Abstract
miR-223-3p is deregulated in several tumor types and plays an important role in tumorigenesis and progression. However, its role in the pathogenesis of testicular germ cell tumor (TGCT) remains uncharacterized. We previously demonstrated that miR-223-3p expression was increased in TGCTs compared with normal testes (NT), suggesting that miR-223-3p may have an oncogenic role in TGCT. Using published dataset and The Cancer Genome Atlas database, we validated higher miR-223-3p expression in TGCTs than NT, and found a negative correlation between miR-223-3p and FBXW7 mRNA expression levels. Using both gain- and loss-of-function experiments, we show that miR-223-3p regulates FBXW7 protein expression, cell growth and apoptosis in TGCT cell lines. Additionally, we demonstrate that ectopic expression of the full-length coding sequence of FBXW7 could rescue the cell growth and apoptotic effects mediated by miR-223-3p. Our findings suggest an oncogenic role for miR-223-3p in TGCT, which promotes cell growth and inhibits apoptosis through repression of FBXW7.
Keywords: miR-223-3p, cell growth, apoptosis, FBXW7, TGCT
Introduction
Testicular germ cell tumor (TGCT) is the most frequent solid malignancy occurring in males between the ages of 15 and 34 years (1), with a steadily rising incidence for the past few decades in the United States and Europe (2). Histopathologically, ~55% of all TGCTs are classified as seminomas, and the remaining cases as non-seminomas (3). The vast majority of TGCTs have an excellent cure rate with cisplatin-based treatment. Nevertheless, a subset of patients develops cisplatin resistance resulting in tumor progression and reduced survival (4). Therefore, a better understanding of the molecular mechanisms of TGCT tumorigenesis is needed for identification of new therapeutic targets and treatment development.
MicroRNAs (miRNAs) are small non-coding RNAs of ~20–24 nucleotides in length, which play important roles in a broad range of cellular processes, including tumor development and drug response (5). Genome-wide miRNA profiling studies have provided evidence of miRNA deregulations in TGCT. For example, the miR-371-373 cluster is frequently overexpressed in malignant TGCTs of all histopathological subtypes (6,7). Other miRNAs, such as the miR-302 cluster and miR-301, are differentially expressed based on the cellular differentiation of the tumor (7,8). To date, very few miRNAs have been functionally characterized in TGCT. miR-372 and miR-373 have been shown to play oncogenic roles in TGCT by targeting the tumor suppressor LATS2 (9). However, the functional roles of other differentially expressed miRNAs in TGCT have yet to be characterized.
We previously identified a subset of miRNAs that were differentially expressed between TGCTs and normal testes (NT) using a deep sequencing approach (10). Among these, miR-223-3p expression was higher in TGCTs as compared to NT. miR-223-3p is known to be deregulated in a broad range of hematological malignancies and solid tumors (11,12). However, its role in TGCT remains uncharacterized. miR-223-3p has been shown to regulate multiple targets in different cancer types. Among them, F-box/WD repeat-containing protein (FBXW7) is the most common target, which has been reported in acute T-cell lymphoblastic leukemia, esophageal squamous cell carcinoma and gastric cancer (13–15). FBXW7 is the substrate-recognition component of the SCF-(SKP1, CUL1, F-box protein)-ubiquitin-ligase complex, which has been demonstrated to function as a tumor suppressor by promoting the degradation of several oncoprotein substrates, including c-Myc, cyclin E, MCL-1, c-JUN, NFkB2 and Notch1 (16,17). Therefore, suppression of FBXW7 by miR-223-3p can promote tumor development and progression.
In this study, we investigated the expression and function of miR-223-3p and FBXW7 in TGCT clinical samples and cell lines. Our data show that miR-223-3p plays an oncogenic role in TGCT by promoting cell proliferation and inhibiting apoptosis via FBXW7.
Materials and methods
Clinical samples and cell lines
Fifteen frozen TGCTs and five NT were provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute, USA. All samples were included in our previous small RNA-sequencing study (10). The study was approved by the Stanford Human Subjects Review Committee.
Two established TGCT cell lines were included in this study: the TCam-2 seminoma cell line and the 2102Ep non-seminoma cell line (18,19). TCam-2 was kindly provided by Dr Leendert H.J. Looijenga (Department of Pathology, Erasmus MC-University Medical Center Rotterdam, The Netherlands) and 2102Ep by Dr Peter Andrews (Department of Biomedical Science, University of Sheffield, UK). TCam-2 cells were grown in RPMI-1640 and 2102Ep cells were cultured in DMEM medium, supplemented with 10% fetal bovine serum. All cells were cultured at 37°C with 5% CO2. Authentication of the cell lines was verified by short tandem repeat profiling in our recent study (10).
Data extraction and analysis from published data and The Cancer Genome Atlas database
For comparison of miR-223-3p expression between TGCTs and NT, we extracted global TaqMan miRNA profiling data from the study of Gillis et al (7), which analyzed 61 germ cell tumors, three NT and five embryonal carcinoma cell lines. We excluded the 10 dysgerminomas (ovarian germ cell tumors), one ovarian embryonal carcinoma, one ovarian york sac carcinoma and five cell lines, and re-analyzed the miR-223-3p expression by normalization to miR-16 in the 49 TGCTs and three NT.
