Abstract
While mammalian somatic cells are incapable of mounting an effective RNA interference (RNAi) response to viral infections, plants and invertebrates are able to generate high levels of viral short interfering RNAs (siRNAs) that can control many infections. In Drosophila, the RNAi response is mediated by the Dicer 2 enzyme (dDcr2) acting in concert with two cofactors called Loqs-PD and R2D2. To examine whether a functional RNAi response could be mounted in human somatic cells, we expressed dDcr2, in the presence or absence of Loqs-PD and/or R2D2, in a previously described human cell line, NoDice/ΔPKR, that lacks functional forms of human Dicer (hDcr) and PKR. We observed significant production of ∼21-nt long siRNAs, derived from a cotransfected double stranded RNA (dsRNA) expression vector, that were loaded into the human RNA-induced silencing complex (RISC) and were able to significantly reduce the expression of a cognate indicator gene. Surprisingly, dDcr2 was able to produce siRNAs even in the absence of Loqs-PD, which is thought to be required for dsRNA cleavage by dDcr2. This result may be explained by our finding that dDcr2 is able to bind the human Loqs-PD homolog TRBP when expressed in human cells in the absence of Loqs-PD. We conclude that it is possible to at least partially rescue the ability of mammalian somatic cells to express functional siRNAs using gene products of invertebrate origin.
Keywords: Dicer, RNA interference, small interfering RNAs, Drosophila
INTRODUCTION
RNA interference (RNAi) functions as an effective antiviral innate immune response in plants, nematodes, and insects (Ding and Voinnet 2007). The RNAi response is induced by the long double-stranded RNA (dsRNA) intermediates that are essential for the replication of the RNA genomes of all RNA viruses, except retroviruses, and that also arise due to the convergent transcription of the compact genomes of DNA viruses. The first step in mounting an RNAi response is the cleavage of viral dsRNAs into ∼22-bp-long duplexes, bearing 2-nt 3′-overhangs at each end, by the processive dsRNA-specific exonuclease Dicer (Ding and Voinnet 2007). One strand of this small interfering RNA (siRNA) duplex is then loaded into the RNA-induced silencing complex (RISC) where it functions to target RISC to the cognate viral RNA genome or anti-genome and/or to viral mRNAs. If the Argonaute (Ago) protein component in RISC is competent for endonucleolytic cleavage—only Ago2 has this activity in mammals—then the target viral RNA is cleaved and destroyed. Because RNAi functions as an effective antiviral response in plants and invertebrates, many viruses that infect these organisms encode factors that can inhibit RNAi responses and enhance viral pathogenicity (Wu et al. 2010).
In contrast to the above, considerable evidence indicates that an antiviral RNAi response does not occur in virally infected somatic cells of mammalian origin (Cullen et al. 2013). Instead, viral dsRNAs function as a pathogen-associated molecular pattern (PAMP) that is recognized by cellular factors such as PKR, RIG-I, and MDA5 and leads to induction of a potent protein-based innate antiviral immune response called the interferon response. The inability of mammalian somatic cells to mount an effective RNAi response reflects the weak ability of mammalian Dicer to digest long dsRNAs into siRNAs, although this enzyme is very effective at digesting pre-miRNA stem–loops to liberate miRNA duplex intermediates (Zhang et al. 2002; Ma et al. 2008; Flemr et al. 2013; Kennedy et al. 2015). This difference in activity appears to correlate with the fact that the mammalian Dicer protein is not processive, despite the presence of an amino-terminal helicase domain (Zhang et al. 2002; Ma et al. 2008). In contrast, in Drosophila, the Dicer-2 (dDcr2) protein is responsible for the generation of siRNAs from long dsRNAs and is highly processive (Cenik et al. 2011). Indeed, dDcr2 has been reported to remain associated with long dsRNAs and to cleave these reiteratively into siRNAs, an activity that requires the dDcr2 helicase domain and ATP hydrolysis (Cenik et al. 2011). In contrast, Drosophila Dicer-1 (dDcr1), like mammalian Dicer, is nonprocessive and is also effective at cleaving pre-miRNA hairpins yet unable to effectively cleave long dsRNAs into siRNAs (Lee et al. 2004; Bogerd et al. 2014).
Analysis of the ability of full-length human Dicer (hDcr) to process long dsRNAs and pre-miRNAs indicates that the amino-terminal helicase domain actually inhibits long dsRNA processing but does not affect, either positively or negatively, the ability of hDcr to process pre-miRNA hairpins (Ma et al. 2008; Kennedy et al. 2015). However, it has been reported that murine germ cells, and potentially embryonic stem (ES) cells, express a second isoform of murine Dicer (mDcr) that initiates from an alternative, retrotransposon-derived promoter located in intron 6 of the murine dicer1 gene, that is only active in germ cells and somatic stem cell populations (Flemr et al. 2013). This amino-terminally truncated mDcr isoform was reported to be significantly more capable of generating siRNAs from long dsRNAs, thus potentially explaining reports arguing for the existence of RNAi in mouse germ cells and ES cells (Wianny and Zernicka-Goetz 2000; Billy et al. 2001; Yang et al. 2001; Babiarz et al. 2008; Tam et al. 2008; Maillard et al. 2013). The genomic human dicer1 gene, in contrast, does not contain a retrotransposon promoter in the equivalent location and no data documenting the existence of a similar, truncated hDcr protein have been reported. Nevertheless, using a human cell line (NoDice) derived by the genomic inactivation of the dicer1 gene in 293T cells by gene editing (Bogerd et al. 2014), we recently demonstrated that an analogous N-terminal truncation of the hDcr protein, called N1hDcr, is also significantly more capable than wild-type hDcr of generating siRNAs from long dsRNAs of synthetic or viral origin, resulting in a readily detectable level of functional siRNAs able to target either an indicator construct or an infecting virus (Kennedy et al. 2015).
