Abstract
Suppressor of Fused (SuFu), one of the most conserved components of the Hedgehog (Hh) signaling, binds Gli transcription factors and impedes activation of target gene expression in mammalian cells. Despite the central importance of SuFu in the Hh pathway, little is known about SuFu regulation. In a previous study, we identified NIMA-related expressed kinase 2A (Nek2A) as a SuFu-interacting protein. Here, we show that Nek2A stabilizes SuFu through impairing ubiquitin/proteasome degradation of SuFu. In addition, Nek2A negatively regulates target genes of Hh signaling as well as Gli2 transcriptional activity. In turn, inhibition of Hh signaling by GANT61 diminishes mRNA and protein levels of Nek2A, and Hh agonist promotes transcription of NEK2A gene. Chromatin immunoprecipitation assays revealed that Gli1 and Gli2 directly bind to the promoter regions of NEK2A gene and induced its transcription. Thus, we uncovered one of the mechanisms by which Nek2A acts as a modulator of the Hh signaling pathway in the context of a novel negative-feedback loop, which may offer new insights into Gli-mediated Hh signaling regulation in development and human diseases.
Keywords: Nek2A, SuFu, Gli, Hedgehog signaling, feedback loop
Introduction
Suppressor of Fused (SuFu) is a key regulator of Hedgehog (Hh) signaling and Gli activities. Ablation of SuFu in mouse embryos leads to severe neural tube defects and embryonic lethality (1,2). It is widely speculated that SuFu inhibits Gli-mediated Hh pathway through sequestering Gli2 protein in the cytoplasm and promoting proteolytic degradation of full-length (FL) Gli3 (3–5). SuFu also traverses into the nucleus to repress transcription by recruiting the SAP18-mSin3 complex to Gli-binding regions (6). However, recent study showed that SuFu can also play a positive role in the maximal activation of Hh signaling likely through protecting FL Gli2 and Gli3 proteins from SPOP-mediated ubiquitination and degradation (7–9). Other studies delineate the role of SuFu in tumorigenesis and implicate SUFU as a tumor suppressor. Taylor et al found that germline mutations or deletions of SUFU lead to medulloblastoma in a subset of children (10). SUFU is also known to be a rare cause of Gorlin syndrome, of which the patients always harbor mutations in PTCH1 (11). Furthermore, deletion of SUFU has been identified in other human tumors, including rhabdomyosarcoma (12), basal cell carcinoma (13) and prostate cancer (14), which further support SUFU as a tumor suppressor gene. Despite the central and conserved roles of SuFu in Hh signaling pathway and tumor, little is known about its regulation. Limited studies showed that SuFu undergoes ubiquitin-proteasomal degradation in response to Shh signaling in freshly isolated mouse embryonic fibroblasts and in embryonic tissues (15). In addition, the recombinant human SuFu was found to be phosphorylated and stabilized by purified PKA (16).
In our previous study (17), we employed a yeast two-hybrid approach to identify human SuFu interacting proteins. We found NIMA (never in mitosis A)-related expressed kinase 2A (Nek2A) as one of the SuFu-interacting proteins. Nek2A belongs to the Nek family of serine/threonine kinases, and is expressed in vertebrates as two main splice variants, Nek2A and Nek2B. The C terminus of Nek2A, but not Nek2B, contains the binding site for protein phosphatase 1 and motifs targeting the protein for ubiquitin-mediated degradation after mitotic entry. As a cell cycle-regulated kinase, Nek2A localizes to centrosomes and exhibits increased activity in S and G2 phases (18). During mitosis, Nek2 contributes to spindle pole formation through phosphorylation of centriolar cohesion proteins, including C-Nap1, rootletin, and Nep which is required for microtubule anchoring and spindle assembly (19–23).
Nek2 has emerged as an important oncogene due to its regulatory role in mitosis and cancer-related signaling pathways. Increased Nek2 expression has been tied to serosal invasion, lymphatic invasion, peritoneal dissemination and poor prognosis of colorectal cancer (24), for which the reason may be that Nek2 was associated with beta-catenin relocalization from membrane to cytoplasma and nucleus (25). In breast cancer studies, high Nek2 expression correlates with poor prognosis, and in various human breast cancer cell lines, Nek2 knockdown induces aneuploidy and cell cycle arrest that leads to cell death (26). In addition, analysis of the gene expression profiles of breast cancer samples revealed that co-elevated levels of Hec1 and Nek2 correlate with the shortest survival (27). Moreover in ovarian cancer, Nek2 mRNA expression is upregulated, especially in drug-resistant cells. The bioinformatic analysis revealed that Nek2 may directly or indirectly interact with a number of genes, proteins, microRNAs associated with drug resistance in ovarian and other types of cancer (28). Aberrant Nek2 expression has also been found in other cancers, such as non-small cell lung cancer and malignant peripheral nerve sheath tumor (29,30).
Given the importance of SuFu and Nek2A in tumorigenesis, we further dissected the functional relationship of these two proteins. We show here that Nek2A impairs ubiquitin/proteasome-mediated SuFu degradation, thus negatively modulates Hh transduction. Interestingly, in response to SuFu stabilization, Hh adjusts the transcription and expression of Nek2A in mammalian cells. Therefore, Nek2A functions as part of a negative feedback loop that modulates Hh activity, which may provide new insights into a dynamic process of Hh/Gli signaling regulation driven by feedback adaptation mechanisms.
