Abstract
The prognosis of advanced hepatocellular carcinoma (HCC) is dismal. Novel molecular targets for diagnosis and therapy is urgently required. This study evaluated expression and functions of the protein arginine methyltransferase 5 (PRMT5) in HCC. Using HCC cell lines, the expression levels of PRMT5 mRNA were determined using the quantitative real-time reverse-transcription polymerase chain reaction, and the effect of a small interfering PRMT5-siRNA on cell phenotype was evaluated. Further, PRMT5 expression was determined in 144 pairs of resected liver tissues to evaluate its clinical significance. Regardless of their differentiated phenotypes, nine HCC cell lines expressed different levels of PRMT5 mRNA. Inhibition of PRMT5 expression significantly decreased the proliferation, invasion, and migration of HCC cell lines. Although the level of PRMT5 mRNA was not influenced by patient's background liver status, it was significantly higher in HCC tissues than in the corresponding noncancerous tissues. High levels of PRMT5 mRNA in HCC tissues were significantly associated with advanced disease stage and adverse prognosis. In conclusion, our results indicate that PRMT5 may act as a putative oncogene in HCC and that the levels of PRMT5 mRNA represent a promising prognostic marker and a potential target of molecular therapy for HCC.
Keywords: protein arginine methyltransferase 5, hepatocellular carcinoma, oncogene, expression, biomarker
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the third most common cause of cancer-related deaths worldwide (1,2). The 5-year overall survival rate of HCC patients ranges from 0 to 14% (3). Although patients frequently experience recurrence after curative resection, the therapeutic alternatives for advanced and recurrent HCC are limited compared with those available for managing other cancers of the digestive tract (4,5). Therefore, it is important to identify new biomarkers for predicting outcomes as well as new molecular targets for treating HCC (6).
Protein arginine methyltransferases (PRMTs) are the major enzymes that methylate arginine residues in histones (7). There are four types of PRMTs, and PRMT5 is a type II PRMT that catalyzes monomethylation and symmetric dimethylation reactions (8–10). Symmetric dimethylation of histones inhibits transcription, and inhibition of the expression of tumor suppressor genes causes tumorigenesis and the progression of cancer (10–12). PRMT5 acts as an oncogene in some neoplasms through inhibition of the transcription of certain tumor suppressor genes, promotes cell proliferation, and inhibits apoptosis (10,11,13,14). Moreover, PRMT5 contributes to epithelial-mesenchymal transition through interactions with E-cadherin and affects RNA processing (7,10,12). Zhang et al indicated that inhibition of PRMT5 induced HCC cell growth via the downregulation of β-catenin (15). However, little is known about the role of PRMT5 in the pathogenesis of HCC. Therefore, the aim of this study was to investigate the expression and functions of PRMT5 in HCC, and the results presented here lead us to propose that PRMT5 is associated with HCC.
Materials and methods
Ethics
This study conformed to the ethical guidelines of the World Medical Association Declaration of Helsinki-Ethical Principles for Medical Research Involving Human Subjects and has been approved by the Institutional Review Board of Nagoya University, Japan. Written informed consent for usage of clinical samples and data, as required by the institutional review board, was obtained from all patients.
Sample collection
The human HCC cell lines Hep3B, HepG2, PLC/PRF/5 and SK-Hep1 were purchased from the American Type Culture Collection (Manassas, VA, USA). HLE, HLF, HuH1 and HuH7 were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan) (16). HuH2 cells were a gift from Aichi Cancer Center (Nagoya, Japan). Cells were stored at −80°C and cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) at 37°C in an atmosphere containing 5% CO2. Primary HCC tissues and corresponding noncancerous tissues appropriate for extraction of total RNA were collected from 144 patients undergoing liver resection for HCC at Nagoya University Hospital between January 1998 and January 2012. None of the patients underwent preoperative treatment including chemotherapy or interventional radiology. Tissue samples were frozen immediately in liquid nitrogen and stored at −80°C. RNA was extracted from HCC tissue samples ~5-mm2 that contained >80% tumor cells and lacked necrotic tissue (17,18). The corresponding noncancerous tissue samples were collected from areas >2 cm away from the tumor edge. Specimens were classified histologically according to the TNM Classification of Malignant Tumors, 7th Edition (19). Clinicopathological parameters and the patient follow-up data were collected from medical records.
Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR)
PRMT5 mRNA levels were determined using qRT-PCR. Total RNAs (10 μg) were extracted from nine HCC cell lines and 144 pairs of clinical samples and were amplified using primers specific for PRMT5 as follows: sense 5′-TCTCATGGTTTCCCATCCTC-3′ in exon 16 and antisense 5′-CCTTCTTGGAATTGCTGCAT-3′ in exon 17, which generate a 102-bp product. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (TaqMan, GAPDH control reagents; Applied Biosystems, Foster City, CA, USA) was quantified in each sample for standardization. qRT-PCR was performed using the SYBR-Green PCR Core reagents kit (Applied Biosystems) as follows: one cycle at 95°C for 10 min, 40 cycles at 95°C for 5 sec, and 60°C for 60 sec. The expression levels of each sample were determined in triplicate, and the data are presented as the PRMT5 value divided by the GAPDH value (20,21). Patients were stratified into PRMT5-low and PRMT5-high groups, respectively, according to the median value of the PRMT5 mRNA expression level in cancer tissues of 144 patients as follows: high (higher than the median value) and low (the median value or lower).
Immunofluorescence staining
The expression and location of PRMT5 protein was investigated by immunofluorescence staining using three HCC cell lines, Hep3B, HLE and HuH7, expressed relatively high levels of PRMT5. HCC cells were fixed on microscope slides by methanol. Slides were cooled at −10°C for 5 min and incubated with mouse monoclonal antibody raised against PRMT5 (sc-376937 FITC; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), diluted 1:50 in phosphate-buffered saline (PBS). The nuclei were counter-stained with 1 μg/ml DAPI. The slides were observed under a fluorescence microscope, FSX100 (Olympus, Tokyo, Japan).
Western blot analysis
Cells were incubated in RIPA lysis buffer, and the lysates were stored at −30°C. Total lysate protein (15 μg/well) was electrophoretically transferred onto nitrocellulose membranes that were blocked using 5% skim milk and incubated at 4°C overnight with a rabbit anti-PRMT5 polyclonal antibody (Cell Signaling Technology, Beverly, MA, USA) diluted 1:1,000. The membrane was then washed and probed with an anti-rabbit secondary antibody conjugated to horseradish peroxidase (Cell Signaling Technology). After the final wash, immune complexes were visualized using an LAS-4010 image analyzer (GE Healthcare, Pittsburgh, PA, USA). β-actin served as an endogenous control (22).
Knockdown of PRMT5 expression using a PRMT5-specific siRNA
Small interfering RNAs (siRNAs) specific for PRMT5 (siPRMT5), siPRMT5-1, 5′-CAGCCACUGAUGGACAAUCUGGAAU-3′ and siPRMT5-2, 5′-CCGGCUACUUUGAGACUGUGCUUUA-3′ (Hokkaido System Science, Sapporo, Japan) were used to transfect HLE and HuH7 cells that express relatively high levels of PRMT5. AccuTarget Negative Control siRNA fluorescein-labeled (Cosmo Bio Co., Ltd., Tokyo, Japan) served as a control nontargeting siRNA (siControl). HLE and HuH7 cells (4×105 and 2×106 cells/dish, respectively) were seeded into 10 cm dishes containing 10 ml of antibiotic-free DMEM with 10% FBS, and were transfected with 400 pmol of siControl or siPRMT5-1 and 2 in the presence of 40-μl LipoTrust EX Oligo (Hokkaido System Science) 24 h after cell seeding. After 72 h incubation following siRNA transfection, total RNA and protein were extracted, and cells were treated with EDTA-trypsin and used for functional assays.
Cell proliferation assay
The proliferation of HLE and HuH7 cells was evaluated using a Premix WST-1 Cell Proliferation assay system (Takara Bio Inc., Kusatsu, Japan). After transfection of siPRMT5, HLE and HuH7 (3×103 cells/well) cells were seeded into 96-well plates in DMEM containing 2% FBS. The optical density of each well was measured 1 h after the addition of 10 μl of WST-1 0, 1 and 3 days after seeding (23). The growth rate indicated an incremental rate of the optical density from the cell seeding day.
Cell invasion assay
The ability of cells to invade Matrigel was determined using BioCoat Matrigel invasion chambers (BD Biosciences, Bedford, MA, USA) according to the manufacturer's protocol. HLE and HuH7 cells (2.5×103 cells/well) were suspended in serum-free DMEM and seeded in the upper chamber. After an appropriate incubation time, cells present on the surface of the membrane were fixed, stained and counted using a microscope in eight randomly selected fields (24).