Given our ability to demonstrate the reconstitution of the RNAi response in human cells using an hDcr mutant, we wondered whether we could achieve a similar effect by ectopic expression of the dDcr2 protein together with its known cofactors loquacious PD (Loqs-PD) and R2D2 (Liu et al. 2003; Zhou et al. 2009; Marques et al. 2010; Hartig and Forstemann 2011; Okamura et al. 2011). We report that dDcr2 can indeed give rise to siRNAs in human cells that can program human RISC to inhibit the function of target mRNAs. However, we were unable to demonstrate inhibition of influenza A virus (IAV) RNA expression by ectopic dDcr2 in infected human cells even though N1hDcr does exert a significant inhibitory effect.
RESULTS AND DISCUSSION
In Drosophila, siRNA biogenesis by dDcr2 is facilitated by two cofactors called Loqs-PD and R2D2, which function at the dsRNA cleavage step and RISC loading step, respectively (Liu et al. 2003; Zhou et al. 2009; Marques et al. 2010; Hartig and Forstemann 2011; Okamura et al. 2011). To ask if we could reconstitute RNAi in mammalian cells, we transfected human NoDice/ΔPKR cells, which lack functional forms of both the dicer1 and e1f2ak2 (pkr) gene (Kennedy et al. 2015), with plasmids expressing the FLAG epitope-tagged version of dDcr2 together with Loqs-PD and/or R2D2. As a positive control, we used the N1hDcr mutant form of hDcr that lacks the amino-terminal helicase domain and that we have previously shown is able to produce significant levels of functional siRNAs when overexpressed in the NoDice/ΔPKR cell line (Kennedy et al. 2015). As a source of dsRNAs for dicing by N1hDcr or dDcr2, we also cotransfected the previously described plasmid pCD-RLuc (Kennedy et al. 2015), which uses two RNA polymerase III (Pol III) promoters to convergently transcribe a 257-bp dsRNA derived from a Renilla luciferase (RLuc) cDNA.
As shown in Figure 1A, we were able to readily detect comparable levels of N1 hDcr, dDcr2, Loqs-PD, and R2D2 in transfected NoDice/ΔPKR cells. Analysis of siRNAs production by Northern blot revealed a substantial accumulation of siRNAs derived from pCD-RLuc in cells cotransfected with pK-N1hDcr (Fig. 1B, lane 2), as previously reported (Kennedy et al. 2015). In contrast, we observed far lower levels of siRNAs in cells coexpressing dDcr2, although a faint band was detected in NoDice/ΔPKR cells coexpressing both Loqs-PD and R2D2 (Fig. 1B, lane 6).
FIGURE 1.
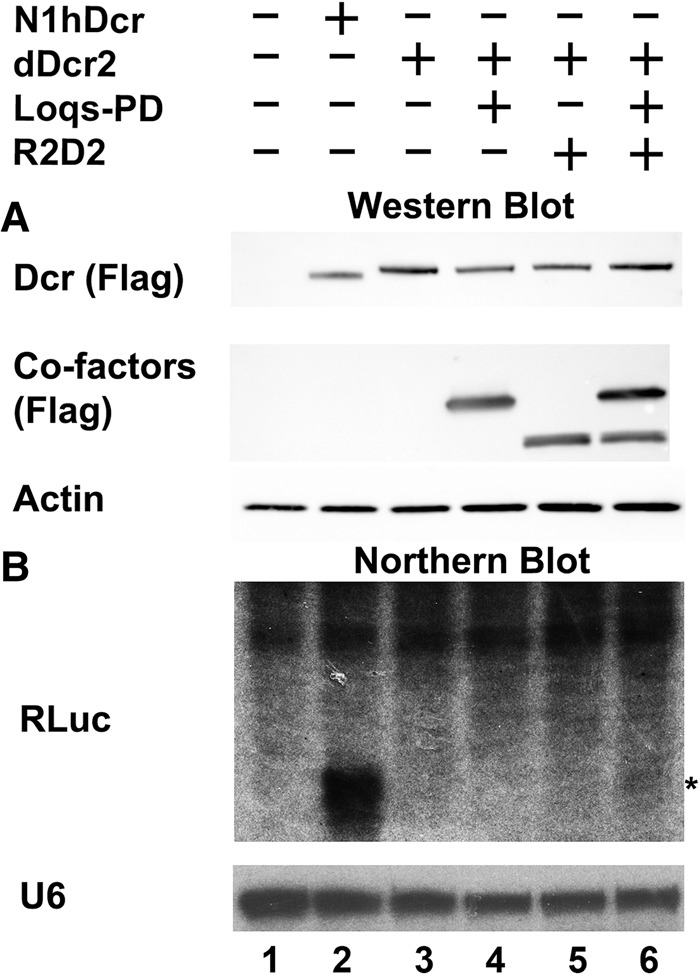
Expression of siRNAs in human somatic cells. NoDice/ΔPKR cells were cotransfected with pCD-RLuc, a previously described (Kennedy et al. 2015) construct that expresses a 257-bp dsRNA derived from RLuc, together with an empty expression vector (Neg) or vectors expressing N1hDcr (positive control) or expression constructs encoding dDcr2 and the Drosophila cofactors Loqs-PD and/or R2D2. (A) Shown in the top two panels are Western blots probing for the Flag epitope tag fused to N1hDcr, dDcr2, Loqs-PD, and R2D2. Endogenous β-actin was used as a loading control. (B) Northern analysis of NoDice/ΔPKR cells transfected as indicated at the top of the panel using a probe specific for the dsRNA insert present in pCD-RLuc. Endogenous U6 RNA was used as a loading control. The asterisk indicates the expected mobility of ∼21 nt siRNAs.