Materials and methods
Reagents, antibodies and small molecular inhibitors
Lubrol-PX and other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA). Lipofectamine 2000 was obtained from Invitrogen Life Technologies (Carlsbad, CA, USA). The sources for monoclonal antibodies are as follows: Sigma-Aldrich (anti-Flag M2, F3165; anti-c-Myc, M4439), Abcam (anti-Pathed/PTCH1, ab55629; anti-SuFu, ab52913), Santa Cruz Biotechnology (normal rabbit IgG, sc-2027; anti-Gli2, sc-271786), BD Pharmingen (anti-Nek2 MAB, 610594), Covance (anti-ubiquitin P4G7, MMS-258R), and Millipore (anti-GAPDH, MAB374). Polyclonal antibodies were purchased from Abcam (anti-SuFu, ab28083; anti-Smo, ab38686; anti-Gli1, ab92611). The working concentrations for small molecular inhibitors and chemicals are as follows: GANT61 (20 mM, G9048; Sigma-Aldrich), MG-132 (25 mM, S2619; Selleck), purmorphamine (20 mM, S3042; Selleck), CHX (100 mg/ml, C7689; Solarbio). DMSO (0231; Amresco) was used as the solvent for the inhibitors and the vehicle control.
Plasmids
For mammalian cell expression, human cDNAs of SuFu (NM_016169) from human fetal brain cDNA library (Takara, Otsu, Japan) were amplified by PCR and cloned into mammalian expression vectors tagged with Flag. C-terminal Myc/his tagged human cDNA of Nek2A (NM_002497) was generated by PCR using the same strategy as described above and subcloned in pcDNA3.1. The miRNAi expression vectors that suppress Nek2A expression were generated by the BLOCK-iT™ Pol II miR-RNAi Expression system (Invitrogen). The oligonucleotide sequences for miRNAi constructs miR-Nek2A-80, -202 and -603 were: 5′-GGA AGA GTG ATG GCA AGA TAT-3′; 5′-GTT CGT TAC TAT GAT CGG ATT-3′; 5′-ATT GGG CTG CTT GCT GTA TGA-3′. The authenticity of the constructs was verified by DNA sequencing.
Cell culture, transfection and Bcl-2 reporter assay
HEK293T, H4 (American Type Culture Collection, Manassas, VA, USA) and CaES-17 (Shanghai Institute of Cell Biology, Shanghai, China) cells were cultured in DMEM, supplemented with 10% fetal calf serum, penicillin (100 U/ml) and streptomycin (100 mg/ml), and maintained at 37°C in an atmosphere of 5% CO2 and 95% humidity. Transient transfection of cells was performed with the standard calcium phosphate technique or with Lipofectamine 2000 according to instructions of the manufacturer. For Bcl-2 luciferase reporter assay, HEK293T cells were seeded in triplicate in 12-well plates and allowed to settle for 12 h. Three hundred nanograms of Bcl-2 luciferase reporter plasmid and the internal control plasmid pRL-TK (10 ng/well) were co-transfected into HEK293T cells. Plasmids for the expression of Gli1 and Gli2 (0.7 mg/well) were introduced to exaggerate the luciferase signal. The luciferase and Renilla signals were measured 48 h after transfection using Dual Luciferase Reporter assay kit (Promega, Madison, WI, USA). Each reporter gene assay was performed in triplicate.
Ubiquitination assay
To assay the effects of Nek2A on the ubiquitination of SuFu, Flag-tagged SuFu and Myc-tagged Nek2A were co-transfected into HEK293T cells. After 48 h of transfection, cells were treated with MG-132 for indicated times (0, 4 and 8 h) before they were lysed in Co-IP buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 50 mM NaF, 2 mM EDTA, 10% glycerol, 0.5% NP-40). Then immuno-precipitation was carried out with anti-Flag M2 Magnetic Beads (M8823; Sigma-Aldrich), and the immunoprecipitated proteins were subjected to 8% SDS-PAGE, followed by western blot analysis.
Western blotting
Cells were harvested 24-48 h after transfection and subjected to western blot analysis as described previously (31). Cells were washed with cold PBS and then suspended in protein lysis buffer at 4°C for 30 min and centrifuged (12,000 rpm, 15 min at 4°C) to maintain the supernatant. Protein concentrations were determined using Pierce™ BCA Protein assay kit (Thermo Scientific™, Rockford, IL, USA) according to the standard protocol of the manufacturer. Proteins were subsequently resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Bio-Rad), and then probed with the specified primary antibody followed by appropriate secondary antibody. The immunostaining was visualized by using Kodak X-ray film, which were subsequently scanned with an Epson 1680 scanner. Quantitative analysis was performed on scanned images of blots using ImageJ software (NIH Image analysis website http://rsb.info.nih.gov/ij/)
N-Shh conditioned medium
HEK293T cells were transfected with 10 mg of the Flag-N-Shh plasmid per 10-cm cell culture dish. Twelve hours after addition of the plasmids, the media were replaced with 10 ml of DMEM with 2% fetal calf serum per dish. The culture media were harvested 24 h after the medium change. Then, cells were incubated for an additional 24 h with 10 ml of DMEM containing 2% fetal calf serum. After that, the media were harvested and combined with the previously collected media. The combined medium was termed N-Shh conditioned medium and stored at 4°C until used.
