Abstract
Large-scale metabolic profiling is expected to develop into an integral part of functional genomics and systems biology. The metabolome of a cell or an organism is chemically highly complex. Therefore, comprehensive biochemical phenotyping requires a multitude of analytical techniques. Here, we describe a profiling approach that combines separation by capillary liquid chromatography with the high resolution, high sensitivity, and high mass accuracy of quadrupole time-of-flight mass spectrometry. About 2,000 different mass signals can be detected in extracts of Arabidopsis roots and leaves. Many of these originate from Arabidopsis secondary metabolites. Detection based on retention times and exact masses is robust and reproducible. The dynamic range is sufficient for the quantification of metabolites. Assessment of the reproducibility of the analysis showed that biological variability exceeds technical variability. Tools were optimized or established for the automatic data deconvolution and data processing. Subtle differences between samples can be detected as tested with the chalcone synthase deficient tt4 mutant. The accuracy of time-of-flight mass analysis allows to calculate elemental compositions and to tentatively identify metabolites. In-source fragmentation and tandem mass spectrometry can be used to gain structural information. This approach has the potential to significantly contribute to establishing the metabolome of Arabidopsis and other model systems. The principles of separation and mass analysis of this technique, together with its sensitivity and resolving power, greatly expand the range of metabolic profiling.
Metabolomics, the comprehensive analysis of metabolites present in a biological sample, has emerged as the third major path of functional genomics beside mRNA profiling (transcriptomics) and proteomics (Fiehn, 2002; Sumner et al., 2003). Metabolomic approaches seek to profile metabolites in a nontargeted way, i.e. to reliably separate and detect as many metabolites as possible in a single analysis. Combined with information on transcript and protein abundance, this would ideally lead to a nearly complete molecular picture of the state of a particular biological system at a given time.
A characteristic of plant life is the production of a vast number of natural compounds, often called secondary metabolites. Secondary metabolites have crucial roles in plant development as well as in the interaction of a plant with its biotic and abiotic environment (Kutchan, 2001). They are “secondary” only in the sense that in different plant species, different sets of metabolites occur (Pichersky and Gang, 2000). Ample evidence has been obtained in the past three decades for a wide range of functions of secondary metabolites. Arrays of antimicrobial compounds help defend against pathogens (Dixon, 2001; Hahlbrock et al., 2003) and deter herbivores. Secondary metabolites can function as signals internally or in communication with a symbiont (Peters et al., 1986). They provide protection against abiotic stresses such as UV light, drought, or high salt concentrations (Jin et al., 2000).
The majority of steps in the biosynthesis of secondary metabolites is assumed to be catalyzed by specific enzymes (Pichersky and Gang, 2000). Appropriately, completion of the Arabidopsis genome sequence revealed that a significant proportion of the approximately 25,000 predicted genes encode proteins assumed to function in secondary metabolism. The Arabidopsis genome contains >250 cytochrome P450 genes (The Arabidopsis Genome Initiative, 2000), >100 acyl transferase genes, >300 glycosyl transferase genes, >300 glycoside hydrolase genes (http://www.Arabidopsis.org/info/genefamily/genefamily.html), and a large number of genes encoding enzymes such as dioxygenases, O-methyl transferases, terpene synthases, or polyketide synthases. There is a huge discrepancy between the number of these genes and the number of known reactions catalyzed by these types of enzymes in Arabidopsis, leading to the conclusion that a large number of metabolites have yet to be identified. Some of them might even belong to compound classes that so far have not been known to occur in Arabidopsis at all (The Arabidopsis Genome Initiative, 2000). Thus, understanding a significant part of Arabidopsis biology requires methods allowing the sensitive detection and quantification as well as the identification of secondary metabolites. Applying such techniques to various genetic backgrounds, and to different environmental and developmental conditions then would help elucidate the function of such compounds and of the genes involved in their biosynthesis.
Profiling schemes for Arabidopsis and other plants have been developed in recent years (Roessner et al., 2000, 2001; Fiehn et al., 2000; Wagner et al., 2003). The main focus of these mostly gas chromatography (GC)-mass spectrometry (MS)-based approaches have been metabolites of the primary metabolism such as sugars, amino acids, organic acids, or sugar alcohols. Several hundred compounds can be robustly and reliably detected. However, these first pioneering reports already emphasize the need for complementary liquid chromatography (LC)-MS-based approaches to allow a more comprehensive profiling of metabolites (Roessner et al., 2000). The coupling of electrospray ionization (ESI) MS with capillary (Cap) electrophoresis (Soga et al., 2002) and hydrophilic interaction chromatography (Tolstikov and Fiehn, 2002) has been successfully applied to metabolomics problems. Every analytical procedure is necessarily limited as to what type of compounds can be separated and detected. GC-MS is predominantly applied to very polar or unpolar substances, and the main application range of LC-MS is more related to compounds of medium polarity. Furthermore, LC coupled to an MS technique providing soft ionization and high mass accuracy has the potential to generate information useful for the identification of unknown compounds because molecular ions and characteristic fragments can be detected (De Hoffmann, 1996; Niessen, 1999).
We initiated a project aiming at exploiting the potential of hybrid mass spectrometers developed in the past few years for the profiling of Arabidopsis metabolites. Here, we introduce an experimental system based on Cap LC coupled to ESI quadrupole time-of-flight MS (CapLC-ESI-QqTOF-MS). Using this approach, we were able to detect more than 800 mass signals (m/z values) in root extracts and more than 1,400 mass signals (m/z values) in leaf extracts of Arabidopsis. Electrospray as a soft ionization method combined with the enhanced resolution and mass accuracy of the TOF instrument and tandem MS make the identification of unknown compounds feasible, and examples are presented.