To get a better sense of the pattern of siRNA production in NoDice/ΔPKR cells expressing dDcr2, we repeated the experiment shown in Figure 1, but instead of harvesting small RNAs for Northern blot analysis, we subjected these RNAs to small RNA deep sequencing, as previously described (Kennedy et al. 2015). As expected, NoDice/ΔPKR cells transfected with pCD-RLuc alone failed to give rise to any siRNAs (Fig. 2A), while cells coexpressing N1hDcr gave rise to a substantial number of siRNA, derived from the pCD-RLuc insert, that were predominantly 20–22 nt in length (Fig. 2B).
FIGURE 2.
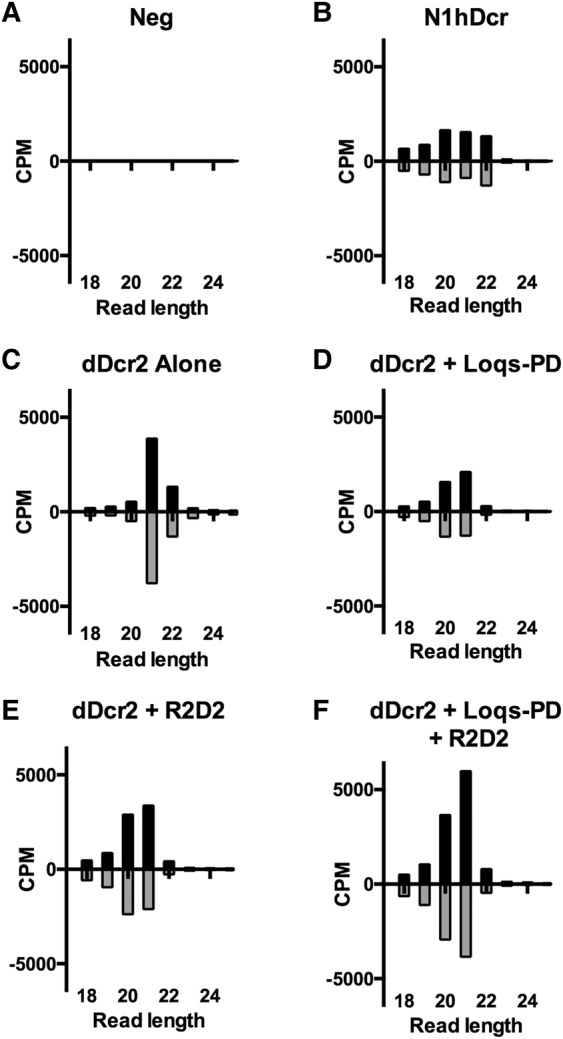
Detection of siRNAs generated by dDcr2 by deep sequencing. Small RNA reads, 18–25 nt in length, derived from the dsRNA encoded by pCD-RLuc, were identified by small RNA-seq. Reads are shown by length distribution, normalized to the total number obtained, and are given in counts per million (CPM). NoDice/ΔPKR cells were transfected with pCD-RLuc and with expression plasmids encoding the following proteins: (A) Negative control (Neg), (B) N1hDcr, (C) dDcr2, (D) dDcr2 plus Loqs-PD, (E) dDcr2 plus R2D2, (F) dDcr2 plus Loqs-PD, and R2D2.
Surprisingly, coexpression of dDcr2 alone gave rise to a strong peak of siRNAs with a size of predominantly 21 nt, although some siRNAs of 22 nt were detected (Fig. 2C). Coexpression of Loqs-PD did not significantly enhance siRNA production but did influence the size of the resultant siRNAs, which now revealed very few 22-nt siRNAs but instead included more siRNAs 20 nt in length (Fig. 2D). Coexpression of R2D2 had a similar effect, again enhancing the expression of siRNAs of 20 nt in length, while reducing the 22-nt siRNA level (Fig. 2E). Finally, coexpression of dDcr2, Loqs-PD, and R2D2 enhanced the overall production of siRNAs, which were again primarily 20 or 21 nt in length. While Northern analysis suggested that dDcr2, in the presence of Loqs-PD and/or R2D2, was significantly less effective at producing stable siRNAs than N1hDcr (Fig. 1B), this was not reflected in the number of siRNA reads obtained by deep sequencing, which was fairly comparable between all transfected NoDice/ΔPKR cultures except the negative control (Fig. 2), although the presence of both dDcr2 cofactors did enhance siRNA production, as determined both by Northern analysis (Fig. 1B) and deep sequencing (Fig. 2F). We hypothesize that this discrepancy may be explained by the fact that N1hDcr efficiently processes the endogenous pre-miRNA hairpins that accumulate in the NoDice cell line while dDcr2 is not capable of effectively processing pre-miRNAs (Lee et al. 2004; Bogerd et al. 2014). As the data shown in Figure 2 are normalized to the total number of small RNA reads obtained, the number of siRNA reads will clearly appear proportionately greater in the absence of any endogenous mature miRNAs.