Quantitative real-time PCR analysis
Total RNA of HEK293T cells was harvested using TRIzol® reagent (Life Technologies™) and evaluated by quantitative real-time PCR. Briefly, 1 mg of total RNA was employed to prepare cDNA via reverse transcription using a PrimeScript® RT reagent kit with gDNA Eraser (DRR047A; Takara). Quantitative real-time PCR was performed using SYBR® Premix Ex Taq™ II (Tli RnaseH Plus) (DRR820A; Takara) in an ABI StepOnePlus™ Real-Time PCR system (Applied Biosystem, Inc.). GAPDH was used as an internal control to normalize the variability in expression levels. The primers are shown in Table I.
Table I.
The real-time PCR primers.
| PCR primer | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| NEK2A | TCTTAATCTTCCATCCTCAG | CTATACAGAAAGGCATGGCT |
| SUFU | CCCAAGCTTGATGGCGGAGCTGCGGCCTAG | CGCGGATCCCTAGTGTAGCGGACTGTCGAAC |
| GLI1 | TCCTACCAGAGTCCCAAGTT | CCCTATGTGAAGCCCTATTT |
| GLI2 | CCTGGCATGACTACCACTATGAG | GGCTTGGCTGGCATGTTG |
| GAPDH | CAGGGCTGCTTTTAACTCTGGT | GATTTTGGAGGGATCTCGCT |
In silico promoter analysis and chromatin immunoprecipitation (ChIP) assay
Identification of transcription factor binding sites in the NEK2A promoter was performed using the MatInspector module of the Genomatix database, together with Matrix Family Library version 8.3 (32). ChIP assay was performed using the EZ-ChIP kit (Upstate Biotechnology) according to the manufacturer's manual. Briefly, cells were cross-linked in 1% formaldehyde, and DNA was sonicated into a range of 200–1000 base pairs in size using a Bioruptor Sonicator (Diagenode) for five cycles of 3 sec on/3 sec off. The extracts were pre-cleared in BSA-blocked protein A/G beads and incubated with antibodies or IgG control overnight. After washes, DNA was eluted and reverse-cross-linked overnight at 65°C, and then purified and amplified by PCR. The primers for PCR are shown in Table II.
Table II.
The PCR primers in ChIP assay.
| Promotor region | Sequence | Forward (5′-3′) | Reverse (5′-3′) | Length (bp) |
|---|---|---|---|---|
| −3062 to −3054 | AGCCACCCA | GTCTGTTGAGCAGGCTGGAGT | TGTGGTGGCACGTCTGTAGTC | 124 |
| −2602 to −2594 | GCCCACTCA | CAGCCTCCTGTGCTACTCTTT | CCAACCCACTCCCTTATCCA | 560 |
| −1880 to −1872 | GACCTCCCA | CTCCTGACCTCGTAATCCACC | AGGCTGCTGCCAGATGCTAC | 213 |
| −449 to −441 | GCCCACCCG | ACTCCTGGGCTCAAGCGACC | GAGCTGAATACAAATTAGAAATACAGAC | 114 |
Statistical analysis
All data were expressed as mean ± SD for experiments performed at least three times. The difference between 2 groups was analyzed using Student's t-test or one-way ANOVA. Significance was defined at P<0.05.
Results
Nek2A prevents SuFu from proteasome-dependent degradation
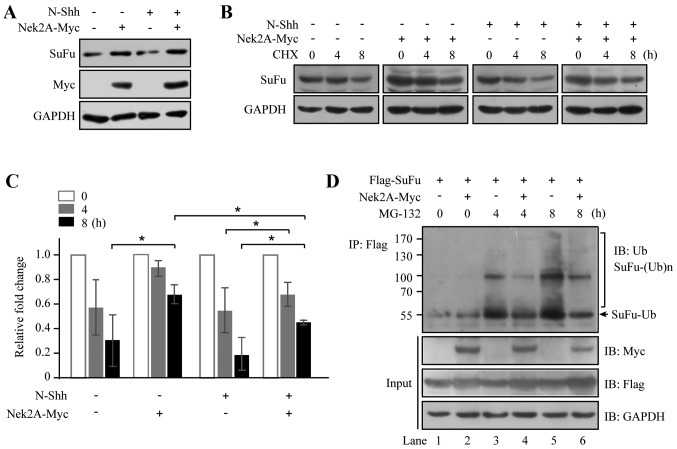
SuFu was shown to be degraded rapidly in some cancer cells (15). In a previous study, we demonstrated that Nek2A is an interacting protein of SuFu and Nek2A stabilizes SuFu (17). Here we further investigated the detailed mechanism of SuFu stabilization induced by Nek2A. We transfected HEK293T cells with Nek2A and then stimulated with N-Shh conditioned medium. As shown in Fig. 1A, SuFu abundance decreased upon the treatment of N-Shh, consistent with a previous study (15). However, Nek2A reversed the effect of N-Shh on SuFu degradation, suggesting that Nek2A stabilizes SuFu. Furthermore, half-life of SuFu was evaluated in the presence of CHX, which inhibits protein synthesis (Fig. 1B). SuFu turnover was attenuated in Nek2A-transfected cells; however, when N-Shh was added, SuFu rapidly degraded and Nek2A reversed the stimulatory effect of N-Shh on SuFu turnover (Fig. 1C), which further supporting that Nek2A may stabilize SuFu. To delineate the mechanism for the interaction of these two proteins, we treated HEK293T cells with Nek2A alone or together with MG-132, an inhibitor of 26S proteasome, for 4 and 8 h. Lysates were immunoprecipitated with Flag-tagged SuFu and the precipitants were probed for ubiquitin. As shown in Fig. 1D, lanes 1 and 2 exhibited basal ubiquitination profiles of SuFu without or with Nek2A respectively, and the polyubiquitinated proteins are hardly detected. Pretreatment of MG-132 in the absence of Nek2A (lanes 3 and 5) resulted in significant accumulation of ubiquitinated SuFu. However, Nek2A treatment diminished SuFu ubiquitination (lanes 4 and 6) in comparison with the control. These results indicate a role of Nek2A in SuFu regulation through ubiquitin/proteasome pathway.