For FBXW7 mRNA, we extracted the microarray gene expression data of 101 TCGTs and five NT from Gene Expression Omnibus (GEO accession no. GSE3218; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3218).
For analysis of correlation between miR-223-3p expression and FBXW7 mRNA levels, we extracted miR-223-3p and FBXW7 mRNA data from The Cancer Genome Atlas (TCGA) testicular cancer database using the UCSC Xena browser (http://xena.ucsc.edu/). These miR-223-3p and FBXW7 expression data had been generated by miRNA expression Illumina HiSeq and exon expression RNAseq, respectively.
RNA extraction
Total RNA was extracted using the mirVana miRNA isolation kit (AM1560; Ambion/Thermo Fisher Scientific, Waltham, MA, USA) and RNA concentration was measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). All RNA samples were stored at −80°C until further use.
TaqMan reverse transcription quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to evaluate the transfection efficiency of miR-223-3p overexpression or inhibition using the StepOnePlus Real-Time PCR system (Applied Biosystems/Thermo Fisher Scientific). cDNA was synthesized from 20 ng of total RNA and used to quantify miR-223-3p (ID 002295) and RNU48 (ID 001093). All reactions were performed in triplicate. The relative expression of miR-223-3p was normalized to RNU48, and the fold change of miR-223-3p in cells transfected with miR-223-3p mimic/inhibitor relative to their respective control was reported as 2−ΔΔCt.
Transfection
For miR-223 overexpression and inhibition, 2×105 cells were transfected with 30 nM of miRNA inhibitor (anti-miR-223, AM12301 or anti-miR negative control no. 1, AM17010; Ambion) or 10 nM of miRNA mimic (pre-miR-223, PM12301 or pre-miR negative control no. 1, AM17110; Ambion) using siPORT NeoFX transfection agent (AM4511; Ambion).
For co-transfection of miR-223 mimic and FBXW7-expressing plasmid, 1.5×105 cells were co-transfected with 500 ng of pCMV-Myc FBXW7 and 10 nM of pre-miR-223 or pre-miR-NC using Lipofectamine 2000 (no. 11668-019; Invitrogen/Thermo Fisher Scientific). Cells co-transfected with an empty vector and pre-miR-NC was used as a control. Cells were collected 48 h after transfection for subsequent analysis. The pCMV-Myc FBXW7 plasmid was obtained from Addgene (no. 16652; Cambridge, MA, USA; https://www.addgene.org/). The empty vector was prepared by cleavage of pCMV-Myc FBXW7 with BglII and NotI to remove the full-length coding sequence of FBXW7.
Annexin V cell apoptosis and EdU (5-ethynyl-2′-deoxyuridine) cell proliferation assays
Cell apoptosis and proliferation were evaluated in TCam-2 and 2102Ep cells 72 h after transfection using Annexin V FITC Apoptosis kit (PHN1018; Invitrogen) and Click-iT EdU Alexa Fluor 488 flow cytometry assay (C10425; Invitrogen), respectively. All experimental conditions were according to the manufacturer's instructions and analyzed by NovoCyte flow cytometer (ACEA Biosciences, San Diego, CA, USA). At least three independent experiments were performed in each cell line.
Trypan blue exclusion assay
Trypan blue exclusion assay was performed in TCam-2 and 2102Ep cells 48 or 72 h after transfection. Cells were stained with 0.4% trypan blue solution and counted by TC10™ Automated Cell Counter (Bio-Rad, Hercules, CA, USA).
WST-1 assay
Cell growth was measured by WST-1 colorimetric assay (no. 11644807001; Roche Diagnostics, Indianapolis, IN, USA) in TCam-2 and 2102Ep cells 72 h after transfection. Cells were plated into a 96-well plate at a concentration of 5×103/well in 100 μl culture medium. At different time intervals (0, 24, 48 or 72 h after transfection), 10 μl of WST-1 reagent was added to each well and incubated for 3 h at 37°C. After incubation, absorbance values were detected at the wavelengths 450 nm (measurement) and 650 nm (reference) using the VERSAmax ELISA Microplate Reader (Molecular Devices, Sunnyvale, CA, USA). Each experimental group was performed in six replicates for each time-point and all experiments were repeated three times independently.
Western blotting
Total protein lysates were extracted using NP-40 cell lysis buffer (FNN0021; Invitrogen), supplemented with 1 mM of phenylmethanesulfonyl fluoride (P7626; Sigma-Aldrich, St. Louis, MQ, USA) and protease inhibitor (P8340; Sigma-Aldrich). Protein concentrations were determined using the Pierce™ BCA Protein assay kit (no. 23227; Pierce Biotechnology, Thermo Fisher Scientific). Thirty micrograms of protein lysates were separated in NuPAGE Novex 4–12% Bis-Tris gels (NP0321BOX; Invitrogen) and transferred to 0.2 μm nitrocellulose membranes (no. 88024; Invitrogen). After blocking with 5% skim milk powder (no. 70166; Sigma-Aldrich) in Tris-buffered saline/0.05% Tween-20, membranes were incubated with anti-FBXW7 (NBP1-59631; Novus Biologicals, Littleton, CO, USA; 1:1,000 dilution), anti-cleaved PARP (ab32064; Abcam, Cambridge, UK; 1:1,000 dilution) or anti-Myc-Tag (no. 2276; Cell Signaling Technologies, Danvers, MA, USA; 1:500 dilution) overnight at 4°C. Anti-rabbit IgG-HRP (no. 170-6515; Bio-Rad Laboratories; 1:3,000 dilution) or anti-mouse IgG-HRP (sc-2005; Santa Cruz Biotechnology, Dallas, TX, USA, 1:10,000 dilution) was used as secondary antibodies. For normalization purpose, the membrane was incubated with anti-GAPDH (sc-47724; Santa Cruz Biotechnology; 1:1,000 dilution). Signals were detected using the Novex ECL HRP chemiluminescent substrate reagent (WP20005; Invitrogen) and LAS-1000 image analyzer (Fujifilm, Tokyo, Japan).