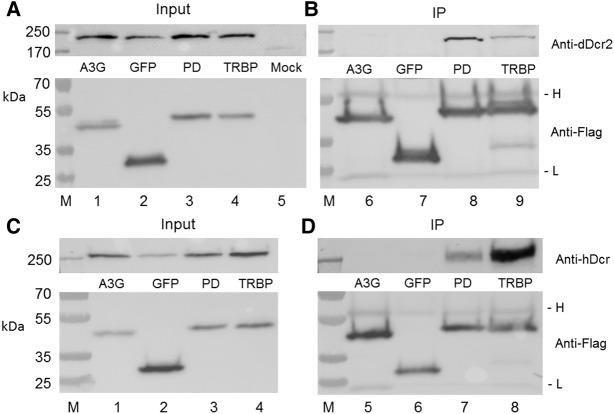
Previously, it has been reported that Loqs-PD is required for the initial dsRNA cleavage step in siRNA production in Drosophila (Zhou et al. 2009; Hartig and Forstemann 2011), so the ability of dDcr2 to produce significant levels of siRNAs in the absence of Loqs-PD was unexpected. We note however that the NoDice/ΔPKR cells used here express normal levels of the hDcr cofactor TRBP, which shows substantial protein sequence homology to both the PD and PB isoforms of Loqs (Hartig et al. 2009; Zhou et al. 2009). We therefore wondered if TRBP might retain the ability to bind to dDcr2 and facilitate its function. We therefore cotransfected NoDice/ΔPKR cells with plasmids expressing a His-tagged form of dDcr2 and plasmids expressing Flag-tagged forms of Loqs-PD as a positive control, green fluorescent protein (GFP) or APOBEC3G as negative controls, or human TRBP. We then immunoprecipitated the Flag-tagged proteins and asked whether dDcr2 would coimmunoprecipitate. As shown in Figure 3A, all of these proteins were expressed at readily detectable levels, as determined by Western blot, and moreover all the Flag-tagged proteins were clearly effectively immunoprecipitated (Fig. 3B, lower panel). As shown in Figure 3B (upper panel), neither GFP nor the RNA binding protein APOBEC3G were able to detectably interact with dDcr2, as expected (lanes 6 and 7). In contrast, we observed efficient coimmunoprecipitation of dDcr2 by Loqs-PD, again as predicted (lane 8). Importantly, we also observed a readily detectable coimmunoprecipitation of dDcr2 with human TRBP (lane 9), consistent with the hypothesis that TRBP is indeed facilitating dDcr2 function in human cells. We next performed the converse experiment, asking whether hDcr would be able to interact with not only TRBP but also Loqs-PD in human cells. As shown in Figure 3D (upper panel), we again observed a specific interaction of hDcr with not only its human cofactor TRBP but also with the Drosophila Loqs-PD (lanes 7 and 8). Therefore, these data clearly demonstrate that the specific interaction of dDcr2 or hDcr with their homologous cofactors Loqs-PD and TRBP has been evolutionarily conserved.
FIGURE 3.
Interaction of Loqs-PD and TRBP with dDcr2 and hDcr. This figure examines the ability of dDcr2 (panels A,B) or hDcr (panels C,D) to interact with Drosophila Loqs-PD (PD) or human TRBP in coexpressing human cells. (A, upper panel) Equivalent expression of His-tagged dDcr2 in cotransfected NoDice/ΔPKR cells (input). (Lower panel) Equivalent expression of Flag-tagged APOBEC3G, GFP, Loqs-PD, and TRBP in the cotransfected cells (input). (B, upper panel) Loqs-PD and TRBP, but not GFP or APOBEC3G, specifically coimmunoprecipitate dDcr2 (IP). (Lower panel) This shows equivalent loading of the Flag-Dynabeads with Flag-tagged APOBEC3G, GFP, Loqs-PD, and TRBP. (C, upper panel) Equivalent expression of hDcr in cotransfected NoDice/ΔPKR cells (input). (Lower panel) Equivalent expression of Flag-tagged APOBEC3G, GFP, Loqs-PD, and TRBP (input). (D, upper panel) Both Loqs-PD and TRBP specifically coimmunoprecipitate hDcr (IP). (Lower panel) Equivalent loading of the Flag-Dynabeads with Flag-tagged APOBEC3G, GFP, Loqs-PD, and TRBP. (M) Protein molecular weight markers with sizes indicated in kilodaltons (kDa) at the left of the figure. (H) Antibody heavy chain, (L) antibody light chain.
R2D2 has been proposed to function by facilitating the loading of siRNAs into Drosophila Ago2, which mediates RNAi, rather than Ago1, which facilitates microRNA (miRNA) function (Marques et al. 2010; Okamura et al. 2011). While mammalian Ago2 is also the only Ago protein able to mediate mRNA cleavage after binding of an siRNA-loaded RISC, evidence suggests that, at least in mammalian somatic cells, miRNAs are loaded equivalently into RISCs containing any of the four mammalian Ago proteins (Wang et al. 2012). We were therefore curious to see if siRNAs produced by dDcr2 would load into human RISCs and whether this was influenced by R2D2 expression. We therefore again repeated the experiment shown in Figures 1 and 2 but this time used a pan-Ago antibody to immunoprecipitate RISC, and its associated siRNAs and miRNAs, from the transfected NoDice/ΔPKR cells prior to deep sequencing (Whisnant et al. 2013; Kennedy et al. 2015). We then quantified the total number of siRNA reads derived from pCD-RLuc and normalized them to the run yield and the number of reads aligning to the miRNA hairpins.
As previously reported (Kennedy et al. 2015), the NoDice/ΔPKR cells gave rise to essentially no detectable siRNAs when transfected with pCD-RLuc alone but substantial numbers of RISC-associated siRNAs when N1hDcr was coexpressed (Fig. 4A). Analysis of RISC-associated small RNAs of pCD-RLuc origin in NoDice/ΔPKR cells expressing dDcr2 alone revealed very few RISC-associated reads. However, this number was slightly enhanced by coexpression of Loqs-PD and somewhat more so by coexpression of R2D2 (Fig. 4A). Finally, coexpression with dDcr2 of both Loqs-PD and R2D2 increased the level of RISC-associated siRNAs by approximately sixfold to ∼40% of the level seen with N1 hDcr (Fig. 4A). This suggests that R2D2 may enhance RISC loading in not only Drosophila cells but also in human cells and is consistent with previous results, from ourselves and others, arguing that not all siRNAs or miRNAs recovered by deep sequencing of the total small RNA population are necessarily RISC associated (Janas et al. 2012; Stalder et al. 2013; Flores et al. 2014).
FIGURE 4.