RESULTS
The major objective of the work described here was to develop a metabolite profiling scheme using the robust and well-established separation of extracts by LC on reversed-phase material in combination with state-of-the-art MS. The basic assumption was that such an approach would very well complement the pioneering GC-MS-based schemes by extending the range of metabolome analysis to those compound classes not amenable to GC analysis. These would include a significant fraction of the plant secondary metabolism.
A few years ago, QqTOF mass spectrometers were introduced. They combine TOF mass analysis with the established technique of ESI, resulting in high sensitivity, high mass resolution and high mass accuracy (Chernushevich et al., 2001), and are therefore principally well suited for comprehensive metabolite profiling. However, to our knowledge, no reports of such an application of hybrid mass spectrometers have yet been published.
Plant Growth, Extraction, and Cap LC
To be able to grow Arabidopsis with high reproducibility and to have easy access to leaf and root material, we established an aseptic hydroponic growth system. This more laborious approach limits throughput to some extent. The major benefit in addition to a more complete analysis of plant biology is that this system allows us to also biochemically profile the responses in a plant organ not directly exposed to a certain change in the environment.
Plant tissues were routinely harvested after 6 weeks and were frozen in liquid nitrogen. Freeze-drying was omitted to minimize loss of potentially susceptible compounds, such as volatiles, glucosinolates, and glycosides. Frozen plant material was homogenized in liquid nitrogen and was extracted twice with 80% (v/v) aqueous methanol. A third extraction did not produce any considerable gain in yield. Extracts were separated by Cap LC using C18 material. Flow rate and composition of the gradient had to be optimized to obtain good separation and maximum stability of the electrospray. A flow rate of 5.5 μL min–1 and a gradient starting from 5% (v/v) acetonitrile were found to produce the best results.
Manual Data Analysis and Reproducibility
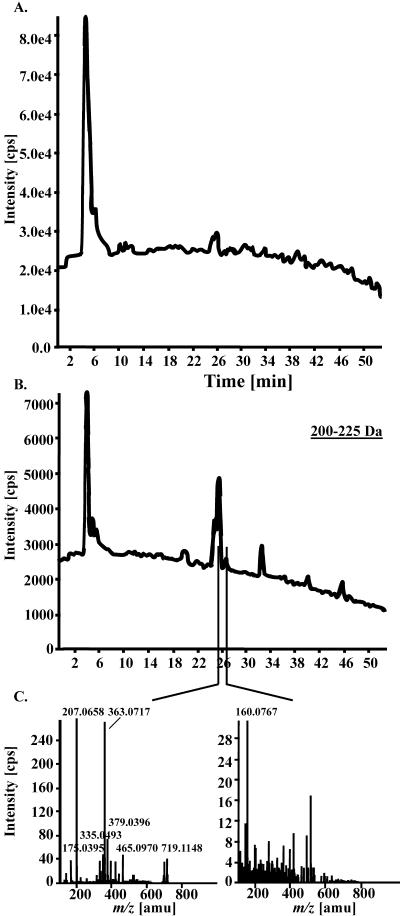
Cap LC was coupled to ESI-QqTOF-MS analysis operated in positive ion mode. Instrument parameters were set in such a way that the mass range from 106 to 1,000 D was monitored simultaneously. The signals generated by ions arriving at the detector were routinely summarized in a spectrum every 2 s for a total of 1,590 spectra during a 53-min run. The resulting total ion chromatogram did not resolve distinct peaks (Fig. 1A). Extensive data deconvolution was required to extract defined mass spectra. This was done manually by dividing the mass range of 106 to 1,000 D into mass windows of 25 D (Fig. 1B). Mass spectra were then extracted from peaks apparent within a particular mass window (Fig. 1C). Several hundred mass signals are extractable in this way from a single run.
Figure 1.
Manual deconvolution of data generated by CapLC-ESI-QqTOF-MS. A 53-min LC-MS analysis generated a total ion chromatogram consisting of 1,590 single MS spectra. A, The total ion chromatogram was manually deconvoluted in 25-D slices resulting in single extracted ion chromatograms (XICs) covering, for example, a mass range from 200 to 225 D. B, Mass spectra were then extracted from peaks apparent within a particular time frame of the XIC. The extracted peak at tR 26.0 min (C, left) represents sinapoyl malate ([M+Na]+ m/z 363.0717;[2 sinapoyl malate+K]+ m/z 719.1148; in-source fragment at m/z 207.0658). The extracted XIC valley at 26.6 min (C, right) represents a typical background mass spectrum.
At this stage of manual data deconvolution, quantifiability of mass signals, as well as the reproducibility of the mass spectrometric analysis, of the extraction and of the biological material were assessed. The isoflavone biochanin A was selected as an internal standard. A calibration curve was recorded, and 5 pmol was determined to lie within the range of linearity between amount and signal intensity. When added to a plant extract, the recovery rate of biochanin A was practically 100%. Twenty-one and 24 mass signals in root and leaf extracts, respectively, across the Cap LC run and representing a wide variety of intensities and masses were then randomly chosen (Tables I and II). Analysis of different dilutions of extracts and inspection of resulting calibration curves showed that, for most signals, the linear range covered almost two orders of magnitude. Up to 50-fold dilution resulted in a linear decrease in intensity. Untreated wild-type Arabidopsis Columbia (Col-O) plants were analyzed in four independent experiments. To obtain maximum mass accuracy, each run was recalibrated against five characteristic fragment or adduct ions of sinapoylmalate (Fig. 1C, left) in the case of leaf samples, and five characteristic fragment or adduct ions of hirsutin in the case of root samples. The selected signals were then quantified by integrating the peak areas using AnalystQS and by normalizing them to the fresh weight of the plant material extracted.
Table I.