Figure 1.
Nek2A impairs ubiquitin/proteasome-mediated degradation of SuFu protein. (A) Nek2A inhibits the degradation of SuFu induced by Shh. HEK293T cells were transfected with Nek2A-Myc for 24 h and stimulated with N-Shh conditioned medium for additional 12 h. SuFu protein levels in total cell lysates were determined by western blot analysis. The levels of Myc and GAPDH were used as loading controls. (B) Nek2A affects the degradation kinetics of SuFu. HEK293T cells were transfected with Nek2A in the presence or absence of N-Shh conditioned medium following CHX (100 mg/ml) treatment for different time spans. (C) Protein expression in (B) was quantified by ImageJ and normalized to GAPDH. Error bars represent the standard deviation of three independent experiments. *P<0.05 compared with the control groups. (D) Nek2A impedes SuFu ubiquitination. HEK293T cells transfected with Nek2A-Myc and Flag-SuFu for 48 h were treated with MG-132 to enrich ubiquitinated proteins, and subsequently subjected to co-immunoprecipitation assay. Precipitated proteins were probed using anti-Ub antibody.
Suppression of Nek2A activates Hh signaling pathway
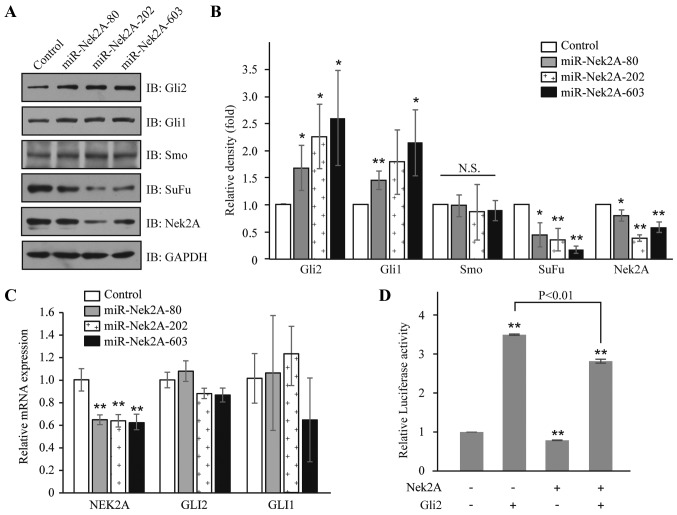
Considering the critical repressive role of SuFu in Hh signaling, we reasoned that Nek2A may regulate activation of Hh pathway through stabilizing SuFu protein. Therefore, we generated Nek2A miRNAi constructs that inhibit Nek2A expression (17). In HEK293T cells, repression of Nek2A by miR-Nek2A-80, -202 or -603 resulted in elevated protein levels of Gli2 and its target Gli1, but no obvious change of Smo, which is a member of the frizzled family of seven-pass transmembrane receptors (33) (Figs. 2A and B). Interestingly, upon the introduction of miR-Nek2A, the mRNA levels of Gli2 and Gli1 had not been affected (Fig. 2C), indicating that Nek2A acts as a modulator for SuFu at post-transcriptional level. Furthermore, to assess the effects of Nek2A on the transcriptional activity of Hh, we transfected Nek2A in HEK293T cells and found that in cells overexpressing Nek2A, the transcription of Bcl-2 (a target of Hh signaling), was significantly decreased compared to control cells with or without Gli2 expression (Fig. 2D). These results suggest that Nek2A negatively regulates Hh signaling.
Figure 2.
Nek2A attenuates transcriptional activity of Gli2. (A) Nek2A regulates Hh components. HEK293T cells were transfected with miR-Nek2A-80, -202 and -603 for 48 h. The components of Hh signaling were detected by western blotting with indicated antibodies. GAPDH serves as a loading control. (B) Quantitation of the data in (A) by ImageJ. GAPDH level is used for data normalization. Error bars represent the standard deviation of three independent experiments. *P<0.05 and **P<0.01 compared with the control groups. (C) mRNA levels of Gli1 and Gli2 in miR-Nek2A treated cells. Total mRNA was extracted and quantified by real-time PCR. Error bars represent the standard deviation of three independent experiments. (D) Overexpression of Nek2A attenuates transfection activity of Gli2. HEK293T cells were co-transfected with Nek2A-Myc and Gli2, together with Bcl-2 luciferase reporter plasmid using Lipofectamine 2000 reagent. TK-Renilla construct was introduced to normalize firefly luciferase activity. The reporter gene activity was assayed by the Dual Luciferase assay system. All experiments were performed in triplicate. Error bars represent the standard deviation of three independent experiments. **P<0.01 compared with the control groups.