Statistical analyses
All statistical analyses were performed using MS Office Excel 2007 or SPSS 22.0 (IBM Corp., Armonk, NY, USA). Comparisons between TGCT and NT were performed by Mann-Whitney U test, and the transfection experiments were assessed by Student's paired t-test. Correlation between miR-223-3p and FBXW7 mRNA expression levels was evaluated using Pearson's correlation analysis. All statistical tests were two-sided and P-values <0.05 were considered as statistically significant.
Results
Expression of miR-223-3p and FBXW7 in TGCTs and NT
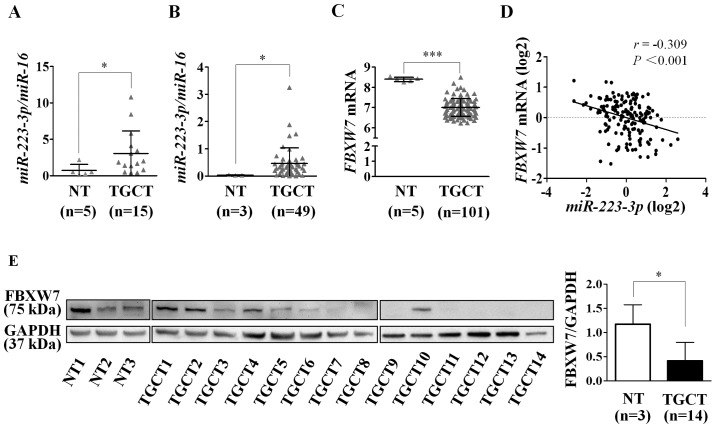
To validate our previous observation of miR-223-3p overexpression in TGCTs, we re-analyzed miR-223-3p expression from the miRNA profiling data of Gillis et al (7), with inclusion of 49 TGCTs and 3 NT. In agreement with our previous finding (10) (Fig. 1A), miR-223-3p was overexpressed in TGCTs compared to NT (P=0.011; Fig. 1B).
Figure 1.
Expression analysis of miR-223-3p and FBXW7 mRNA and protein in testicular germ cell tumors (TGCTs) and normal testes (NT). (A and B) The graphs depict miR-223-3p expression between NT and TGCTs in Özata et al (10) (A) and Gillis et al (7) (B). In both cohorts, miR-223-3p expression was quantified by RT-qPCR and miR-16 was used for normalization. (C) The graph shows the normalized expression data of FBXW7 mRNA in NT (n=5) and TGCTs (n=101), which were extracted from the microarray gene expression of GEO database accession no. GSE3218. (D) miR-223-3p expression and FBXW7 mRNA data were obtained from the TCGA database. Correlation was assessed using the Pearson's correlation analysis. (E) Western blot analysis of FBXW7 protein in NT (n=3) and TGCTs (n=14). GAPDH was used as a loading control. Data represent mean ± SD. All comparisons were evaluated using Mann-Whitney U test. *P<0.05; ***P<0.001.
To determine whether FBXW7 could be a candidate target of miR-223-3p in TGCT, we analyzed FBXW7 expression from the microarray gene expression data of 101 TGCTs and 5 NT in the Gene Expression Omnibus (GEO) database (accession no. GSE3218). Indeed, we found that FBXW7 mRNA expression was decreased in TGCTs as compared to NT (P<0.001; Fig. 1C). We further assessed the correlation between miR-223-3p and FBXW7 expression levels using miRNA and gene expression profiles from the TCGA testicular cancer datasets. The analysis revealed an inverse correlation (r=−0.309; P<0.001; Fig. 1D), supporting the miRNA-target relationship.
Additionally, we also quantified FBXW7 protein expression in 3 NT and 14 TGCT samples by western blot analysis. As shown in Fig. 1E, the expression of FBXW7 was low or undetectable in 10/14 TGCTs (71.4%), and moderate or high in the remaining four tumors. By contrast, all three NT showed moderate to high expression of FBXW7. Consistent with the mRNA expression pattern, the FBXW7 protein level in TGCTs was lower than in NT (P=0.023).