Mass signals from root extracts used for assessing the variance of the CapLC-ESI-QqTOF-MS analysis
A total of 21 signals and the internal standard biochanin A were chosen. Listed here are their high-resolution (HR) masses and retention times (tR) as identifiers. Based on the mass, elemental compositions were calculated and the corresponding compounds were tentatively identified.
| HR Mass | tR | Elemental Composition (if available) | Compound | Substance Class |
|---|---|---|---|---|
| m/z | min | |||
| 138.1277 | 46.5 | C9H16N | 9-Methylthiononanenitrile [M + H-CH3SH]+ | Nitrile |
| 138.1277 | 26.5 | C9H16N | 9-Methylsulphinylnonanenitrile [M + H-CH4SO]+ | Nitrile |
| 160.0756 | 38.8 | C10H10NO | Methoxy-substituted indole | Indole |
| 160.0756 | 30.1 | C10H10NO | Methoxy-substituted indole | Indole |
| 179.0722 | 24.2 | |||
| 202.1251 | 26.5 | C10H20NOS | 9-Methylsulphinylnonanenitrile [M + H]+ | Nitrile |
| 234.0986 | 35.9 | C10H20NOS2 | 8-Methylsulphinyloctyl ITC (hirsutin [M + H]+) | Isothiocyanate |
| 256.0798 | 35.9 | C10H19NOS2Na | 8-Methylsulphinyloctyl ITC (hirsutin [M + Na]+) | Isothiocyanate |
| 277.1416 | 43.4 | |||
| 285.0756 | 40.7 | C16H13O5 | Biochanin A (internal standard) | Isoflavone |
| 317.1200 | 4.6 | |||
| 381.0858 | 4.7 | |||
| 429.2454 | 4.7 | |||
| 438.1396 | 24.0 | |||
| 441.1396 | 24.2 | C20H25O11 | Substituted ferulic acid glucoside? | |
| 457.1134 | 24.2 | C23H21O10 | Substuituted phenylpropane glucoside? | |
| 489.1708 | 35.9 | C20H38N2O2S4Na | 8-Methylsulphinyloctyl ITC (hirsutin. [2M + Na]+) | Isothiocyanate |
| 537.3151 | 43.4 | |||
| 553.2743 | 43.4 | |||
| 699.3608 | 38.8 | |||
| 810.2553 | 27.9 | |||
| 844.2019 | 28.0 |
Table II.
Mass signals from leaf extracts used for assessing the variance of the CapLC-ESI-QqTOF-MS analysis
A total of 24 signals and the internal standard biochanin A were chosen. Listed here are their HR masses and tR as identifiers. Based on the mass, elemental compositions were calculated and the corresponding compounds tentatively were identified.
| HR Mass | tR | Elemental Composition (if available) | Compound | Substance Class |
|---|---|---|---|---|
| m/z | min | |||
| 114.0371 | 23.8 | C5H8NS | 4-Methylsulphinylbutyl-ITC (sulforaphan, [M + H-CH4SO]+ | Isothiocyanate |
| 116.0706 | 4.9 | C5H10NO2 | Proline [M + H]+ | Amino acid |
| 170.0997 | 35.9 | C9H16NS | 8-Methylsulphinyloctyl ITC (hirsutin, [M + H-CH4SO]+ | Isothiocyanate |
| 200.0180 | 23.8 | C6H12NOS2Na | 4-Methylsulphinylbutyl-ITC (sulforaphan, [M + Na]+ | Isothiocyanate |
| 207.0652 | 25.4 | C11H11O4 | Sinapoyl malate [CVF: sinapic acid + H-H2O]+ | Phenylpropanoid |
| 285.0756 | 40.7 | C16H13O5 | Biochanin A (internal standard) | Isoflavone |
| 321.1960 | 30.3 | [M + H]+ or [M + H-H2O]+ | ||
| 349.2379 | 31.2 | |||
| 363.0687 | 25.4 | C15H16O9Na | Sinapoyl malate [M + Na]+ | Phenylpropanoid |
| 365.1536 | 31.2 | [M + H]+/2 | Glycoside | |
| 423.2202 | 49.9 | [M + H]+/2 | ||
| 431.2107 | 50.4 | [M + H]+/2 | ||
| 494.2567 | 45.0 | [M + H]+/2 | Glycoside | |
| 502.2501 | 45.0 | [M + H]+/2 | Glycoside | |
| 510.2358 | 45.0 | [M + H]+/2 | Glycoside | |
| 579.1708 | 23.7 | C27H31O14 | Kaempferol-3-O-a-l-rhamnopyranoside-7-O-a-l-rhamnopyranoside | Flavone glycoside |
| [M + H]+ | ||||
| 675.2524 | 31.4 | |||
| 703.1481 | 25.4 | C30H32O18Na | [2 Sinapoyl malate + Na]+ | Phenylpropanoid |
| 719.1220 | 25.4 | C30H32O18K | [2 Sinapoyl malate + K]+ | Phenylpropanoid |
| 735.0755 | 25.3 | |||
| 802.4232 | 47.6 | |||
| 815.4580 | 46.6 | |||
| 829.4661 | 50.3 | |||
| 965.5394 | 45.0 | [M + H]+ | Glycoside | |
| 984.5030 | 45.0 | [M + H]+ | Glycoside |
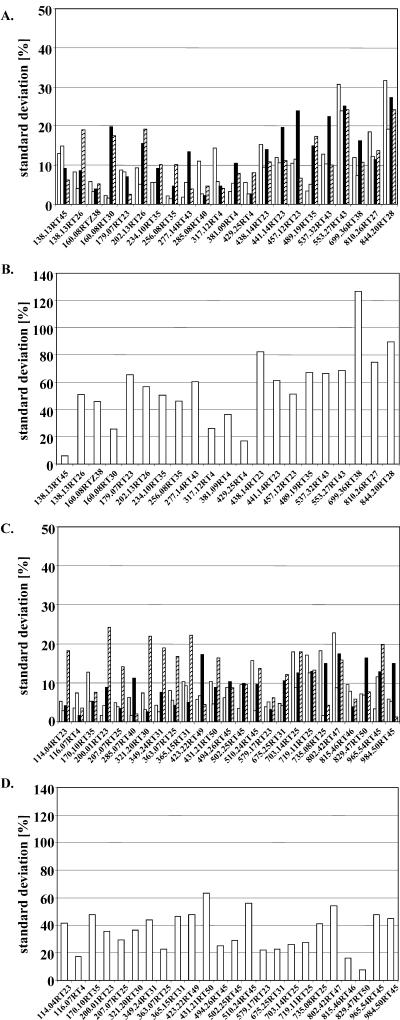
The reproducibility of the CapLC-ESI-QqTOF analysis was determined by extracting the same material three times and by running three times the same extract. The cumulative sd for repeated analysis of the same extract was 11.1% (Fig. 2, A and C). When different extracts of the same material were compared, an average variability of 25% ± 17.9% was found for leaves, and 21.6% ± 13.4% for roots. The numbers indicate that the technical reproducibility varies considerably, indicating a dependence on the mass signal in question. When four independent experiments were analyzed, an average biological variation was measured of 35.5% ± 14.0% for leaf signals and 55.9% ± 26.0% for root signals (Fig. 2, B and D). Again, the degree of variation was dependent on the mass signal.