Wound healing assay
The migration of HLE and HuH7 cells was determined using a wound-healing assay. After transfection, HLE (2×104 cells/well) and HuH7 (5×104 cells/well) were seeded into each well of a 35 mm dish with culture insert (Ibidi GmbH, Martinsried, Germany) in DMEM containing 2% FBS. After 24 h, the insert was removed, and the width of the wound was measured at 100-μm intervals (20/well, ×40 magnification) at cell-dependent time intervals.
Statistical analysis
Differences between data of two groups were evaluated using the Mann-Whitney test. The χ2 test was used to analyze the significance of the association between the expression levels of PRMT5 mRNA and the patient clinicopathological parameters. Overall survival rates were calculated using the Kaplan-Meier method, and the differences in survival curves were evaluated using the log-rank test. All statistical analyses were performed using JMP 10 software (SAS Institute Inc., Cary, NC, USA). A p-value <0.05 was considered statistically significant.
Results
Expression of PRMT5 in HCC cell lines
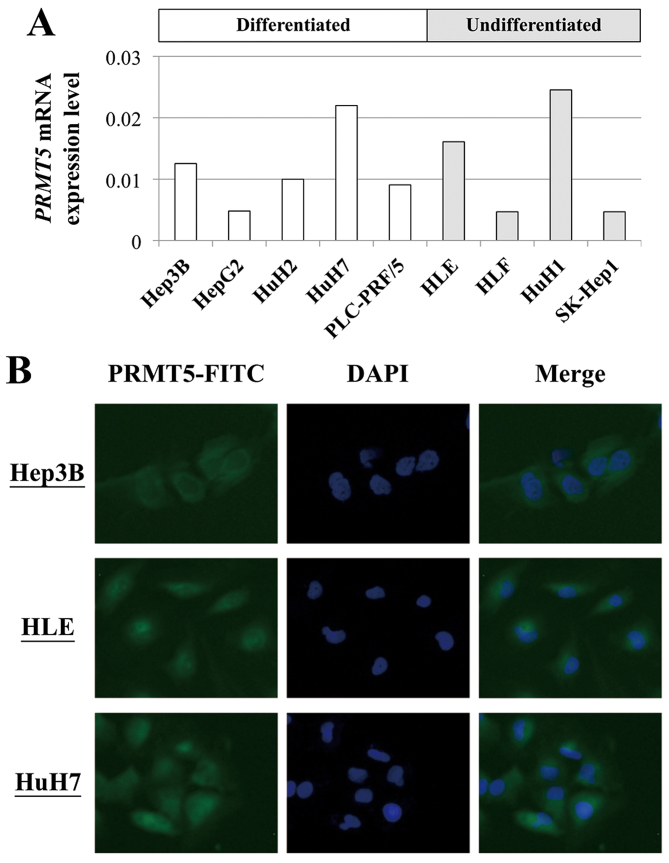
The levels of PRMT5 mRNA differed among the nine HCC cell lines regardless of their differentiated phenotypes (Fig. 1A). HuH7 (differentiated) and HLE (undifferentiated) cells expressed relatively high levels of PRMT5 mRNA and were used in the functional analyses described below. As shown in Fig. 1B, PRMT5 protein was distributed in the cytoplasm, especially in the perinuclear compartment.
Figure 1.
Analysis of the expression and function of PRMT5 in HCC cell lines. (A) The levels of PRMT5 mRNA in HCC cell lines according to their differentiated phenotypes. (B) Immunofluorescence staining for PRMT5 protein in HCC cell lines.