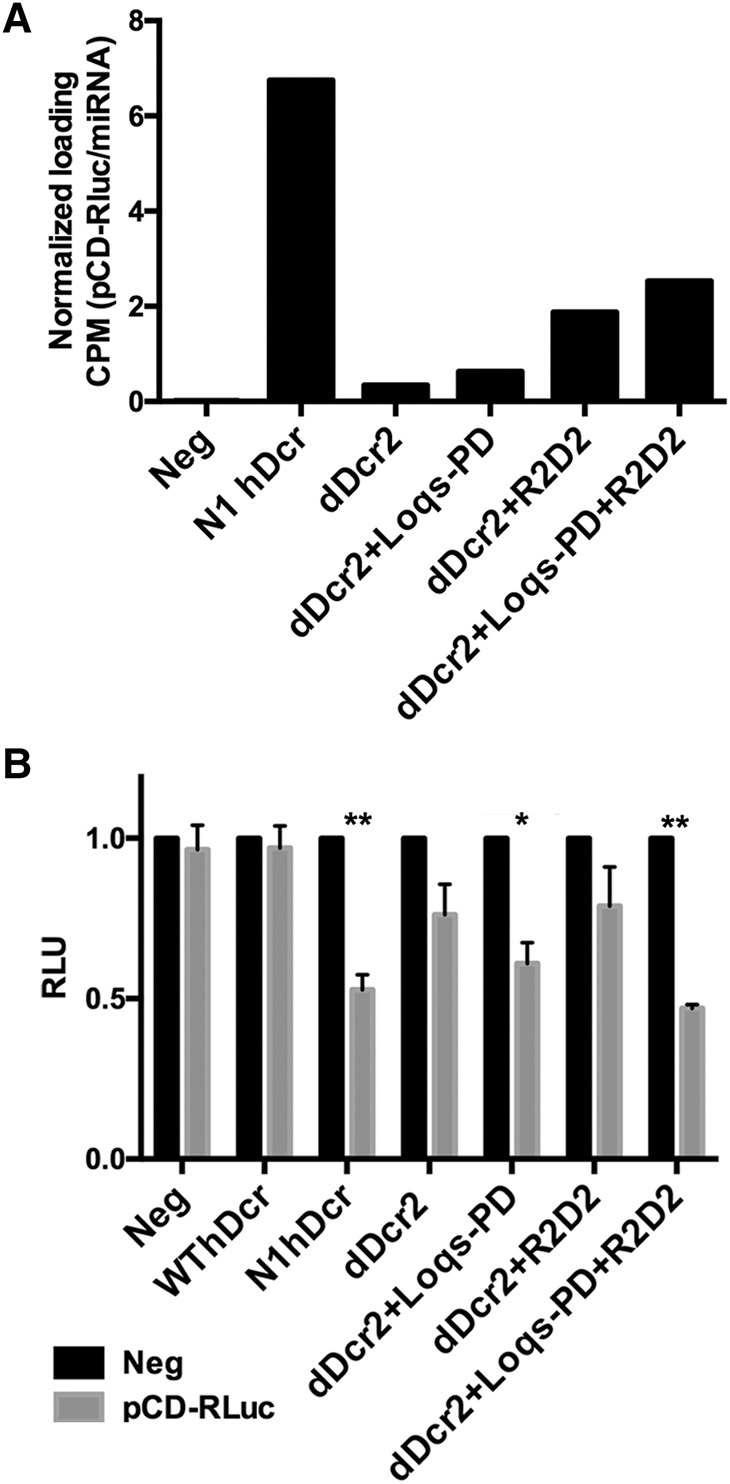
siRNAs generated by dDcr2 are loaded into human RISC and are functional. (A) RISC-associated small RNAs were recovered from NoDice/ΔPKR cells cotransfected with pCD-RLuc and pK-N1hDcr or combinations of pK-dDcr2 and vectors encoding dDcr2 cofactors. Immunoprecipitation was performed using a pan-Ago antiserum and the recovered RISC-associated RNAs subjected to small RNA deep sequencing. Reads that aligned to the pCD-RLuc dsRNA insert were computationally identified and normalized to the total number of reads of miRNA origin obtained, given as CPM. (B) Dual luciferase assays were performed with the indicator plasmid psiCheck2, which encodes RLuc and an FLuc internal control (the dsRNA expression vector pCD-RLuc targets RLuc). Assays were conducted as described previously (Kennedy et al. 2015) with the indicated combinations of expression constructs encoding N1hDcr or dDcr2, plus the dDcr2 cofactors. Average of three independent experiments with SD indicated. Statistical significance was determined using an unpaired, two-tailed t-test. (*) Significant repression (P < 0.05), (**) highly significant repression (P < 0.005).
We next wished to determine whether the RISC-loaded siRNAs produced by dDcr2 were functional. For this purpose, we again repeated the experiment shown in Figures 1, 2, and 4A but this time included an indicator plasmid, psiCheck, that encodes both RLuc and firefly luciferase (FLuc). As the dsRNA insert in pCD-RLuc is derived from an RLuc cDNA, we would expect any siRNAs produced to inhibit RLuc expression by RNAi while the FLuc mRNA, expressed from the same plasmid, would function as an internal control (Kennedy et al. 2015).
As shown in Figure 4B, and as previously reported (Kennedy et al. 2015), overexpression of the wild-type hDcr protein in NoDice/ΔPKR cells expressing the pCD-RLuc dsRNA insert did not induce repression of RLuc expression, when compared to the empty vector negative control, while a highly significant (P = 0.004) level of inhibition of RLuc expression was detected in cells coexpressing the N1hDcr mutant, which shows a significantly enhanced ability to generate siRNAs from dsRNA substrates (Kennedy et al. 2015).