Figure 2.
Technical and biological variance of the CapLC-ESI-QqTOF-MS analysis assessed by analyzing 25 mass signals from leaf extracts and 22 mass signals from root extracts. Signals are identified by mass and tR. Technical reproducibility of the LC-MS analysis of methanolic root (A) and leaf extracts (C) was determined by extracting the same material three times as well as performing three repetitions of the LC-MS measurements. Quantification of 22 selective ion traces in root extracts and 25 ion traces in leaf extracts was carried out. The sd of the three independent LC-MS analyses of a single extract is represented in the white, gray, and black bars of the charts. The cumulative sd for repeated analysis of the same extract showed a value of 11.1%. Comparison of the three different extracts of the same material (striped bar) resulted in an average variability of 25% ± 17.9% for leaves and 21.6% ± 13.4% for roots. When four independent experiments were analyzed, an average biological variation was measured of 35.5% ± 14.0% for leaf signals (D) and 55.9% ± 26.0% for root signals (B).
Signals were identified based on tR and mass. The stability of tR was determined for six mass peaks eluting between 25 and 51 min. Seventeen analyses of six different extracts were performed over a period of several days. The maximum sd was 0.3 min and the average sd was 0.25 min, indicating the stability of the Cap LC separation. The mass accuracy was evaluated by averaging the mass difference for 11 signals from known compounds. Signal intensities covered a range of about two orders of magnitude. The average mass difference through 18 runs was 9.0 ± 3.3 ppm.
Thus, we concluded at this stage that CapLC-ESI-QqTOF-MS can be a valuable tool for the robust and reproducible simultaneous detection and quantification of several hundred mass signals.
Identification of Known Compounds
As mentioned above, it has to be assumed that most of the Arabidopsis metabolites are as yet unknown. Moreover, unlike in the analysis of the major primary metabolites, standards are available for only very few compounds monitored by LC-MS and, in contrast to GC-MS, extensive mass spectral libraries do not exist for LC-MS. Therefore, being able to directly gain structural information on metabolites detected in profiling experiments is a key requirement for metabolomics. The QqTOF-MS technology has the potential to provide this information because the HR mass of a single ion can be used for calculation of a number of elemental compositions corresponding to those within a window of less than 30 ppm, usually 5 to 15 ppm. As shown, this mass accuracy is routinely achievable in profiling experiments with complex samples. A first evaluation of the potential of QqTOF-MS was carried out with the list of mass signals selected for the assessment of reproducibility. Elemental compositions were calculated. To further restrict the number of possible elemental compositions, the isotopic patterns of the ions of interest were checked for the presence of characteristic heteroatoms, such as sulfur, by comparing the calculated with the measured isotopic distribution. The possible presence of nitrogen can be deduced from even-mass numbered [M+H]+ and/or [M+Na]+ ions, respectively. With this information, a literature search for known natural products could now be performed using commercially available sources such as SciFinder, the Chapman & Hall Dictionary of Natural Products on CD-ROM, the NIST database, etc. Tables I and II list the information obtained on these peaks based on the exact mass and literature data. In leaf extracts, nine out of 24 signals could tentatively be identified, and in root extracts, 10 out of 21 signals could tentatively be identified. For six additional peaks in leaf extracts, it was found that they represent glycosides.
Automatic Data Deconvolution and Analysis
Using manual data deconvolution, we were able to demonstrate the potential of CapLC-QqTOF-MS for metabolomics with respect to sensitivity and robustness, as well as the ability to quantify and identify compounds. However, manual data deconvolution is time consuming and therefore obviously not acceptable for any type of metabolomic analysis that is aiming at a considerable throughput. To overcome this limitation, we tested and optimized MetaboliteID, a metabolite processing software (Applied Biosystems, Foster City, CA). MetaboliteID allows us to automatically extract mass spectra from the total ion chromatogram and to generate peak lists displaying tR, and mass and intensity of a peak. We first evaluated MetaboliteID by comparing its output with the results of manual data deconvolution. Following the optimization of data extraction parameters such as minimum signal strength (50 counts s–1, 2.5-fold higher than background) and XIC window width (0.35), we analyzed the same set of mass signals as before in the reproducibility experiments. The average variability calculated from the same data sets with the unsupervised method was only slightly higher than that determined manually. The technical variability changed from 25.0% to 34.1% for leaf extracts and from 21.6% to 26.9% for root extracts. Values for the biological variability changed from 35.5% to 40.7% and from 55.9% to 67.5% for leaves and roots, respectively. Thus, the cumbersome manual data deconvolution could be replaced by unsupervised automatic deconvolution without significant losses in reproducibility.