Effect of miR-223-3p modulation on FBXW7 in TGCT cell lines
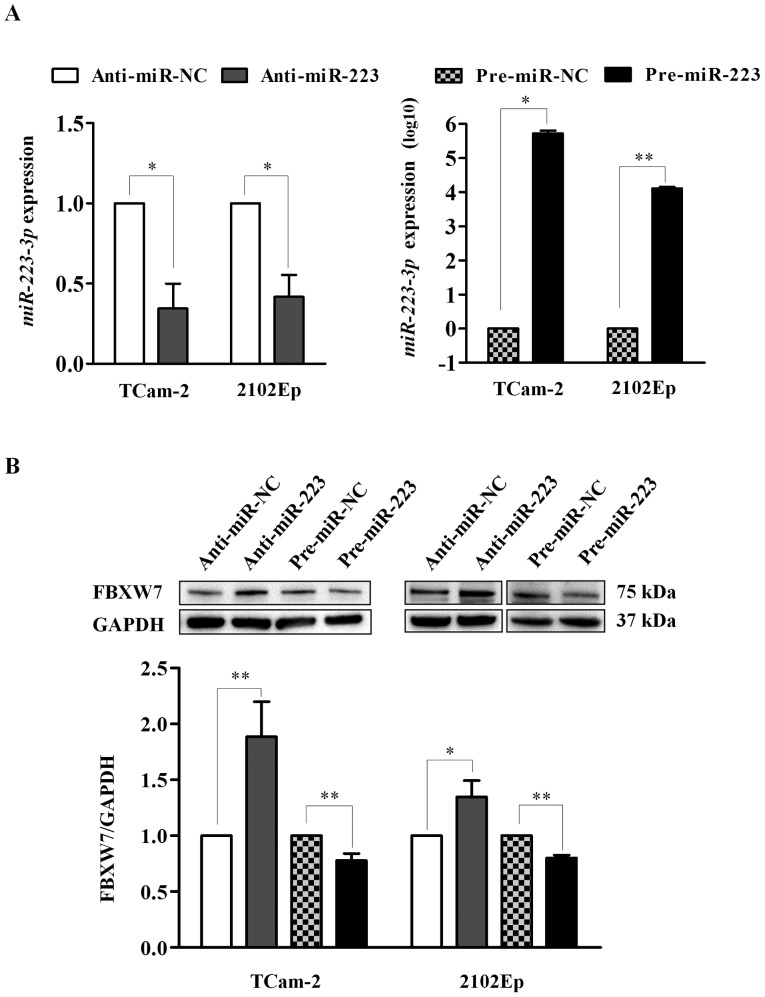
To further determine whether miR-223-3p could regulate FBXW7 in TGCT, we performed miR-223-3p overexpression and inhibition in two TGCT cell lines and evaluated the effect on FBXW7 protein expression using western blot analysis. As shown in Fig. 2A, cells transfected with anti-miR-223 showed significantly lower miR-223-3p expression than the anti-miR-NC-treated cells in both cell lines (P=0.018 for both). Similarly, miR-223-3p expression was significantly increased in cells transfected with pre-miR-223 relative to its negative control (TCam-2: P=0.014 and 2102Ep: P=0.003). The data support the efficiency of transfection.
Figure 2.
Effect of miR-223-3p regulation on FBXW7 protein expression in TGCT cells. (A) TCam-2 and 2102Ep cells were transfected with anti-miR-223 or pre-miR-NC and their respective negative controls. After 72 h of transfection, miR-223-3p expression was quantified by RT-qPCR and normalized to RNU48. (B) FBXW7 protein expression was measured in both TGCT cell lines transfected with anti-miR-223 or pre-miR-223 and controls using western blotting. GAPDH was used as a loading control. Data represent mean ± SD from at least three independent experiments. P-values were calculated by paired t-test. *P<0.05; **P<0.01.
Furthermore, inhibition of miR-223-3p led to a significant increase of FBXW7 expression in TCam-2 (1.9-fold; P=0.003) and 2102Ep (1.3-fold, P=0.015) cells. Similarly, overexpression of miR-223-3p significantly reduced FBXW7 expression in both cell lines (0.8-fold and P<0.01 for both) (Fig. 2B). The findings indicate that miR-223-3p suppresses FBXW7 expression in human TGCT cells.