Effects of inhibiting PRMT5 expression
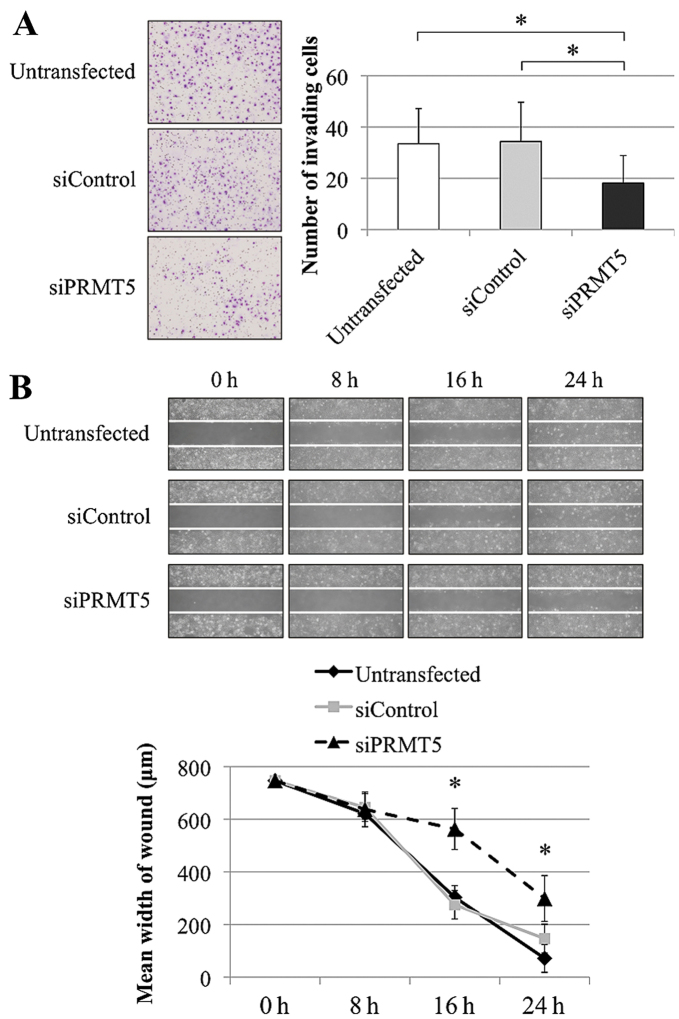
The specificity of siPRMT5s were checked using BLAST in the NCCI database (25). Then, the total scores and E-values of the two siPRMT5s were 50.1 and 5e−6, and the query cover rates of transcriptomes other than PRMT5 mRNA were <70%. The knockdown effects of siPRMT5-1 and 2 were determined by qRT-PCR and western blot analysis (Fig. 2A). The knockdown effect by single siPRMT5 sequence was not enough for functional analysis, we transfected HCC cells with both the siPRMT5s in following analysis. To determine the functions of PRMT5 in HCC, we used HLE and HuH7 cells transfected with siPRMT5-1 and -2 sequences for evaluation of the proliferation, invasion, and migration. In HuH7 cells, PRMT5 knockdown significantly decreased cell proliferation on days 1 and 3 compared with the untransfected and siControl groups (Fig. 2B). Second, the number of cells that invaded Matrigel markedly decreased when PRMT5 expression was inhibited (Fig. 3A). Third, the migration of HuH7 cells decreased significantly after transfection with the siPRMT5 compared with the untransfected and siControl-transfected cells at 16 and 24 h (Fig. 3B). Similarly, invasion and migration of HLE cells were markedly reduced in siPRMT5-transfected cells (data not shown).
Figure 2.

Confirmation of knockdown of PRMT5 expression and function of PRMT5 in HCC cell lines. (A) qRT-PCR and western blot analysis to confirm the activity of PRMT5 siRNAs. (B) Cell proliferation assay of HuH7 cells. The PRMT5-specific siRNA significantly decreased cell growth. *p<0.05.
Figure 3.
Phenotypes of HuH7 cells transfected with a PRMT5-specific siRNA. Knockdown of PRMT5 expression inhibited the invasiveness (A) and migration (B) of HCC cells. *P<0.05.
Patient characteristics
The ages of the 144 patients ranged from 34 to 84 years (median, 65.5 years), the male-to-female ratio was 121:23, and 37 and 80 patients were infected with hepatitis B or C virus, respectively. The numbers of patients with normal liver function, chronic hepatitis and cirrhosis were 10, 82 and 52, respectively. The median duration of patient follow-up was 40.1 months (range, 2.3–145 months); 90, 37 and 17 patients were diagnosed with stage I, II or III HCC, respectively.