Next, the number of detectable mass signals was determined using the optimized MetaboliteID parameter settings. Through 72 runs of 24 extracts from four independent experiments, on average, 1,415 signals were detected in leaf extracts and 827 in root extracts. The sd was ±204 for the number of leaf signals and was ±180 for the number of root signals. When the same extract was analyzed three times, the numbers varied on average by only 5.8% for leaves and 5.5% for roots.
Tools are required for the processing of such large data sets. We decided to use Excel macros and programming in Visual Basic to establish automatic analysis of peak lists generated by MetaboliteID. As a first step, a procedure for establishing master lists for each analysis was developed. This was necessary to summarize ions that were detected more than once; for instance, because of problematic peak shape. The correct assignment of corresponding signals is essential for comparing different CapLC-ESI-QqTOF-MS runs. Identifiers of signals are the mass and the tR. The demonstrated accuracy and robustness of the analysis allowed us to define narrow mass windows of 0.02 to 0.06 m/z. Because the mass error increases slightly with increasing mass, we divided the mass range into three areas: m/z = 106 to 300 (mass window 0.02), m/z = 301 to 600 (mass window 0.03), and m/z = 601 to 1,000 (mass window 0.06). An adjustment of tR was introduced based on the simplifying assumption of a linear shift across a particular LC run. A routinely used tR window for signal identification is 0.4 min. When these windows were applied, it was possible to correctly assign corresponding signals from different CapLC-ESI-QqTOF-MS runs. Based on this, signal intensities can be averaged and compared between different experiments.
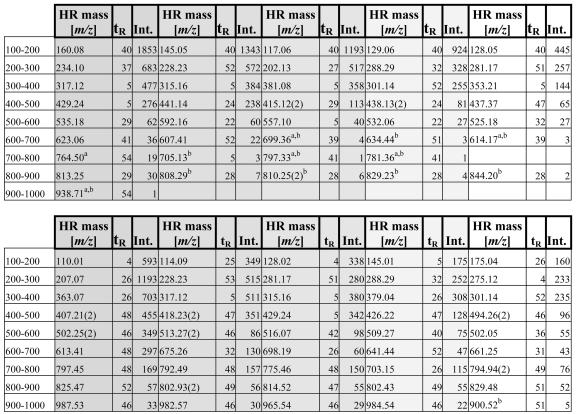
To provide a reference list for future use of this technology, we listed in Table III the five strongest signals in root and leaf extracts for each 100-D mass window between 100 and 1,000 D. Thirty-six MetaboliteID analyses of the reproducibility experiments were scanned for the mass signals that showed highest intensity (given as area per milligram) and were present in at least 75% of the LC-MS runs. For the mass ranges above 600 D, few signals are detectable in root extracts. In these cases, we included manually integrated signals that were below the routinely used MetaboliteID threshold of 50 counts s–1.
Table III.
List of the most abundant mass signals from root (top) and leaf (bottom) extracts incremented in 100-Da steps
From left to right and indicated by the shading, the five strongest signals are listed. HR mass (m/z) and tR (min) are used as identifiers. Intensities (Int.) are given as area per milligram of fresh weight. Doubly charged ions are marked with the charge state in brackets (2).
Example for Detection of Metabolic Differences between Samples
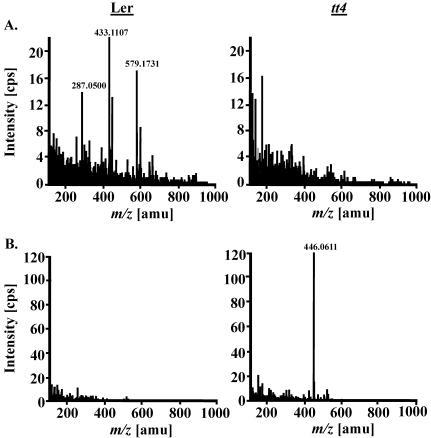
Having established rapid unsupervised automatic data extraction and processing, we assessed the applicability of our profiling scheme by analyzing a well-characterized mutant with a metabolic defect. Landsberg erecta (Ler) tt4 plants lack a functional chalcone synthase (Shirley et al., 1995) and are therefore deficient in flavonoid biosynthesis. Kaempferol glycosides are the main flavonoids biosynthesized by Arabidopsis plants under normal laboratory conditions (Veit and Pauli, 1999). When Ler and tt4 mutant plants were compared by CapLC-ESI-QqTOF-MS, the expected lack of kaempferol and its glycosides rha-rha-kaempferide and rha-kaempferide in leaves of tt4 plants was detected (Fig. 3A; Table IV). Additionally, two methoxy-substituted indole carboxaldehydes, probably the 1- and 4-methoxy isomers, 8-methylthiooctyl amine (m/z 176.1472; Kawabata et al., 1989), and probably a methoxy-substituted indole-glutathione conjugate, such as l-γ-glutamyl-S-[(1-methoxy-1H-indol-3-yl) methyl]-l-cysteinyl-Gly (m/z 467.1600; Bjergegaard et al., 2000), were found to be reduced in amount. However, both substances have not been obtained from natural sources. To also test the feasibility of handling very diverse data sets, we performed a comparison of metabolite composition of root and leaf tissue. Through nine samples each, the average overlap was only 8.7% (±1.1%).
Figure 3.
