;6:1227. doi: 10.3389/fmicb.2015.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Välimaa AL, Tilsala-Timisjärvi A, Virtanen E. Rapid detection and identification methods for Listeria monocytogenes in the food chain – a review. Food Control. 2015;55:103–114. [Google Scholar]
- 4.Vazquez-Boland JA, Kuhn M, Berche P, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14(3):584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Wang Y, Ma A, et al. The novel multiple inner primers-loop-mediated isothermal amplification (MIP-LAMP) for rapid detection and differentiation of Listeria monocytogenes. Molecules. 2015;20(12):21515–21531. doi: 10.3390/molecules201219787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maertens de Noordhout C, Devleesschauwer B, Angulo FJ, et al. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14(11):1073–1082. doi: 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drevets DA, Bronze MS. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol Med Microbiol. 2008;53(2):151–165. doi: 10.1111/j.1574-695X.2008.00404.x. [DOI] [PubMed] [Google Scholar]
- 8.Warriner K, Namvar A. What is the hysteria with Listeria? Trends Food Sci Technol. 2009;20(6–7):245–254. [Google Scholar]
- 9.Joelsson AC, Brown AS, Puri A, et al. Comparative evaluation of veriflow(R) Listeria monocytogenes to USDA and AOAC culture based methods for the detection of Listeria monocytogenes in food. J AOAC Int. 2015;98(5):1325–1334. doi: 10.5740/jaoacint.15-122. [DOI] [PubMed] [Google Scholar]
- 10.Qi H, Zhong Z, Zhou H-X, et al. A rapid and highly sensitive protocol for the detection of Escherichia coli O157: H7 based on immunochromatography assay combined with the enrichment technique of immunomagnetic nanoparticles. Int J Nanomedicine. 2011;6:3033–3039. doi: 10.2147/IJN.S25684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma H, Mutharasan R. hlyA gene-based sensitive detection of Listeria monocytogenes using a novel cantilever sensor. Anal Chem. 2013;85(6):3222–3228. doi: 10.1021/ac303561c. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Zhi D, Zhao Y, et al. Lateral flow test strip based on colloidal selenium immunoassay for rapid detection of melamine in milk, milk powder, and animal feed. Int J Nanomedicine. 2014;9:1699–1707. doi: 10.2147/IJN.S58942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Wang Y, Ma AJ, et al. Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci Rep. 2015;5:11902. doi: 10.1038/srep11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Wang Y, Zhang L, et al. Multiplex, rapid, and sensitive isothermal detection of nucleic-acid sequence by endonuclease restriction-mediated real-time multiple cross displacement amplification. Front Microbiol. 2016;7:753. doi: 10.3389/fmicb.2016.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, Ainsworth AJ, Austin FW, Lawrence ML. Use of PCR primers derived from a putative transcriptional regulator gene for species-specific determination of Listeria monocytogenes. Int J Food Microbiol. 2004;91(3):297–304. doi: 10.1016/j.ijfoodmicro.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Wang Y, Ma A, Li D, Ye C. Rapid and sensitive detection of Listeria monocytogenes by cross-priming amplification of lmo0733 gene. FEMS Microbiol Lett. 2014;361(1):43–51. doi: 10.1111/1574-6968.12610. [DOI] [PubMed] [Google Scholar]
- 17.Milillo SR, Friedly EC, Saldivar JC, et al. A review of the ecology, genomics, and stress response of Listeria innocua and Listeria monocytogenes. Crit Rev Food Sci Nutr. 2012;52(8):712–725. doi: 10.1080/10408398.2010.507909. [DOI] [PubMed] [Google Scholar]
- 18.Patra JK, Baek KH. Novel green synthesis of gold nanoparticles using Citrullus lanatus rind and investigation of proteasome inhibitory activity, antibacterial, and antioxidant potential. Int J Nanomedicine. 2015;10:7253–7264. doi: 10.2147/IJN.S95483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howes PD, Rana S, Stevens MM. Plasmonic nanomaterials for biodiagnostics. Chem Soc Rev. 2014;43(11):3835–3853. doi: 10.1039/c3cs60346f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge Y, Wu B, Qi X, et al. Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device. PLoS One. 2013;8(8):e69941. doi: 10.1371/journal.pone.0069941. [DOI] [PMC free article] [PubMed] [Google Scholar]