Nek2A is regulated by Hh signaling
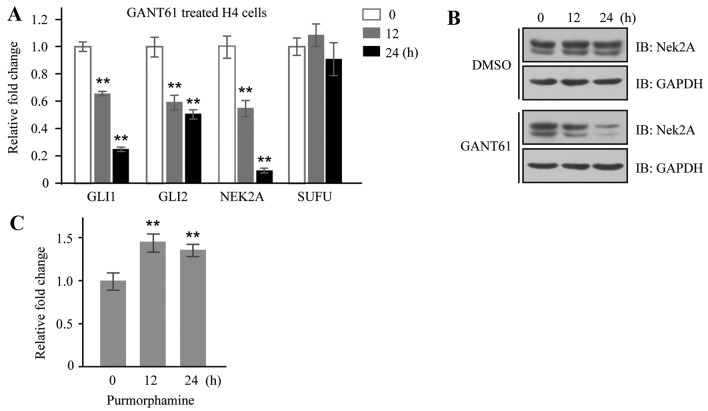
Nek2 has been found expressed aberrantly in multiple cancers and emerged as a possible oncogene due to its regulatory role in mitosis and cancer-related signaling pathways. Given the important roles of SuFu and Nek2 in cancer and the fact that these two proteins interact with each other, we set out to investigate the relationship of Nek2A and Hh signaling. GANT61, an inhibitor of Gli transcription factors, was employed to inhibit Hh signaling in H4 cells, and mRNA expressions of GLI1 and GLI2 were downregulated as expected; interestingly, the expression of NEK2A was also reduced (Fig. 3A). Moreover, the inhibitory effect of GANT61 on Nek2A expression was further verified in H4 cells by western blotting (Fig. 3B). On the other hand, we detected NEK2A mRNA expression in the same cells stimulated by purmorphamine, a purine derivative found to activate the Hh pathway by directly targeting Smo (34), and found that mRNA level of NEK2A was significantly increased in treated cells (Fig. 3C). The fact that Nek2A can be regulated by Hh signaling indicates that Nek2A may be a new target gene of Hh pathway.
Figure 3.
Nek2A is regulated by Hh signaling. (A) mRNA levels of Nek2A in GANT61 treated H4 cells. H4 cells were treated with GANT61 for 0, 12 and 24 h, respectively. Total mRNA was extracted and quantified by real-time PCR. Error bars represent the standard deviation of three independent experiments. (B) Protein levels of Nek2A in GANT61 treated cells. H4 cells were treated with GANT61 for 0, 12 and 24 h, respectively. Total proteins were extracted and Nek2A was detected by western blotting with monoclonal antibody. (C) mRNA levels of Nek2A in purmorphamine treated H4 cells. Total mRNA was extracted and quantified by real-time PCR. Error bars represent the standard deviation of three independent experiments. **P<0.01 compared with the control groups.
NEK2A is a target gene of transcription factor Gli
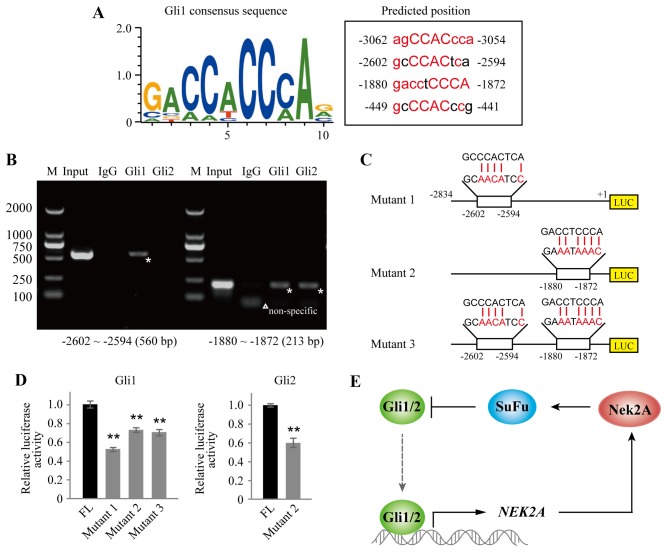
To further elucidate the upstream regulation of Nek2A and its relationship with Hh signaling, we used the Genomatix software MatInspector module for in silico analysis of putative transcription factor Gli binding sites, which were presented within the −3500/+1 region of the human Nek2A promoter. Four possible binding sites were found in the promoter region of Nek2A as shown in Fig. 4A. We next conducted a set of ChIP assays in CaES-17 cells and found that Gli1 displayed a strong binding enrichment to the specific regions: −2602 to −2594, and −1880 to −1872, of the Nek2A promoter, whereas Gli2 only bound to region of −1880 to −1872, which are indicated with stars in Fig. 4B.
Figure 4.