Clinical implications of PRMT5 expression levels
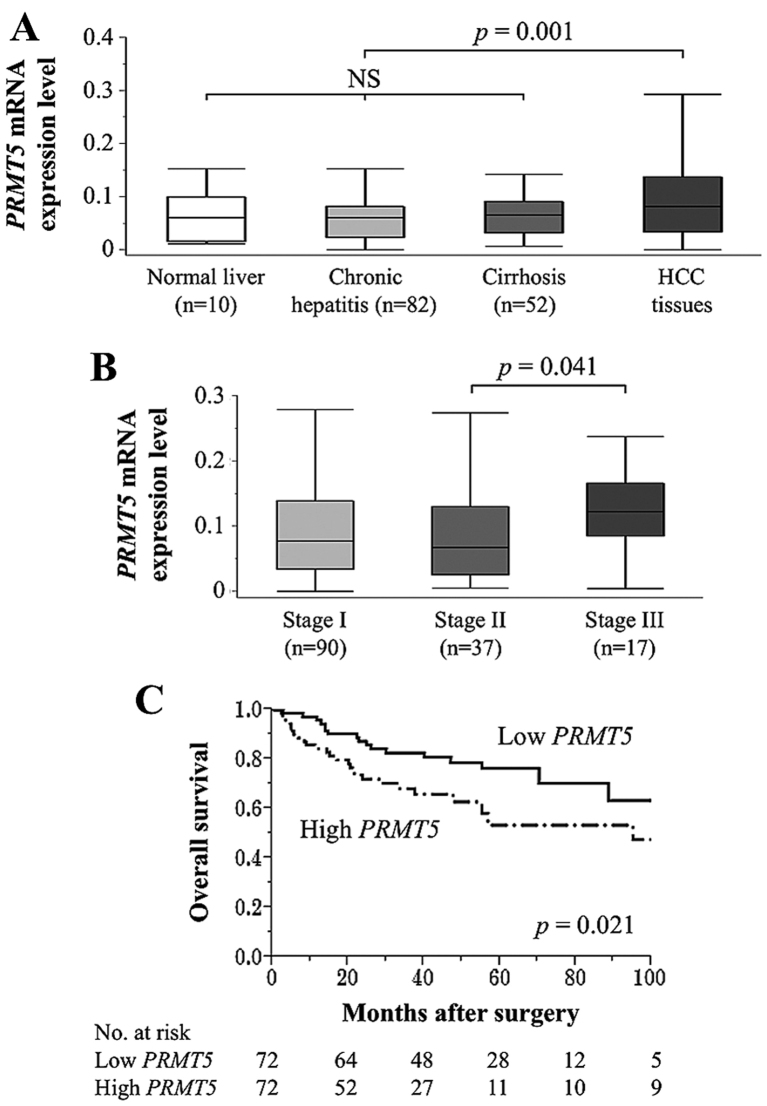
There were no significant differences in PRMT5 mRNA levels in noncancerous tissue samples from patients with normal liver function, chronic hepatitis, and cirrhosis. In contrast, the mean level of PRMT5 mRNA in HCC tissues was significantly higher compared with that of the corresponding noncancerous tissues (p=0.001) (Fig. 4A). When patients were divided according to TNM stage, PRMT5 mRNA expression levels in patients with stage III HCC were significantly higher compared with patients with stage II HCC (Fig. 4B).
Figure 4.
Expression of PRMT5 mRNA in resected tissues. (A) There were no significant differences of PRMT5 mRNA levels among noncancerous tissues categorized according to the status of the background uninvolved liver. PRMT5 mRNA was expressed at higher levels in HCC tissues compared with the corresponding noncancerous tissues. NS, not significant. (B) Analysis of PRMT5 mRNA levels as a function of HCC stage. (C) Patients with HCC with high levels of PRMT5 expressions experienced significantly shorter overall survival.
To evaluate the prognostic significance of PRMT5 expression, patients were categorized into low- and high-PRMT5 groups (n=72 each) according to their median levels of PRMT5 mRNA expression in HCC tissue described in Materials and methods. High levels of PRMT5 mRNA were significantly associated with advanced TNM stage, independent of background liver status, tumor size or vascular invasion (Table I). The members of the high-PRMT5 group were more likely to have a worse prognosis compared with those in the low PRMT5 group (5-year overall survival rates, 53 and 76%, respectively; p=0.021) (Fig. 4C). However, there were no significant differences between the groups in relapse-free survival rate, and high levels of PRMT5 were not an independent prognostic factor in multivariate analysis.
Table I.
Association between expression levels of PRMT5 mRNA and clinicopathological parameters in 144 patients with hepatocellular carcinoma (HCC).