Mass spectra of two metabolites differentially accumulating in Arabidopsis wild-type (Ler WT) and mutant plants (Ler tt4 mutant). The flavonoid glycoside Rha-Rha-kaempferide ([M+H]+ at m/z 579.1731, in-source fragments at m/z 433.1107 and 287.0500) was not detected in leaves of the tt4 mutant line (A), whereas the glucosinolate gluconasturtiin ([M+Na]+ at m/z 446.0611) accumulated to much higher amounts in the roots of tt4 plants (B).
Table IV.
Mass signals differentially expressed between Ler wild-type and tt4 mutant plants Tentative identification is listed whenever possible.
| HR Mass | tR | Average Fold Change | Molecular Formula (if available) | Substance Class | Compound | |
|---|---|---|---|---|---|---|
| m/z | min | |||||
| Leaf, reduced | 287.0550 | 23.7 | 17 (+/-7) | C15H11O6 | Flavone Glycoside | Kaempferol [M + H]+ [m/z 579-2×146(Rha)] |
| 579.1708 | 23.8 | 88 (+/-12) | C27H31O14 | Flavone Glycoside | Rrha-rha-kaempferide [M + H]+ | |
| 433.1129 | 24.0 | 11 (+/-7) | C21H21O10 | Flavone Glycoside | Rha-kaempferide [M + H]+ [m/z 579-146 (Rha)] | |
| Leaf, higher | 661.3178 | 38.8 | 4.2 (+/-1.2) | |||
| 789.4299 | 49.4 | 4.2 (+/-1.5) | ||||
| Root, reduced | 116.0500 | 33.6 | 3 (+/-0.1) | C8H6N | Methoxy-subst. indole | Methoxyindolecarboxaldehyde |
| 133.0521 | 33.6 | 3.4 (+/-0.4) | C8H7NO | Methoxy-subst. indole | Methoxyindolecarboxaldehyde | |
| 144.0443 | 33.6 | 4.2 (+/-0.9) | C9H6NO | Methoxy-subst. indole | Methoxyindolecarboxaldehyde | |
| 176.0711 | 33.6 | 3.2 (+/-0.1) | Methoxy-subst. indole | Methoxyindolecarboxaldehyde | ||
| 176.1472 | 22.9 | 3.5 (+/-0.9) | C9H22NS | Amine | 8-Methylthiooctylamine | |
| 136.0579 | 5.3 | 4.7 (+/-0.1) | ||||
| 467.1600 | 23.7 | 5.1 (+/-0.9) | C20H27N4O7S | Methoxy-subst. indolyl-GSH conjugate | l-γ-glutamyl-S-[(1-methoxy-1H-indol-3-yl) methyl]-l-cysteinyl-glycine [M + H]+ | |
| Root, higher | 419.2765 | 52.1 | 11.8 (+/-3.2) | |||
| 423.0106 | 6.1 | 7.8 (+/-3.2) | S-Containing compound | |||
| 446.0555 | 26.6 | 581.5 (+/-418) | C15H21NO9S2Na | 2-Phenylethyl glucosinolate (gluconasturtiin [M + Na]+) |
Identification of Compounds by Tandem MS
Frequently, specific metabolites display changes correlated to a particular genetic background or elicited by an environmental or developmental stimulus. After detection of such changes by nontargeted metabolomic analysis, the identification of the respective metabolites is of key importance for biochemical phenotyping and gene function analysis. Additionally, identification is desirable to extend the catalog of known metabolites for Arabidopsis or any other plant species under study.
In-source fragmentation of compounds can already provide valuable structural information sufficient for identification. This was demonstrated above for the mass signals used in the reproducibility experiments (see Tables I and II). Tandem MS represents an additional powerful option. The first quadrupole of the mass spectrometer can be used as a mass filter and the second quadrupole can be used as a collision cell. Collision-induced dissociation (CID) yields fragments that can be used for elucidation of structures. In the following three examples from the method evaluation experiments are documented.
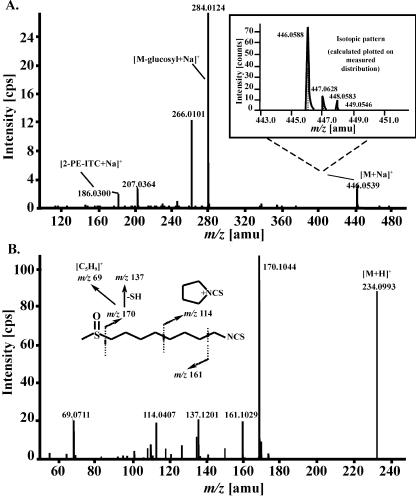
Roots of tt4 mutants were found to accumulate high amounts of gluconasturtiin, whereas only minor amounts were found in the corresponding root extracts of the Ler plants (Fig. 3B). The isotopic pattern of the observed ion at m/z 446.0623 ([M+Na]+) clearly indicates the presence of two sulfur atoms (see Fig. 4A). The CID spectrum is dominated by fragment ions at m/z 284 {[M+Na-162(glucosyl)]+}, m/z 266 ([m/z 284-H2O]+), whereas m/z 186 ([phenylethylisothiocyanate+Na]+) represents the phenylethyl moiety. Hirsutin was used for our root analysis as a stably occurring reference compound. It is known to arise as a myrosinase-catalyzed degradation product from its parent compound, 8-methylsulphinyloctyl glucosinolate (glucohirsutin). Both metabolites were originally obtained from the seeds of rock cress (Arabis hirsuta) by Kjær and Christensen (1958). EI-MS spectra were given by Kjær et al. (1963) as well as by Spencer and Daxenbichler (1980). The ESI-TOF mass spectrum displays a [M+H]+ ion at m/z 234 (see Fig. 4B). The CID spectrum displays a prominent key ion at m/z 170 ([M+H-CH4SO]+), also appearing as an intense in-source fragment, which is a group-characteristic ion of methylsulphinyl isothiocyanates (ITCs). Additional typical ions are m/z 161 ([M+H-CH3-NCS]+), which is a key fragment of mustard oils, and the stable cyclic immonium ion at m/z 114.