Nek2A is the target gene of transcription factor Gils. (A) Prediction of Gli binding sites in Nek2A promoter region using Genomatix software. Gli1 consensus sequence is shown on the left, and the predicted binding sites are on the right. Nucleotide in red, consensus index value >60. Capitals, core sequence, highest conserved, consecutive positions of the matrix. (B) ChIP analysis of Glis bound to the NEK2A promoter. The lysate from CaES-17 cells was immunoprecipitated with antibodies against Gli1, Gli2 or normal IgG. The inputs (10% of the cell lysates) and ChIP products were amplified by PCR reactions. PCR products then conduct electrophoresis in 1% agarose gel stained with ethidium bromide and visualized under UV light. Lane M, marker. *Indicates specific binding domains; △Indicates non-specific band. (C) Construction of NEK2A promoter-reporter fusion mono-site and dual-site mutants. The nucleotides in red are indicated as the substitution. (D) Luciferase assay of NEK2A promoter mutants. HEK293T cells were transfected with Gli1 or Gli2, together with Nek2A promoter-reporter fusion mutant and Bcl-2 luciferase reporter plasmid using Lipofectamine 2000 reagent. TK-Renilla construct was introduced to normalize firefly luciferase activity. All experiments were performed in triplicate. Error bars represent the standard deviation of three independent experiments. **P<0.01. (E) Schematic representation of a feedback regulation between Nek2A and Gli1/2 in mammalian cells.
To verify these binding domains, we constructed luciferase reporter vectors driven by FL or mutant Nek2A promoter (Fig. 4C). The FL or mutants of Nek2A (mutants 1, 2 and 3) were co-transfected with Gli1 or Gli2 in HEK293T cells as indicated. As shown in Fig. 4D, luciferase activities of single mutants (mutant 1 or 2) and double mutant (mutant 3) diminished significantly when compared with FL control in Gli1 overexpressed cells. In Gli2 transfected cells, mutant 2 also showed attenuated luciferase activity. Together, these data support that Nek2A is the target gene of Gli transcription factors. A working model is proposed to illustrate the feedback regulatory loop between Nek2A and SuFu (Fig. 4E). In brief, Gli1/2 directly activates NEK2A. Increased Nek2A protein then interacts with SuFu and promotes its stabilization. Stabilized SuFu subsequently inhibits Gli1/2 entering into the nucleus and suppresses Gli1/2 transcriptional function on NEK2A.
Discussion
The present study casts new insight into the regulation of Hh/Gli signaling. In particular, Nek2A stabilizes SuFu partly through the inhibition of SuFu ubiquitination; moreover, Nek2A impedes protein expression and transcriptional activity of Gli2. We also demonstrate that Nek2A is a target gene of Gli transcription activity, revealing a feedback loop of Nek2A in regulating Gli-mediated Hh signaling (Fig. 4E).
Hh signaling pathway was first identified by a genetic screen in Drosophila for mutants affecting body patterning in the early 1980s (35). Additional proteins have been found to be involved in regulating the Hh pathway since then (36). Here, our study introduces Nek2A as an important regulator of Hh signaling. Moreover, we demonstrate that besides their role in the activation of Hh downstream signaling, Gli1 and Gli2 both bind to the promoter region of Nek2A gene and activate Nek2A transcription. These findings reveal a new regulatory mechanism of Hh/SuFu/Gli signaling pathway.
Feedback loops are the building blocks of system dynamics in biological systems. It may operate via an indirect and integrated inhibitor/stimulator feedback loop influencing the production of the other. Many feedback loops have been reported in different signaling pathways and biological processes. For instance, Hh-dependent GLI1 induction constitutes a positive feedback loop, while Hh-dependent regulation of PTCH1, HHIP1, CDON and BOC constitutes negative feedback loops (37,38). We present here a novel, negative feedback loop in which Nek2A stabilizes SuFu, generally a key inhibitory regulator in Hh signaling, through impairing ubiquitin/proteasome degradation of SuFu, and simultaneously requires for the activating effects of Gli1/2, positive regulators in Hh signaling. Since the binding of SuFu and Glis prevents Gli1/2 from turning on transcription in the nucleus, resulting in diminished expression of Gli target genes (e.g. Nek2A), our results uncover a negative feedback loop. Under the normal physiology condition, constituents of Hh signaling and Nek2A possess a homeostasis condition. When Hh signaling is aberrantly activated by excessive Shh stimulation under pathological condition, suppression effect of SuFu to Gli1/2 would be diminished resulting from SuFu degradation, and we considered this process a positive feedback regulation of Hh signaling pathway. In order to prevent further dysfunction of the biosystem, this positive feedback loop is supposed to be rescued with some intrinsic mechanisms, of which stabilization effect of Nek2A on SuFu may be one.
Many proteins are variously modified, and one prominent intersection between post-translational modifications is phosphorylation and ubiquitination. In our previous study (17), SuFu is demonstrated to be stabilized by Nek2A through phosphorylation on two specific sites of T225 and S352. In the present study, we identify a new mechanism for SuFu regulation as Nek2A impedes ubiquitin/proteasome proteolysis of SuFu. It is known that phosphorylation can promote or block ubiquitination. In this study, we propose that SuFu phosphorylation inhibits its ubiquitination, likely through the following possible mechanisms: one modification masks the recognition site for a second post-transcriptional modification; phosphorylation negatively regulates the activity of the E3 ligase responsible for ubiquitin transfer; phosphorylation influences ubiquitination by regulating the substrate-ligase interaction at the level of subcellular compartmentalization.