| Clinicopathological parameters | High PRMT5 group (n=72) | Low PRMT5 group (n=72) | p-value |
|---|---|---|---|
| Age (years) | 0.615 | ||
| <65 year | 31 | 34 | |
| ≥65 year | 41 | 38 | |
| Gender | 0.254 | ||
| Male | 58 | 63 | |
| Female | 14 | 9 | |
| Background liver | 0.391 | ||
| Normal liver | 6 | 4 | |
| Chronic hepatitis | 37 | 45 | |
| Cirrhosis | 29 | 23 | |
| Pugh-Child's classification | 0.511 | ||
| A | 68 | 66 | |
| B | 4 | 6 | |
| Hepatitis virus | 0.558 | ||
| Absent | 14 | 13 | |
| HBV | 21 | 16 | |
| HCV | 37 | 43 | |
| AFP (ng/ml) | 1.000 | ||
| ≤20 | 39 | 39 | |
| >20 | 33 | 33 | |
| PIVKA II (mAU/ml) | 0.497 | ||
| ≤40 | 31 | 27 | |
| >40 | 41 | 45 | |
| Tumor multiplicity | 0.228 | ||
| Solitary | 53 | 59 | |
| Multiple | 19 | 13 | |
| Tumor size | 0.475 | ||
| <3.0 cm | 25 | 21 | |
| ≥3.0 cm | 47 | 51 | |
| Differentiation | 0.079 | ||
| Well | 22 | 13 | |
| Moderate to poor | 50 | 59 | |
| Growth type | 0.370 | ||
| Expansive growth | 58 | 62 | |
| Invasive growth | 14 | 10 | |
| Serosal infiltration | 0.331 | ||
| Absent | 57 | 52 | |
| Present | 15 | 20 | |
| Formation of capsule | 0.213 | ||
| Absent | 27 | 20 | |
| Present | 45 | 52 | |
| Infiltration to capsule | 0.503 | ||
| Absent | 35 | 31 | |
| Present | 37 | 41 | |
| Septum formation | 0.484 | ||
| Absent | 23 | 27 | |
| Present | 49 | 45 | |
| Vascular invasion | 0.122 | ||
| Absent | 50 | 58 | |
| Present | 22 | 14 | |
| UICC pathological stage | 0.012a | ||
| I | 42 | 48 | |
| II | 16 | 21 | |
| III | 14 | 3 |
Statistically significant difference (p<0.05). HBV, hepatitis B virus; HCV, hepatitis C virus; AFP, α-fetoprotein; PIVKA, protein induced by vitamin K antagonists; UICC, Union for International Cancer Control.
Discussion
In the present study, we investigated the function and expression of PRMT5. We show here that PRMT5 mRNA was differentially expressed among HCC cell lines, and that PRMT5 protein was expressed mainly at the cytoplasm of HCC cells. Inhibition of PRMT5 expression in HCC cell lines using a PRMT5-specific siRNA decreased not only the proliferation and migration was reported in a previous study (15), but also invasiveness. These findings suggest that PRMT5 contributes to the pathogenesis of HCC.
Post-translational protein modification is involved at all levels of cellular regulation. PRMTs catalyze the addition of methyl groups to the guanidinium nitrogen atoms of arginine amino acid residues. PRMT5 is a type II arginine methyltransferase that catalyzes the symmetric dimethylation of the arginine residues of the histone proteins H3 and H4, which alters chromatin structure to promote the inhibition of the transcription of target genes (7,8,13). These modifications play an important role in cell differentiation and tumor cell growth. PRMT5 promotes tumor progression by stimulating cell cycle progression as well as inhibiting proliferation and apoptosis (12,14,26). Moreover, PRMT5 plays an important role in stem cell maintenance through its suppression of genes that control differentiation (7,12,27).
PRMT5 acts as an oncogene in ovarian cancer, glioblastoma, colorectal cancer, and lung cancer, and PRMT5 interacts with many other genes (10–14,28). PRMT5 methylates histone residues H3R8 and H4R3 to induce silencing of tumor suppressor genes such as ST7 and NM23 (8). Further, PRMT5 contributes to diverse cellular functions through its interactions with E2F-1, IL-2, cyclin E1, the CDK4 complex, and E-cadherin (28–30). In particular, the inhibitory function of PRMT5 on signaling pathways that inhibit the activities of p53 and the TRAIL receptors may represent important mechanisms underlying its putative oncogenic activities (8). From these previous studies, it can be speculated that high PRMT5 expression was associated with an elevated proliferation ability of HCC cells via an apoptosis restraint through suppression of p53, TRAIL receptors and E2F-1 (8,29,30). Increased migration and invasion abilities of tumor cells by overexpression of PRMT5 may be attributed to suppression of E-cadherin (7). Further analyses of PRMT5-related pathways will be desirable to elucidate molecular mechanisms of PRMT5 in HCC.