Figure 4.
Identification of significant metabolites of Arabidopsis by HR-ESI-MS/MS (product ion scan). A, Positive ion CID-MS of gluconasturtiin (in the box: calculated and measured isotopic pattern of the [M+Na]+ ion at m/z 446). B, Positive ion ESI-CID-MS of hirsutin.
We could confirm the expected reduction of kaempferol-3-O-α-l-rhamnopyranoside-7-O-α-l-rhamnopyranoside in tt4 mutants compared with Ler control plants. Its ESI-CID mass spectrum showed a subsequent loss of the two rhamnosyl moieties from the [M+H]+ ion at m/z 579 generating fragments at m/z 433 and 287, respectively, and was found to be identical with that of a standard sample (data not shown). The structure of the aglycon was established by comparing the CID-MS of m/z 287 with the [M+H]+ ion of authentic kaempferol. Thus, characteristic aglycon fragments are formed by subsequent losses of water and carbon monoxide, respectively, or from the retro-Diels-Alder reaction.
DISCUSSION
The current status of metabolomics can be viewed as being in some ways equivalent to the situation of sequencing programs such as the Human Genome Project around 1990 (Sumner et al., 2003). The enormous potential of comprehensive biochemical phenotyping for the functional analysis of biological systems is realized and numerous projects have been initiated. However, major technological limitations need to be overcome. For instance, the chemical diversity of the metabolome necessitates the use of different analytical techniques to cover the wide range of polarities found among the metabolites occurring in a cell.
Metabolomic approaches aim at monitoring the biochemical status of an organism by simultaneously measuring as many metabolites as possible. A robust and reproducible analysis that provides qualitative and quantitative data and allows high sample throughput is desired. Maybe equally important at this stage is to contribute to cataloging the metabolome of an organism. No metabolome is completely known as of yet. Systematic identification of the metabolites occurring in a species is particularly relevant for plants, given the wealth of natural products they produce. Sequence data indicate that Arabidopsis expresses a large number of enzymes for which substrates and products are unknown (The Arabidopsis Genome Initiative, 2000).
LC coupled to HR-MS has great potential to play an important role in metabolomics as a complement to GC-MS. However, reports of such a use are few, and thus far, most concern targeted profiling approaches (Lange et al., 2001; Huhman and Sumner, 2002). Our objective was to explore Cap LC coupled to state-of-the-art ESI-QqTOF-MS for the nontargeted profiling of Arabidopsis extracts. Reversed-phase LC on C18 was chosen because this was found to be suitable and highly reproducible for secondary metabolite profiling in a number of plant species including Arabidopsis (Graham, 1991). In principle, other techniques such as Cap electrophoresis could also be coupled to ESI-QqTOF-MS to further expand the range of metabolite profiling. Hallmarks of TOF-MS are the HR and high mass accuracy that make it an invaluable tool for the identification of compounds and its use for the sensitive simultaneous detection of large numbers of metabolites separable by LC on reversed-phase material. Major question marks concerning this approach were the robustness of the analysis, its reproducibility, and its suitability for high-throughput and quantitative analysis. Our results demonstrate that CapLC-ESI-QqTOF-MS principally meets all the necessary criteria for powerful metabolic analysis.
tRs of compounds eluting from the Cap LC column were found to show only little variation. Optimized flow rate and gradient composition produced a stable ion spray. Evaluation of the mass accuracy showed that the target value in the range of 5 ppm error (Chernushevich et al., 2001) can be reached for many signals even in a complex mixture of compounds, allowing, as discussed below, the calculation of elemental compositions and thereby the tentative identification of compounds. Obviously, the accuracy is correlated with signal strength and the error is higher for signals with intensities closer to background. Robustness of the analysis is illustrated by the fact that the average deviation in intensity for a range of mass signals was no higher than 11% when the same extract was run several times. Reproducibility of tR and mass accuracy provided a sufficient basis for the unequivocal detection and identification of mass signals.
Quantification of metabolites is a serious hurdle for metabolomics (Sumner et al., 2003). There are inherent limitations to the dynamic range of MS and, as emphasized by Trethewey et al. (1999), for nontargeted approaches, compromises have to be made concerning the quantification of metabolites. Dilution series of Arabidopsis extracts indicated a satisfactory linear range of the CapLC-ESI-QqTOF-MS analysis for many of the signals tested. However, the linear range is dependent on the metabolite in question, and there is currently no feasible way of exactly assessing it for every signal. As predicted by Fiehn (2002), we therefore see the immediate application of CapLC-ESI-QqTOF-MS predominantly in the highly sensitive and rapid detection of qualitative and also pronounced quantitative metabolic differences.
The potential for such a use is very high because of the large number of mass signals that can be resolved. On average, about 1,400 signals were detected in leaf extracts and about 800 signals were detected in root extracts. Given the limited overlap between root and leaf metabolites, a total of about 2,000 different signals can be detected in extracts from Arabidopsis plants grown under control conditions. Most likely, the numbers will be significantly higher once the analysis is extended to other tissues such as flowers (Chen et al., 2003), to different developmental stages, and to plants exposed to environmental stimuli. Increases in the number of detectable mass signals can be achieved by extending the analysis to the negative mode, which is less effective yet allows the measuring of metabolites not seen in positive mode, such as the intact glucosinolates or salicylic acid (data not shown).