Analysis of NoDice/ΔPKR cells cotransfected with pCD-RLuc, psiCheck, and the dDcr2 expression plasmid did not reveal a statistically significant repression of RLuc activity (Fig. 4B). Similarly, we did not observe significant repression of RLuc in cells coexpressing dDcr2 with R2D2, although a significant (P = 0.01) repression of RLuc expression was observed in NoDice/ΔPKR cells coexpressing dDcr2 and Loqs-PD. Finally, in the culture coexpressing dDcr2, Loqs-PD, and R2D2, we saw a highly significant (P < 0.001) repression of RLuc expression, consistent with the increased level of total (Figs. 1B, 2F) and RISC-associated (Fig. 4A) siRNAs seen when dDcr2 was expressed with both cofactors.
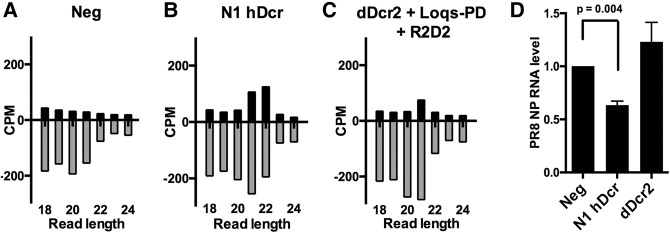
Previously, we have reported that ectopic expression of N1hDcr in NoDice/ΔPKR cells not only results in the production of a significant level of siRNAs of viral origin upon infection by the segmented, negative-sense RNA virus influenza A virus (IAV) but also induces a modest but significant reduction in IAV replication, as measured by viral RNA production. In contrast, ectopic expression of WT hDcr in NoDice/ΔPKR cells resulted in the production of substantially fewer siRNAs of IAV origin and did not lead to a detectable inhibition in IAV RNA expression (Kennedy et al. 2015). The ability of N1hDcr to generate siRNAs of viral origin in IAV infected NoDice/ΔPKR cells is reproduced in Figure 5B, and the resultant inhibition in IAV replication is confirmed in Figure 5D by qRT-PCR analysis of the level of expression of transcripts derived from the IAV nucleoprotein (NP) RNA segment. While expression of dDcr2, together with Loqs-PD and R2D2, did lead to the production of a low but statistically significant (P < 0.003, χ2 test) level of ∼21 nt siRNAs of IAV origin in infected NoDice/ΔPKR cells (Fig. 5C), this was not sufficient to produce any inhibition of IAV RNA expression (Fig. 5D).
FIGURE 5.
Analysis of IAV siRNA expression and function. Small RNA reads, 18–25 nt in length, derived from the plus or minus strands of the IAV genome, were identified by small RNA-seq. Reads are shown by length distribution, normalized to the total number of small RNA reads obtained, and are given in counts per million (CPM). NoDice/ΔPKR cells were mock transfected (A), transfected with pK-N1hDcr (B), or with plasmids expressing dDcr2, R2D2, and Loqs-PD (C). The cells were then infected with IAV at an MOI of 1 and total RNA harvested 24 h later, as previously described (Kennedy et al. 2015). (D) A qRT-PCR analysis of the relative level of expression of transcripts, of both plus and minus orientations, derived from the IAV nucleoprotein (NP) RNA segment at this time point. Average of three independent experiments with SD indicated.
The only previous paper that, to our knowledge, has examined whether dDcr2 could produce siRNAs in mammalian cells looked at the effect of dDcr2 and R2D2 (but not Loqs-PD) expression on the production and function of siRNAs of viral origin in wild-type 293T cells infected with Sindbis virus (SINV) (Girardi et al. 2015). This report identified a low number of potential siRNAs of viral origin that derived almost entirely from the SINV RNA plus strand and that were not of a discrete size. While the level of association of these putative siRNAs with RISC was not assessed, the authors reported that coexpression of dDcr2 and R2D2 did not result in a detectable reduction in SINV RNA expression. Here, we demonstrate, in contrast, that dDcr2 is capable of generating siRNAs from a coexpressed dsRNA in human cells lacking the hDcr enzyme (Fig. 2) and that the production and RISC-loading of siRNAs by dDcr2 is enhanced by coexpression of the Drosophila cofactors Loqs-PD and, especially, R2D2 (Figs. 2, 4A). However, the previously described (Kennedy et al. 2015) amino-terminal N1 truncation mutant of hDcr clearly generated a higher level of both total (Fig. 1B) and RISC-associated (Fig. 4A) siRNAs than did these Drosophila proteins, even though the observed expression levels were comparable (Fig. 1). Nevertheless, we were able to demonstrate, for the first time, that siRNAs produced by dDcr2 from an artificial dsRNA substrate in human cells, in the presence of Loqs-PD and R2D2, are not only loaded into RISC (Fig. 4A) but also functional (Fig. 4B), although the observed inhibition was a relatively modest approximately twofold. We did not, however, detect a significant inhibition of viral RNA expression in IAV-infected NoDice/ΔPKR cells upon expression of dDcr2 together with R2D2 and Loqs-PD, though a small number of siRNAs of viral origin were detected (Fig. 5). Therefore, although dDcr2 can produce functional siRNAs in human cells, this process is far less efficient than in insect cells even in the presence of both known dDcr2 cofactors, Loqs-PD and R2D2. The low level of siRNA production is clearly not due to the low level of expression of dDcr2 and its cofactors (Fig. 1), and it therefore appears likely that full reconstitution of RNAi in human cells using insect RNAi mediators will require additional components, for example the Drosophila Ago2 protein, and may indeed not be feasible given our currently incomplete understanding of how siRNAs are generated and function in virus-infected cells. Nevertheless, this report does document that an at least partial reconstitution of a functional RNAi response in human cells can be achieved using RNAi factors of insect origin.