Although the mean level of PRMT5 mRNA was significantly higher in HCC tissues compared with those of the corresponding noncancerous tissues, it was independent of chronic inflammation and hepatic fibrosis, suggesting that suppression of PRMT5 expression may represent a specific event that occurs in the final stage of the initiation of HCC. Zhang et al showed that patients with high expression of PRMT5 protein in cytoplasm had shorter survival duration in the cohort with 54 HCC cases (15). Our results also indicated that high expression of PRMT5 mRNA contributed to worse prognosis. It is important that the influence of PRMT5 expression level on patient prognosis was consistent with another large cohort. Consistent with the results of our functional analyses, high levels of PRMT5 expression in HCC tissues were an adverse prognostic factor after curative hepatectomy. Further, higher levels of PRMT5 mRNA were significantly associated with advanced disease stage.
Taken together, our findings suggest that analysis of PRMT5 expression will enhance the clinical management of HCC. For example, determining the levels of PRMT5 mRNA in liver biopsies or resected tissues will facilitate risk stratification of patients with HCC and may serve as a criterion for determining the most appropriate therapy tailored for individual patients. Moreover, our findings show promise for developing new therapies for HCC that employ small molecules, and antibodies, or both targeting PRMT5 expression, and activity. Further research on signaling pathways that act through or are regulated by PRMT5 may reveal other targets for the treatment of HCC.
The present study has certain limitations. Overexpression experiments of PRMT5 and global expression analyses of proteins that potentially interact with PRMT5 as well as apoptosis assays will be required for further understanding of the biological functions of PRMT5 in HCC. External validation of the reproducibility of the expression analyses and their standardization across laboratories are required as well. Further, this study was limited by the relatively small sample size.
Our results indicate that PRMT5 plays oncogenic roles in HCC by enhancing the malignant phenotype of tumor cells, though further analysis are required for clarifying the specific mechanism of PRMT5 in HCC. PRMT5 shows promise as a biomarker for patient stratification and is a potential target of molecular therapy in HCC.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Kanda M, Sugimoto H, Kodera Y. Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10584–10597. doi: 10.3748/wjg.v21.i37.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 4.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 6.Kanda M, Sugimoto H, Nomoto S, Oya H, Hibino S, Shimizu D, Takami H, Hashimoto R, Okamura Y, Yamada S, et al. B cell translocation gene 1 serves as a novel prognostic indicator of hepatocellular carcinoma. Int J Oncol. 2015;46:641–648. doi: 10.3892/ijo.2014.2762. [DOI] [PubMed] [Google Scholar]
- 7.Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72:2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka H, Hoshikawa Y, Oh-hara T, Koike S, Naito M, Noda T, Arai H, Tsuruo T, Fujita N. PRMT5, a novel TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis via nuclear factor-kappaB activation. Mol Cancer Res. 2009;7:557–569. doi: 10.1158/1541-7786.MCR-08-0197. [DOI] [PubMed] [Google Scholar]
- 9.Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, Surani MA. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholas C, Yang J, Peters SB, Bill MA, Baiocchi RA, Yan F, Sïf S, Tae S, Gaudio E, Wu X, et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1) PLoS One. 2013;8:e74710. doi: 10.1371/journal.pone.0074710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han X, Li R, Zhang W, Yang X, Wheeler CG, Friedman GK, Province P, Ding Q, You Z, Fathallah-Shaykh HM, et al. Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J Neurooncol. 2014;118:61–72. doi: 10.1007/s11060-014-1419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Chitnis N, Nakagawa H, Kita Y, Natsugoe S, Yang Y, Li Z, Wasik M, Klein-Szanto AJ, Rustgi AK, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5:288–303. doi: 10.1158/2159-8290.CD-14-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao X, Zhao S, Liu T, Liu Y, Liu Y, Yang X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J Histochem Cytochem. 2013;61:206–217. doi: 10.1369/0022155413475452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim R, Matsubara D, Osman W, Morikawa T, Goto A, Morita S, Ishikawa S, Aburatani H, Takai D, Nakajima J, et al. Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum Pathol. 2014;45:1397–1405. doi: 10.1016/j.humpath.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, Dong S, Li Z, Lu L, Zhang S, Chen X, Cen X, Wu Y. Targeting protein arginine methyltransferase 5 inhibits human hepatocellular carcinoma growth via the downregulation of beta-catenin. J Transl Med. 2015;13:349. doi: 10.1186/s12967-015-0721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezaka K, Kanda M, Sugimoto H, Shimizu D, Oya H, Nomoto S, Sueoka S, Tanaka Y, Takami H, Hashimoto R, et al. Reduced expression of adherens junctions associated protein 1 predicts recurrence of hepatocellular carcinoma after curative hepatectomy. Ann Surg Oncol. 2015;22(Suppl 3):S1499–S1507. doi: 10.1245/s10434-015-4695-9. [DOI] [PubMed] [Google Scholar]
- 17.Kanda M, Nomoto S, Okamura Y, Hayashi M, Hishida M, Fujii T, Nishikawa Y, Sugimoto H, Takeda S, Nakao A. Promoter hypermethylation of fibulin 1 gene is associated with tumor progression in hepatocellular carcinoma. Mol Carcinog. 2011;50:571–579. doi: 10.1002/mc.20735. [DOI] [PubMed] [Google Scholar]
- 18.Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Hishida M, Suenaga M, Yamada S, Inokawa Y, Nishikawa Y, et al. Downregulation of DENND2D by promoter hypermethylation is associated with early recurrence of hepatocellular carcinoma. Int J Oncol. 2014;44:44–52. doi: 10.3892/ijo.2013.2165. [DOI] [PubMed] [Google Scholar]
- 19.Sobin LH, Gospodarowicz MK, Wittekind CH, editors. International Union Against Cancer: TNM Classification of Malignant Tumors. 7th edition. Wiley-Blackwell; New York: 2009. [Google Scholar]
- 20.Kanda M, Nomoto S, Oya H, Hashimoto R, Takami H, Shimizu D, Sonohara F, Kobayashi D, Tanaka C, Yamada S, et al. Decreased expression of prenyl diphosphate synthase subunit 2 correlates with reduced survival of patients with gastric cancer. J Exp Clin Cancer Res. 2014;33:88. doi: 10.1186/s13046-014-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanda M, Nomoto S, Oya H, Shimizu D, Takami H, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S, et al. Dihydropyrimidinase-like 3 facilitates malignant behavior of gastric cancer. J Exp Clin Cancer Res. 2014;33:66. doi: 10.1186/s13046-014-0066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanda M, Nomoto S, Okamura Y, Nishikawa Y, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Detection of metallothionein 1G as a methylated tumor suppressor gene in human hepatocellular carcinoma using a novel method of double combination array analysis. Int J Oncol. 2009;35:477–483. doi: 10.3892/ijo_00000359. [DOI] [PubMed] [Google Scholar]
- 23.Kanda M, Shimizu D, Fujii T, Sueoka S, Tanaka Y, Ezaka K, Takami H, Tanaka H, Hashimoto R, Iwata N, et al. Function and diagnostic value of Anosmin-1 in gastric cancer progression. Int J Cancer. 2016;138:721–730. doi: 10.1002/ijc.29803. [DOI] [PubMed] [Google Scholar]
- 24.Oya H, Kanda M, Sugimoto H, Shimizu D, Takami H, Hibino S, Hashimoto R, Okamura Y, Yamada S, Fujii T, et al. Dihydropyrimidinase-like 3 is a putative hepatocellular carcinoma tumor suppressor. J Gastroenterol. 2015;50:590–600. doi: 10.1007/s00535-014-0993-4. [DOI] [PubMed] [Google Scholar]
- 25.BLAST Basic Local Alignment Search Tool. http://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed March 10, 2016.
- 26.Smil D, Eram MS, Li F, Kennedy S, Szewczyk MM, Brown PJ, Barsyte-Lovejoy D, Arrowsmith CH, Vedadi M, Schapira M. Discovery of a dual PRMT5-PRMT7 inhibitor. ACS Med Chem Lett. 2015;6:408–412. doi: 10.1021/ml500467h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan F, Alinari L, Lustberg ME, Martin LK, Cordero-Nieves HM, Banasavadi-Siddegowda Y, Virk S, Barnholtz-Sloan J, Bell EH, Wojton J, et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014;74:1752–1765. doi: 10.1158/0008-5472.CAN-13-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Günesdogan U, Zylicz JJ, Hackett JA, Cougot D, Bao S, Lee C, Dietmann S, Allen GE, Sengupta R, et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol Cell. 2014;56:564–579. doi: 10.1016/j.molcel.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J, Zhu Y, Nilsson M, Sundfeldt K. TGF-β isoforms induce EMT independent migration of ovarian cancer cells. Cancer Cell Int. 2014;14:72. doi: 10.1186/s12935-014-0072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho EC, Zheng S, Munro S, Liu G, Carr SM, Moehlenbrink J, Lu YC, Stimson L, Khan O, Konietzny R, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012;31:1785–1797. doi: 10.1038/emboj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]