Similar to other kinases involved in spindle assembly or duplication (39), overexpression of Nek2 has been implicated in several neoplastic diseases, such as breast carcinomas (40), lung adenocarcinomas (41), testicular seminomas (42) and diffuse large B cell lymphomas (43). On the other hand, in Hh driven cancers, the therapies of Gli inhibitors are often reported to be related to drug resistance (44,45). However, the underlying mechanism is unclear. Our results on the relationship of Nek2A and Hh signaling may provide one possible explanation for the drug resistance of Gli inhibitors. We found that GANT61 (Gli1/2 inhibitor) inhibited the mRNA expression and the protein synthesis of Nek2A (Fig. 3A and B). Therefore, the diminished Nek2A renders a more fragile SuFu suspectable to degradation, and thus releases the inhibition of Hh signaling, which may counteract the tumor suppressive effect of Gli inhibitors. Future studies will be required to determine precisely whether or how Nek2A may contribute to drug-resistance in certain cancer cells. Furthermore, given that Nek2A is a marker of cell cycle and its abundance increases through S and G2 and reaches peak in late G2/M, we propose that Nek2A may be a key signal indicating the events of excessive proliferation, thus initiating the feedback regulatory process to attenuate the aberrant cell proliferation.
In conclusion, our results identify Nek2A as both a regulator of Hh signaling pathway through impairing ubiquitin/proteasome proteolysis of SuFu and a target gene of Gli1/2. Therefore, we illustrated that Nek2A fine-tunes Hh patterning through a feedback loop, which may be a critical player for the rigorous maintenance of cellular homeostasis.
Acknowledgments
This study was supported in part by grants from the National Natural Science Foundation of China (no. 81560457 to Y.W., no. 31460305 to S.L.), the Natural Science Foundation of Jiangxi Province (no. 20151BAB205037 to Y.W.).
References
- 1.Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, Bale AE. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- 2.Svärd J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergström A, Ericson J, Toftgård R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Stone DM, Murone M, Luoh S, Ye W, Armanini MP, Gurney A, Phillips H, Brush J, Goddard A, de Sauvage FJ, et al. Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J Cell Sci. 1999;112:4437–4448. doi: 10.1242/jcs.112.23.4437. [DOI] [PubMed] [Google Scholar]
- 4.Barnfield PC, Zhang X, Thanabalasingham V, Yoshida M, Hui CC. Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation. 2005;73:397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- 5.Kise Y, Morinaka A, Teglund S, Miki H. Sufu recruits GSK3beta for efficient processing of Gli3. Biochem Biophys Res Commun. 2009;387:569–574. doi: 10.1016/j.bbrc.2009.07.087. [DOI] [PubMed] [Google Scholar]
- 6.Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci USA. 2002;99:5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Heydeck W, Zeng H, Liu A. Dual function of suppressor of fused in Hh pathway activation and mouse spinal cord patterning. Dev Biol. 2012;362:141–153. doi: 10.1016/j.ydbio.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 8.Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137:2001–2009. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 11.Pastorino L, Ghiorzo P, Nasti S, Battistuzzi L, Cusano R, Marzocchi C, Garrè ML, Clementi M, Scarrà GB. Identification of a SUFU germline mutation in a family with Gorlin syndrome. Am J Med Genet A. 2009;149A:1539–1543. doi: 10.1002/ajmg.a.32944. [DOI] [PubMed] [Google Scholar]
- 12.Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgård R, Undén AB. Deregulation of the hedgehog signalling pathway: A possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208:17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 13.Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Seguin N, Modrusan Z, Januario T, Tsui V, Durham AB, Dlugosz AA, Haverty PM, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27:327–341. doi: 10.1016/j.ccell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng T, Li C, Zhang X, Chi S, He N, Chen K, McCormick F, Gatalica Z, Xie J. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3:29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue S, Chen Y, Cheng SY. Hedgehog signaling promotes the degradation of tumor suppressor Sufu through the ubiquitin-proteasome pathway. Oncogene. 2009;28:492–499. doi: 10.1038/onc.2008.403. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Yue S, Xie L, Pu XH, Jin T, Cheng SY. Dual phosphorylation of suppressor of fused (Sufu) by PKA and GSK3beta regulates its stability and localization in the primary cilium. J Biol Chem. 2011;286:13502–13511. doi: 10.1074/jbc.M110.217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Li Y, Hu G, Huang X, Rao H, Xiong X, Luo Z, Lu Q, Luo S. Nek2A phosphorylates and stabilizes SuFu: A new strategy of Gli2/Hedgehog signaling regulatory mechanism. Cell Signal. 2016;28:1304–1313. doi: 10.1016/j.cellsig.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237:155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 19.Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17:470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayor T, Hacker U, Stierhof YD, Nigg EA. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J Cell Sci. 2002;115:3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- 21.Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rapley J, Baxter JE, Blot J, Wattam SL, Casenghi M, Meraldi P, Nigg EA, Fry AM. Coordinate regulation of the mother centriole component nlp by nek2 and plk1 protein kinases. Mol Cell Biol. 2005;25:1309–1324. doi: 10.1128/MCB.25.4.1309-1324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi Y, Iwaya T, Sawada G, Kurashige J, Matsumura T, Uchi R, Ueo H, Takano Y, Eguchi H, Sudo T, et al. Up-regulation of NEK2 by microRNA-128 methylation is associated with poor prognosis in colorectal cancer. Ann Surg Oncol. 2014;21:205–212. doi: 10.1245/s10434-013-3264-3. [DOI] [PubMed] [Google Scholar]