Functional consequences of miR-223-3p regulation in TGCT cells
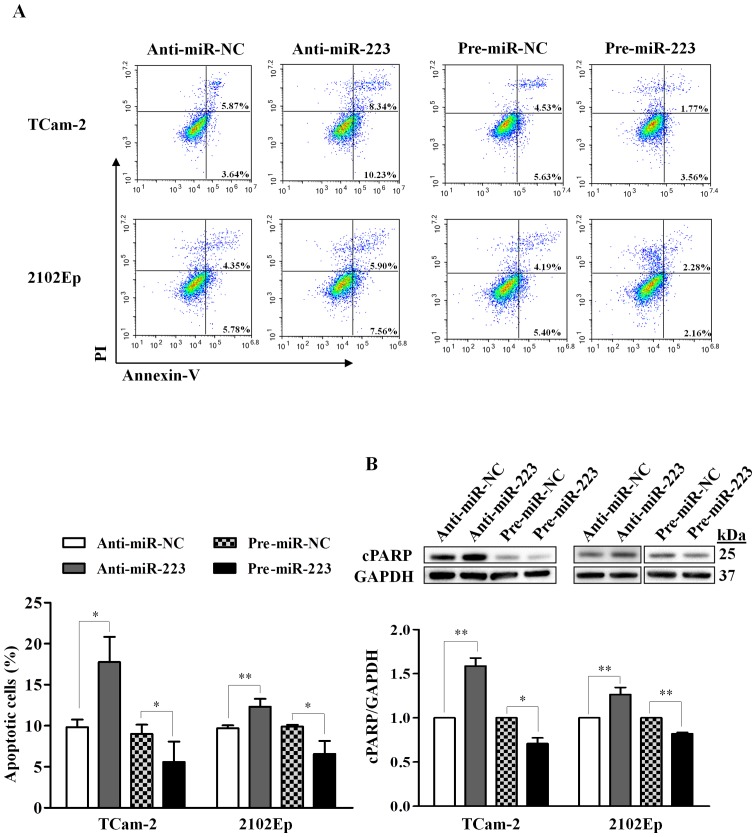
To explore the functional role of miR-223-3p on apoptosis, we investigated the effect using flow cytometric detection of Annexin V-stained cells as well as by western blot analysis of cleaved PARP (cPARP, 25 kDa), which is an apoptosis marker. For the Annexin V assay, we observed that inhibition of miR-223-3p in TCam-2 cells significantly increased apoptotic cells by 80% (P=0.018), while overexpression of miR-223-3p reduced apoptotic cells by 38% (P=0.038), relative to their respective negative controls (Fig. 3A). Similar effects were also observed in 2102Ep cells, however, the effect was less pronounced compared to TCam-2 cells (27% increase in the miR-223-3p inhibition, P=0.009; 34% decrease in the miR-223-3p overexpression, P=0.016; Fig. 3A).
Figure 3.
Modulation of cell apoptosis by miR-223-3p in TGCT cells. (A) Representative flow cytometric images of cells co-stained with Annexin V-FITC and propidium iodide (PI) upon silencing or overexpression of miR-223-3p. The early and late apoptotic cells are presented by Annexin V+/PI− (lower right) and Annexin V+/PI+ (upper right) cells, respectively. Quantification of total apoptotic cells (Annexin V+) is shown below. The percentage of apoptotic cells represents both early and late apoptotic cells. (B) Western blot analysis of the 25-kDa cleaved PARP (cPARP, an apoptosis marker) in both cell lines with miR-223-3p inhibition or overexpression. The relative expression of cPARP was normalized to GAPDH. Data represent mean ± SD from at least three independent experiments. P-values were calculated by paired t-test. *P<0.05; **P<0.01.
For the cPARP detection, silencing of miR-223-3p led to a significant increase of cPARP expression in both TCam-2 (1.6-fold, P=0.003) and 2102Ep (1.3-fold, P=0.007) cells, while overexpressing miR-223-3p resulted in a significant decrease of cPARP expression (TCam-2: 0.7-fold, P=0.016; 2102Ep: 0.8-fold, P=0.002) (Fig. 3B). These results indicate that miR-223-3p inhibits apoptosis in TGCT cells.
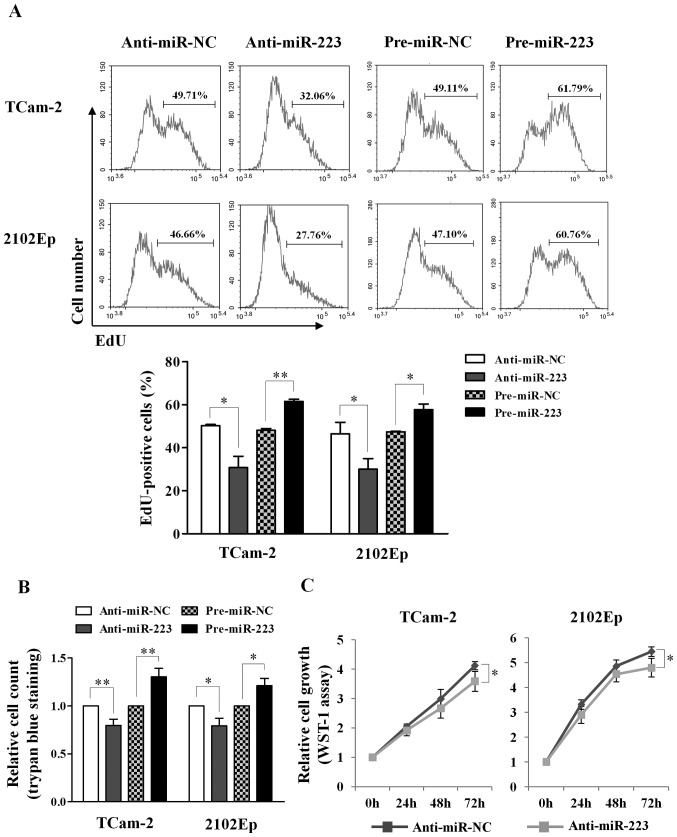
For cell proliferation, we applied three different assays: Click-iT EdU, WST-1 and trypan blue exclusion. Using the EdU assay, we observed reduction of EdU-positive cells upon silencing of miR-223-3p in both TCam-2 (50.3 vs. 30.8%, P=0.026) and 2102Ep cells (46.5 vs. 30.0%, P=0.045), and increase of EdU-positive cells upon overexpression of miR-223-3p (TCam-2: 48.1 vs. 61.6%, P=0.003; 2102Ep: 47.4 vs. 57.7%, P=0.027) (Fig. 4A). Similarly, the trypan blue exclusion assay revealed reduction of cell count upon silencing of miR-223-3p (TCam-2: 0.8-fold, P=0.004; 2102Ep: 0.8-fold, P=0.016) and increase of cell number upon overexpression of miR-223-3p (TCam-2: 1.3-fold, P=0.006; 2102Ep: 1.2-fold, P=0.011) (Fig. 4B). The WST-1 assay also showed that silencing of miR-223-3p reduced cell growth at 72-h post-transfection in both TCam-2 (P=0.043) and 2102Ep (P=0.041) cells (Fig. 4C). Taken together, the results support that miR-223-3p promotes cell proliferation in TGCT cell lines.