MATERIALS AND METHODS
Molecular clones and cell culture
The dDcr2 expression vector (pK-dDcr2) was derived from a previously published construct (Bogerd et al. 2014). Briefly, an N-terminal FLAG epitope tag was fused in-frame with the dDcr2 open reading frame and cloned into the pK expression plasmid. A plasmid expressing a form of dDcr2 bearing an amino-terminal His tag was constructed using a similar approach. A plasmid expressing a FLAG-tagged form of the APOBEC3G protein was derived by substitution of the HA tag for the FLAG tag on a previously described APOBEC3G expression plasmid (Wiegand et al. 2004). Expression plasmids for the dDcr2 cofactors Loqs-PD and R2D2 have been previously described (Bogerd et al. 2014). The hDcr expression plasmid pKH3-Dcr-T7 has been previously described (Yi et al. 2005). The wild-type hDcr and N-terminally truncated N1hDcr expression constructs pK-hDcr and pK-N1hDcr have also been described previously (Kennedy et al. 2015). All RNAi indicator experiments were performed using the psiCheck-2 dual luciferase expression plasmid (Promega). The dsRNA expression construct pCD-RLuc, capable of generating siRNAs homologous to the RLuc mRNA, was described in Kennedy et al. (2015).
The human NoDice/ΔPKR cell line, obtained by inactivation of the dicer1 and eif2ak2 (pkr) genes in 293T cells by gene editing, was described previously (Bogerd et al. 2014, Kennedy et al. 2015). These cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (Gemini) and Gentamicin (Gibco).
Functional knockdown assays with dsRNA substrates
NoDice/ΔPKR cells were plated at 40% confluency in 10 cm2 dishes and cotransfected using polyethyleimine (PEI) with 3.75 µg of effector (pK, pK-hDcr, pK-N1hDcr, or pK-dDcr2), 3.75 µg of the RNAi substrate (pCD-Rluc) or a vector control (pSuper), 3.75 µg of a vector expressing a dDcr2 cofactor (Loqs-PD or R2D2) or a control vector (pcDNA3), and 3.75 µg of a plasmid expressing the Blasticidin resistance gene (pcDNA-Blast). Media was changed at 24 h post-transfection and Blasticidin selection initiated 24 h later. Following selection for 48 h, the surviving NoDice/ΔPKR cells were split into 12-well plates at 40% confluency and transfected with 10 ng of the psiCheck-2 RLuc and FLuc reporter using PEI. In addition, 250 ng of Dicer effector, the pCD-RLuc RNAi substrate, and dDcr2 cofactors were cotransfected along with the reporters. Cells were lysed 48 h later and luciferase levels quantified using a Dual-Luciferase Reporter Assay Kit (Promega).
Western and Northern blots
Expression of N1hDcr, dDcr2, and the dDcr2 cofactors was verified by Western blot using a mouse monoclonal anti-Flag antibody (Sigma-Aldrich) as previously described (Bogerd et al. 2014). siRNAs generated from the dsRNA insert encoded by pCD-RLuc were detected by Northern blot using a random primed probe generated from the insert in pCD-RLuc (Roche Random Primed Probe Kit). Briefly, NoDice/ΔPKR cells were plated in 15-cm2 dishes at 40% confluency and then cotransfected with 15 µg Dicer effector, 15 µg pCD-RLuc expression vector or pSuper control, and 15 µg of a pK-based vector encoding N1hDcr or dDcr2 in the presence or absence of dDcr2 cofactors, or a pcDNA3 vector control, using PEI. At 48 h post-transfection, cells were harvested and subjected to the mirVana small RNA isolation protocol (Ambion). Northern blots were then performed as previously described (Kennedy et al. 2015).
Dicer coimmunoprecipitation experiments
Of note, 4 × 106 NoDice/ΔPKR cells were transfected in a 10 cm2 dish with 7.5 µg of pcDNA-His-dDcr2 or pK-H3-Dcr-T7 and 7.5 µg of pcDNA-Flag-APOBEC3G, pK-FLAG-GFP, pcDNA-Flag-LoqsPD or pcDNA-FLAG-TRBP using PEI. Cells were harvested 72 h post-transfection and lysed in Binding Buffer (50 mM TRIS pH7.4, 150 mM NaCl and 0.5% NP40) supplemented with Protease Inhibitor Cocktail Tablet EDTA-free (Roche). Insoluble material was removed by centrifugation, RNase A (100 µg/mL) was added to the clarified cell lysate and an aliquot of lysate reserved as sample input.
Dynabeads Protein G (Invitrogen) were washed two times with Binding Buffer and incubated with 10 µL anti-Flag antibody (Sigma-Aldrich, F3165) per 50 µL beads per sample for 2 h. Beads were then washed three times with Binding Buffer to remove unbound anti-Flag antibody and the washed Flag-Dynabeads then incubated with the lysate samples while rotating overnight at 4°C. The next morning the samples were washed three times with Binding Buffer and bound proteins eluted by boiling after the addition of Laemmli sample buffer supplemented with 5% 2-mercaptoethanol. Input and IP samples were then analyzed by Western Blot. His-dDcr2 was detected using a mouse α-His monoclonal antibody (27-4710-01, GE Healthcare Life Sciences). The various flag-tagged proteins were detected using a mouse monoclonal anti-Flag antibody (F3165/Sigma-Aldrich). hDcr was detected using a rabbit polyclonal anti-Dicer antibody (H-212/Santa Cruz). After washing, the blots were incubated with anti-mouse-HRP (A9044, Sigma-Aldrich) or anti-rabbit HRP (NA934, GE Healthcare Life Sciences), washed again, and then incubated with the WesternBright Sirius Western blotting detection kit (K-12043 D20, Advansta). Chemiluminesence was visualized using the G:BOX Imaging System and GeneSys software (Syngene).