- 25.Neal CP, Fry AM, Moreman C, McGregor A, Garcea G, Berry DP, Manson MM. Overexpression of the Nek2 kinase in colorectal cancer correlates with beta-catenin relocalization and shortened cancer-specific survival. J Surg Oncol. 2014;110:828–838. doi: 10.1002/jso.23717. [DOI] [PubMed] [Google Scholar]
- 26.Cappello P, Blaser H, Gorrini C, Lin DC, Elia AJ, Wakeham A, Haider S, Boutros PC, Mason JM, Miller NA, et al. Role of Nek2 on centrosome duplication and aneuploidy in breast cancer cells. Oncogene. 2014;33:2375–2384. doi: 10.1038/onc.2013.183. [DOI] [PubMed] [Google Scholar]
- 27.Hu CM, Zhu J, Guo XE, Chen W, Qiu XL, Ngo B, Chien R, Wang YV, Tsai CY, Wu G, et al. Novel small molecules disrupting Hec1/Nek2 interaction ablate tumor progression by triggering Nek2 degradation through a death-trap mechanism. Oncogene. 2015;34:1220–1230. doi: 10.1038/onc.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Upregulation of NEK2 is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;31:745–754. doi: 10.3892/or.2013.2910. [DOI] [PubMed] [Google Scholar]
- 29.Zhong X, Guan X, Liu W, Zhang L. Aberrant expression of NEK2 and its clinical significance in non-small cell lung cancer. Oncol Lett. 2014;8:1470–1476. doi: 10.3892/ol.2014.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stricker TP, Henriksen KJ, Tonsgard JH, Montag AG, Krausz TN, Pytel P. Expression profiling of 519 kinase genes in matched malignant peripheral nerve sheath tumor/plexiform neurofibroma samples is discriminatory and identifies mitotic regulators BUB1B, PBK and NEK2 as overexpressed with transformation. Mod Pathol. 2013;26:930–943. doi: 10.1038/modpathol.2012.242. [DOI] [PubMed] [Google Scholar]
- 31.Luo SW, Zhang C, Zhang B, Kim CH, Qiu YZ, Du QS, Mei L, Xiong WC. Regulation of heterochromatin remodelling and myogenin expression during muscle differentiation by FAK interaction with MBD2. EMBO J. 2009;28:2568–2582. doi: 10.1038/emboj.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi H, Nord SH, Frigessi A, Børresen-Dale AL, Kristensen VN. Overrepresentation of transcription factor families in the genesets underlying breast cancer subtypes. BMC Genomics. 2012;13:199. doi: 10.1186/1471-2164-13-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Struhl G. In vivo evidence that Patched and Smoothened constitute distinct binding and transducing components of a Hedgehog receptor complex. Development. 1998;125:4943–4948. doi: 10.1242/dev.125.24.4943. [DOI] [PubMed] [Google Scholar]
- 34.Sinha S, Chen JK. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat Chem Biol. 2006;2:29–30. doi: 10.1038/nchembio753. [DOI] [PubMed] [Google Scholar]
- 35.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 36.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 37.Holtz AM, Peterson KA, Nishi Y, Morin S, Song JY, Charron F, McMahon AP, Allen BL. Essential role for ligand-dependent feedback antagonism of vertebrate hedgehog signaling by PTCH1, PTCH2 and HHIP1 during neural patterning. Development. 2013;140:3423–3434. doi: 10.1242/dev.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10:647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 40.Hayward DG, Clarke RB, Faragher AJ, Pillai MR, Hagan IM, Fry AM. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004;64:7370–7376. doi: 10.1158/0008-5472.CAN-04-0960. [DOI] [PubMed] [Google Scholar]
- 41.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbagallo F, Paronetto MP, Franco R, Chieffi P, Dolci S, Fry AM, Geremia R, Sette C. Increased expression and nuclear localization of the centrosomal kinase Nek2 in human testicular seminomas. J Pathol. 2009;217:431–441. doi: 10.1002/path.2471. [DOI] [PubMed] [Google Scholar]
- 43.Andréasson U, Dictor M, Jerkeman M, Berglund M, Sundström C, Linderoth J, Rosenquist R, Borrebaeck CA, Ek S. Identification of molecular targets associated with transformed diffuse large B cell lymphoma using highly purified tumor cells. Am J Hematol. 2009;84:803–808. doi: 10.1002/ajh.21549. [DOI] [PubMed] [Google Scholar]
- 44.Kobune M, Takimoto R, Murase K, Iyama S, Sato T, Kikuchi S, Kawano Y, Miyanishi K, Sato Y, Niitsu Y, et al. Drug resistance is dramatically restored by hedgehog inhibitors in CD34+ leukemic cells. Cancer Sci. 2009;100:948–955. doi: 10.1111/j.1349-7006.2009.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amable L, Fain J, Gavin E, Reed E. Gli1 contributes to cellular resistance to cisplatin through altered cellular accumulation of the drug. Oncol Rep. 2014;32:469–474. doi: 10.3892/or.2014.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]