Figure 4.
Effect of miR-223-3p regulation on cell growth in TGCT cells. (A) Representative flow cytometric images of proliferating cells with EdU incorporation in anti-miR-223 or pre-miR-223 cells and their respective negative controls at 72 h post-transfection (upper). The graph shows the changes of EdU-positive cells in both cell lines upon silencing or overexpression of miR-223-3p from three independent experiments (lower). (B) Total live cells were counted using trypan blue dye exclusion assay (n=4). (C) Relative cell growth was examined at different time-points in both cell lines transfected with miR-223-3p inhibitor or negative control using WST-1 assay (n=3 for each time-point). Data represent mean ± SD. P-values were calculated by paired t-test. *P<0.05; **P<0.01.
miR-223-3p mediates regulation of cell growth and apoptosis through FBXW7 in TGCT
Given that FBXW7 expression is a well-characterized target of miR-223-3p, we tested whether ectopically expressed FBXW7 could rescue the miR-223-3p-mediated apoptotic and proliferative effects. We co-transfected TCam-2 cells with pre-miR-223 together with a plasmid expressing the entire open reading frame of FBXW7 without the miR-223-3p binding site (pCMV-Myc FBXW7) or a vector control. The effects on cell apoptosis and proliferation were determined using western blot analysis of cPARP and trypan blue exclusion assay, respectively. As shown in Fig. 5A, the endogenous FBXW7 was reduced in both cells co-transfected with pre-miR-223 and Myc-FBXW7 or vector control as compared with the negative control-transfected cells, indicating the suppression of endogenous FBXW7 by miR-223-3p overexpression.
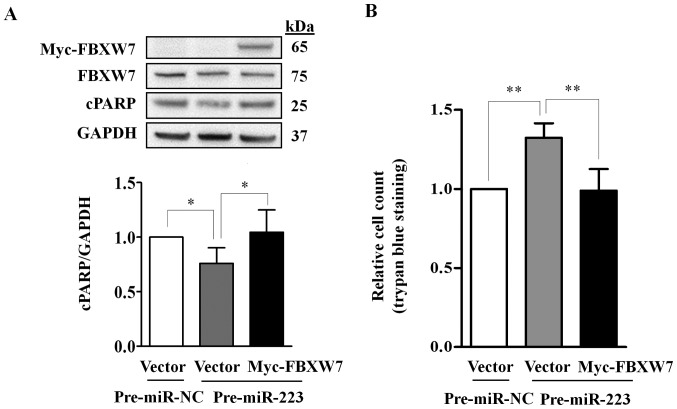
Figure 5.
Rescue of miR-223-3p mediated apoptosis and cell growth effects by ectopically expressed FBXW7. TCam-2 cells were co-transfected with miR-223-3p and Myc-FBXW7 or an empty vector, and compared to cells co-transfected with pre-miR-NC and empty vector. After 48 h of transfection, cells were harvested for apoptosis assay using western blot analysis of cPARP (A) and cell growth using trypan blue exclusion assays (B). (A) Exogenous Myc-FBXW7 and endogenous FBXW7 were detected by immunoblotting with anti-Myc-tag and anti-FBXW7, respectively. The relative cPARP expression was normalized to GAPDH. (B) Relative cell growth was determined by counting trypan blue negative cells. Data represent mean ± SD from four independent experiments. P-values were calculated by paired t-test. *P<0.05; **P<0.01.
Ectopic expression of miR-223-3p significantly reduced the abundance of cPARP (P=0.043) and increased the number of live cells (P=0.006) as compared to their respective controls; the effects were abolished by the ectopically expressed FBXW7 (Fig. 5). Together, our data indicate that miR-223-3p regulates cell growth and apoptosis in TGCT cells through FBXW7.
Discussion
miR-223-3p expression was found higher in TGCTs than NTs in our previous study (10), and here, we validated the findings in independent cohorts using previously published dataset (7). Deregulation of miR-223-3p has been observed in a variety of tumor types. Overexpression was found in T-cell acute lymphoblastic leukemia (11), oral (12), esophageal (14), gastric (20,21), bladder (22), and pancreatic (23) cancers, while its reduced expression has been reported in osteosarcoma (24), chronic lymphocytic leukemia (25), and intrahepatic cholangiocarcinoma (26). These findings indicate that miR-223-3p plays vital roles in a variety of tumor types, either as an oncogene or tumor suppressor depending on the cellular contexts. Consistent with its dual role, miR-223-3p has been shown to function as an oncogene in T-cell acute lymphoblastic leukemia, gastric and lung cancers (13,15,21,27,28), and as a tumor suppressor in cutaneous T-cell lymphoma and prostate cancer (29,30). Given its diverse function in different cancer types, we characterized the functional role of miR-223-3p in TGCT cells. miR-223-3p was shown to promote cell proliferation in TGCT cell lines in all three methods applied and which are based on different principles: the Click-iT EdU assay allows the detection of the thymidine analog EdU incorporated into cellular DNA during replication; the WST-1 assay is based on the metabolic activity of cells for conversion of the tetrazolium salt WST-1 into a colored dye, and the trypan blue exclusion assay provides direct counting of the number of live cells. Our findings support its oncogenic role in TGCT by promoting cell growth and inhibiting apoptosis in TGCT cell lines.