Small RNA deep sequencing
For the siRNA expression experiments, NoDice/ΔPKR cells were transfected with the pK-dDcr2 plasmid, in the presence or absence of plasmids expressing Loqs-PD and/or R2D2, or the pK-N1hDcr expression vector, plus the dsRNA substrate expression plasmid pCD-RLuc, at a ratio of 1:1. At 48 h post-transfection, cells were harvested and small RNAs purified using a mirVana miRNA isolation kit (Ambion). RIP-seq for detection of RISC-associated small RNAs was performed as described previously (Whisnant et al. 2013; Kennedy et al. 2015). All small RNA input was then processed with the TruSeq Small RNA Sample Preparation Kit (Illumina). cDNA was generated as previously described (Whisnant et al. 2014). Briefly, adapter-ligated RNA was reverse transcribed using SuperScript III (Life Technologies) and amplified using GoTaq green PCR master mix (Promega) with the TruSeq 3′ indices. DNA bands corresponding to ∼145 bp were isolated by gel electrophoresis on 10% polyacrylamide gels and sequenced on an Illumina HiSeq 2000.
Deep sequencing analysis
Small RNA reads were analyzed as previously described (Whisnant et al. 2014). Initial reads were quality filtered with cassava 1.8.2. Reads >15 nt were collapsed into FASTA format with the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) using the following pipeline: fastq_quality_filter –Q 33| fastq_to_fasta –Q 33 | fastx_clipper -a TruSeq-Index# -l 15 –c | fastx_collapser. All reads were then subject to alignment using Bowtie v.0.12.7 (Langmead et al. 2009) with the following options: –a --best --strata –m 25. The reads were sequentially aligned and filtered to adapter sequences, miRbase v. 20 annotations of miRNA hairpins or mature miRNA sequences (Kozomara and Griffiths-Jones 2014), Ensembl noncoding RNA database release 70 (Flicek et al. 2014), human functional RNA database version 3.4 (Mituyama et al. 2009), the vector or viral sequence of interest, and finally human genome 19. No mismatches were allowed for alignments to the different references except the human genome for which two mismatches were allowed. Bowtie alignments were then further analyzed with in-house Perl scripts to calculate read size distributions per run. Duplex partner analysis was also completed with a Perl script as described previously (Kennedy et al. 2015), and output of these scripts was used to calculate coverage normalized to reads per million.
Analysis of IAV RNA expression
To determine the relative levels of IAV RNA in the presence of Dicer effectors, NoDice/ΔPKR cells were first plated in 10 cm2 dishes at 40% confluency and transfected with pcDNA3-Blast and either pK (negative control), pK-N1hDcr, or pK-dDcr2 together with plasmids expressing the dDcr2 cofactors Loqs-PD and R2D2. The pcDNA3 base vector was used as the control for the plasmids expressing dDcr2 cofactors. Forty-eight hours post-transfection, the cells were subjected to Blasticidin selection. Following selection, transfected cells were split equally into six-well plates at a confluency of ∼50% and infected with the PR8 strain of IAV at a multiplicity of infection (MOI) of 1.0 for 4 h, as previously described (Kennedy et al. 2015). At that time, the supernatant media were removed and fresh media added. At 24 h post-infection, cells were harvested and subjected to TRIzol extraction of total RNA. qRT-PCR analysis of the relative level of expression of transcripts derived from the viral NP RNA segment was performed as previously described (Kennedy et al. 2015) using the ΔΔCt method.
DATA DEPOSITION
Sequencing data obtained during this research have been deposited with GEO and have been assigned accession number GSE83433.
ACKNOWLEDGMENTS
This research was funded in part by National Institutes of Health grant R21-AI113098. We thank Qinghua Liu, Nicholas Heaton, and Klaus Förstemann for reagents used in this research. We also thank the Duke Center for AIDS Research (CFAR) Biostatistics and Computational Biology Core for helpful discussion.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.059345.116.
REFERENCES
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci 98: 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. 2014. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA 20: 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, Zamore PD. 2011. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell 42: 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR, Cherry S, tenOever BR. 2013. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe 14: 374–378. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130: 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. 2013. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155: 807–816. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2014. Ensembl 2014. Nucleic Acids Res 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Kennedy EM, Skalsky RL, Cullen BR. 2014. Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res 42: 4629–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, Lefevre M, Chane-Woon-Ming B, Paro S, Claydon B, Imler JL, Meignin C, Pfeffer S. 2015. Cross-species comparative analysis of Dicer proteins during Sindbis virus infection. Sci Rep 5: 10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Forstemann K. 2011. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res 39: 3836–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Esslinger S, Bottcher R, Saito K, Förstemann K. 2009. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J 28: 2932–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas MM, Wang B, Harris AS, Aguiar M, Shaffer JM, Subrahmanyam YV, Behlke MA, Wucherpfennig KW, Gygi SP, Gagnon E, et al. 2012. Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA 18: 2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Whisnant AW, Kornepati AV, Marshall JB, Bogerd HP, Cullen BR. 2015. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc Natl Acad Sci 112: E6945–E6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. 2008. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol 380: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. 2013. Antiviral RNA interference in mammalian cells. Science 342: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW. 2010. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol 17: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K. 2009. The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res 37: D89–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Robine N, Liu Y, Liu Q, Lai EC. 2011. R2D2 organizes small regulatory RNA pathways in Drosophila. Mol Cell Biol 31: 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, Fritzsche A, Aeschimann F, Pfanzagl V, Basselet P, et al. 2013. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J 32: 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O'Loughlin E, Lee T, Houel S, O'Carroll D, Tarakhovsky A, Ahn NG, Yi R. 2012. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev 26: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH, Cullen BR. 2013. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio 4: e000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisnant AW, Kehl T, Bao Q, Materniak M, Kuzmak J, Lochelt M, Cullen BR. 2014. Identification of novel, highly expressed retroviral microRNAs in cells infected by bovine foamy virus. J Virol 88: 4679–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol 2: 70–75. [DOI] [PubMed] [Google Scholar]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J 23: 2451–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Wang X, Ding SW. 2010. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 8: 12–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tutton S, Pierce E, Yoon K. 2001. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol 21: 7807–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. 2005. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 11: 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ. 2009. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 15: 1886–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]