Additionally, miR-223-3p has been shown to modulate drug response in several cancer types (31–35). Importantly, miR-223-3p regulates cisplatin sensitivity in gastric and esophageal cancers (31,32). Given that most TGCTs are responsive to cisplatin treatment, it is intriguing to speculate that miR-223-3p may play an important role in cisplatin sensitivity in TGCT. Further investigations are warranted to evaluate the role of miR-223-3p in cisplatin response in TGCT.
FBXW7 has been demonstrated as a direct target of miR-223-3p using luciferase reporter assays (13,15). Here, we show that FBXW7 expression is lower in TGCTs than NT and inversely correlated with miR-223-3p, and miR-223-3p regulates FBXW7 protein expression using both gain- and loss-of-function studies. Most importantly, ectopic expression of the FBXW7 open reading frame can rescue the cell growth and apoptosis effects mediated by miR-223-3p. Together, our findings suggest that miR-223-3p regulates FBXW7 in TGCT and this regulatory pathway plays an important role in TGCT pathogenesis.
As afore-mentioned, FBXW7 is an E3 ubiquitin ligase that degrades several proto-oncogenes involved in cell growth, apoptosis, cell cycle regulation and differentiation (16). Therefore, the functional phenotypes observed in this study could due to the loss of FBXW7-mediated degradation of its substrates. Furthermore, numerous cancer-associated mutations in FBXW7 have been found in many cancer types (36), and loss of FBXW7 function can lead to chromosomal instability and tumorigenesis (37,38). These findings support the tumor suppressor function of FBXW7 in human cancers. Although nothing is known about its role in TGCT, FBXW7 is expressed specifically in the undifferentiated spermatogonia and suppresses cell proliferation of spermatogonial stem cell in mice (39). It is tempting to speculate that loss of FBXW7 expression could lead to uncontrolled cell growth in TGCT.
In conclusion, we report deregulation of miR-223-3p and FBXW7 in human TGCT. Our findings also reveal an oncogenic role of miR-223-3p through repression of the FBXW7 tumor suppressor, suggesting that this regulation is important for cell proliferation and apoptosis in TGCT. This study provides additional evidence of miRNA function in testicular germ cell tumorigenesis.
Acknowledgments
We thank Dr Leendert H.J. Looijenga and Dr Peter Andrews for TGCT cell lines; the Cooperative Human Tissue Network for frozen tissue samples; and the members of the sRNA Group for valuable discussions and suggestions. This study was supported by the Swedish Research Council (523-2009-3517 and 521-2010-3518), the Swedish Cancer Society, the Cancer Research Funds of Radiumhemmet, Karolinska Institutet and Stockholm County Council. J. Liu and H. Shi are the recipients of China Scholarship Council training grants.
References
- 1.Mannuel HD, Mitikiri N, Khan M, Hussain A. Testicular germ cell tumors: Biology and clinical update. Curr Opin Oncol. 2012;24:266–271. doi: 10.1097/CCO.0b013e32835167fc. [DOI] [PubMed] [Google Scholar]
- 2.Nigam M, Aschebrook-Kilfoy B, Shikanov S, Eggener S. Increasing incidence of testicular cancer in the United States and Europe between 1992 and 2009. World J Urol. 2015;33:623–631. doi: 10.1007/s00345-014-1361-y. [DOI] [PubMed] [Google Scholar]
- 3.McGlynn KA, Cook MB. Etiologic factors in testicular germ-cell tumors. Future Oncol. 2009;5:1389–1402. doi: 10.2217/fon.09.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koychev D, Oechsle K, Bokemeyer C, Honecker F. Treatment of patients with relapsed and/or cisplatin-refractory metastatic germ cell tumours: An update. Int J Androl. 2011;34:e266–e273. doi: 10.1111/j.1365-2605.2011.01145.x. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Palmer RD, Murray MJ, Saini HK, van Dongen S, Abreu-Goodger C, Muralidhar B, Pett MR, Thornton CM, Nicholson JC, Enright AJ, et al. Children's Cancer and Leukaemia Group Malignant germ cell tumors display common microRNA profiles resulting in global changes in expression of messenger RNA targets. Cancer Res. 2010;70:2911–2923. doi: 10.1158/0008-5472.CAN-09-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillis AJ, Stoop HJ, Hersmus R, Oosterhuis JW, Sun Y, Chen C, Guenther S, Sherlock J, Veltman I, Baeten J, et al. High-throughput microRNAome analysis in human germ cell tumours. J Pathol. 2007;213:319–328. doi: 10.1002/path.2230. [DOI] [PubMed] [Google Scholar]
- 8.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 10.Özata DM, Li X, Lee L, Liu J, Warsito D, Hajeri P, Hultman I, Fotouhi O, Marklund S, Ährlund-Richter L, et al. Loss of miR-514a-3p regulation of PEG3 activates the NF-kappa B pathway in human testicular germ cell tumors. Cell Death Dis. doi: 10.1038/cddis.2016.464. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiaretti S, Messina M, Tavolaro S, Zardo G, Elia L, Vitale A, Fatica A, Gorello P, Piciocchi A, Scappucci G, et al. Gene expression profiling identifies a subset of adult T-cell acute lymphoblastic leukemia with myeloid-like gene features and over-expression of miR-223. Haematologica. 2010;95:1114–1121. doi: 10.3324/haematol.2009.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manikandan M, Deva Magendhra Rao AK, Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R, Munirajan AK. Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer. 2016;15:28. doi: 10.1186/s12943-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, Khan AA, Setty M, Rondou P, Vandenberghe P, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43:673–678. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K, Baba H. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer. 2012;106:182–188. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Guo Y, Liang X, Sun M, Wang G, De W, Wu W. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol. 2012;138:763–774. doi: 10.1007/s00432-012-1154-x. [DOI] [PubMed] [Google Scholar]
- 16.Welcker M, Clurman BE. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget. 2014;5:2000–2015. doi: 10.18632/oncotarget.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno Y, Gotoh A, Kamidono S, Kitazawa S. Establishment and characterization of a new human testicular germ cell tumor cell line (TCam-2) Nippon Hinyokika Gakkai Zasshi. 1993;84:1211–1218. doi: 10.5980/jpnjurol1989.84.1211. In Japanese. [DOI] [PubMed] [Google Scholar]
- 19.Andrews PW, Goodfellow PN, Shevinsky LH, Bronson DL, Knowles BB. Cell-surface antigens of a clonal human embryonal carcinoma cell line: Morphological and antigenic differentiation in culture. Int J Cancer. 1982;29:523–531. doi: 10.1002/ijc.2910290507. [DOI] [PubMed] [Google Scholar]
- 20.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 22.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Yao Q, Hou Y, Xu M, Liu S, Yang L, Zhang L, Xu H. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed Pharmacother. 2013;67:381–386. doi: 10.1016/j.biopha.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 26.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52:297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 27.Mansour MR, Sanda T, Lawton LN, Li X, Kreslavsky T, Novina CD, Brand M, Gutierrez A, Kelliher MA, Jamieson CH, et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J Exp Med. 2013;210:1545–1557. doi: 10.1084/jem.20122516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang H, Yan X, Pan Y, Wang Y, Wang N, Li L, Liu Y, Chen X, Zhang CY, Gu H, et al. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol Cancer. 2015;14:58. doi: 10.1186/s12943-015-0327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGirt LY, Adams CM, Baerenwald DA, Zwerner JP, Zic JA, Eischen CM. miR-223 regulates cell growth and targets proto-oncogenes in mycosis fungoides/cutaneous T-cell lymphoma. J Invest Dermatol. 2014;134:1101–1107. doi: 10.1038/jid.2013.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurozumi A, Goto Y, Matsushita R, Fukumoto I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, et al. Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 2016;107:84–94. doi: 10.1111/cas.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streppel MM, Pai S, Campbell NR, Hu C, Yabuuchi S, Canto MI, Wang JS, Montgomery EA, Maitra A. MicroRNA 223 is upregulated in the multistep progression of Barrett's esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res. 2013;19:4067–4078. doi: 10.1158/1078-0432.CCR-13-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Jin W, Jia H, Yan J, Zhang G. MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J Exp Clin Cancer Res. 2015;34:28. doi: 10.1186/s13046-015-0145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eto K, Iwatsuki M, Watanabe M, Ishimoto T, Ida S, Imamura Y, Iwagami S, Baba Y, Sakamoto Y, Miyamoto Y, et al. The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. Int J Cancer. 2015;136:1537–1545. doi: 10.1002/ijc.29168. [DOI] [PubMed] [Google Scholar]
- 34.Kumar V, Palermo R, Talora C, Campese AF, Checquolo S, Bellavia D, Tottone L, Testa G, Miele E, Indraccolo S, et al. Notch and NF-κB signaling pathways regulate miR-223/FBXW7 axis in T-cell acute lymphoblastic leukemia. Leukemia. 2014;28:2324–2335. doi: 10.1038/leu.2014.133. [DOI] [PubMed] [Google Scholar]
- 35.Li R, Wu S, Chen X, Xu H, Teng P, Li W. miR-223/FBW7 axis regulates doxorubicin sensitivity through epithelial mesenchymal transition in non-small cell lung cancer. Am J Transl Res. 2016;8:2512–2524. [PMC free article] [PubMed] [Google Scholar]
- 36.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 38.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 39.Kanatsu-Shinohara M, Onoyama I, Nakayama KI, Shinohara T. Skp1-Cullin-F-box (SCF)-type ubiquitin ligase FBXW7 negatively regulates spermatogonial stem cell self-renewal. Proc Natl Acad Sci USA. 2014;111:8826–8831. doi: 10.1073/pnas.1401837111. [DOI] [PMC free article] [PubMed] [Google Scholar]