The robustness of the analysis is again indicated by the stability of mass signal numbers when the same extract was analyzed repeatedly. The average variation was no greater than 6%. Because of in-source fragmentation and the formation of adduct ions, the number of mass signals detected is definitely higher than the number of metabolites. It is impossible at this stage to reliably estimate the actual number of metabolites detected.
Technical variance (as the sum of variation introduced by the extraction and by chromatography and mass spectrometric analysis) was below 30% for all but two of the chosen mass signals. This provides a solid basis for the detection of differences between samples. The biological variability of around 40% is similar to what has been reported for other large-scale profiling techniques. Technical and biological variance are dependent on the mass signal in question. Possible causes are, for instance, a coelution of ions that affects quantification through ion suppression and high metabolic fluctuation. Processing of a large number of profiling experiments will be required to eventually define the range of variability for individual mass signals.
The efficient use of CapLC-ESI-QqTOF-MS for gene function analysis implies a considerable throughput of samples. Automatic unsupervised deconvolution of MS data and processing of the extracted information is a must. After extensive optimization, we found the MetaboliteID software able to function sufficiently well for deconvolution. A comparison with the results from manual stepwise extraction and integration of a range of mass signals showed only small differences. Peak lists generated by MetaboliteID are further processed using Excel macros and Visual Basic programs. These allow us to assign peaks correctly based on tR and mass. The tools are tailored to discover qualitative and quantitative differences between samples and sets of samples. We tested the procedures by analyzing Ler wild-type and tt4 mutant plants. The expected difference in kaempferol glycosides due to the lack of a functional chalcone synthase was detected. In addition, we found several as yet unknown differences. Comparison between root and shoot samples demonstrated that very diverse data sets can also be handled. The limited overlap we found is further proof for the fundamental difference between root and leaf metabolism.
CapLC-QqTOF-MS is to be used for the detection and elucidation of metabolic changes elicited by environmental or developmental stimuli, as well as for the determination of metabolic differences attributable to a particular genetic background. Furthermore, substantial progress in establishing the metabolome of Arabidopsis and other model species is envisioned. All of these uses will rely on the potential to identify metabolites. We explored a nontargeted and a targeted way of gaining structural information. The highly accurate mass of the molecular ion and in-source fragments obtained during routine analysis can be used to tentatively identify metabolites with a good success rate. By systematically analyzing the mass data generated, we predict the ability to identify a large number of compounds that to date have not been known to occur in Arabidopsis. In selected cases of mass signals showing a change of immediate biological interest, CID can be used to generate fragments that, in many cases, provide a basis for an assignment of structure.
As is the case for other profiling schemes, aqueous methanol is used here for the extraction of Arabidopsis metabolites. Obviously, the choice of solvent limits the scope of the analysis, and the use of additional extraction methods is likely to result in the detection of further substance classes. A survey of the compounds tentatively identified in this study shows that the relevant known classes of Arabidopsis secondary metabolites such as indole-derived compounds (e.g. indole acetic acid derivatives), degradation products of glucosinolates (sulfinylnitriles and isothiocyanates), phenylpropanoids (sinapoylmalate), and flavonoids as well as their glycosides (e.g. kaempferol-3-O-α-L-rhamnopyranosid-7-O-α-L-rhamnopyranosid) can be detected by the method described. Indole derivatives, for instance, can be clearly classified by LC-ESI-MS/MS measurements according to their substitution pattern at the ring skeleton (e.g. methoxyindoles). The same is true for the ITCs (e.g. hirsutin). The example of gluconasturtiin, which contains two sulfur atoms, demonstrates the usefulness of the isotopic pattern as an additional feature to support the proposed elemental composition. Arabidopsis secondary metabolites not covered by this method are mono- and sesquiterpenoids, triterpenoid alcohols, phytosterols, waxes, and carotenoids.
MATERIALS AND METHODS
Plant Growth
Surface-sterilized seeds of the Arabidopsis ecotypes Col-O and Ler as well as of the tt4 mutant line were sown on agarose plugs (0.5%, w/v) and were grown hydroponically in one-tenth Hoagland nutrient solution No. 2 (pH 5.3–5.5; Sigma, St. Louis). The medium was supplemented with iron, chelated by N,N′-di-(2-hydroxybenzoyl)-ethylenediamine-N,N′-diacetic acid to a final concentration of 5 μm Fe-N,N′-di-(2-hydroxybenzoyl)-ethylenediamine-N,N′-diacetic acid (Chaney, 1988). Seedlings were grown for 6 weeks in hydroponic greenhouse boxes containing 10 plantlets each. The nutrient solution in the boxes was changed on a weekly basis and was aerated through 0.2-μm filters. Light conditions in the growth cabinet were fixed to 230 to 240 μE m–2 s–1 and a photoperiod of 8 h of light/16 h of dark at 23°C, day and night. Leaves and roots of plants were harvested separately approximately 1 h into the light period, pooled, and stored at –80°C.
Extraction and CapLC-ESI-QqTOF-MS
For the metabolite LC-MS analysis, freshly ground plant material (about 100 mg) was subjected twice to the following extraction procedure: mixing of the plant material with 200 μL of 80% (v/v) MeOH, sonication for 15 min (20°C–22°C), and centrifugation at 19,000g for 10 min. The extracts were combined, filtered (polytetrafluoroethylene filter, pore size of 0.2 μm), and analyzed by LC-MS. Positive LC-ESI-TOF mass spectra were recorded on an API QSTAR Pulsar Hybrid Quadrupole TOF instrument (Applied Biosystems) coupled to a capillary HPLC system (Ultimate; Dionex, Sunnyvale, CA). Typical MS instrument settings were an electrospray voltage of 5.5 kV with nebulizer gas being N2 and collision gas being N2, as well. The LC separation was performed on a Fusica C18 column (3 μm, 0.3 × 150 mm, PepMap; Dionex) applying a gradient system starting from 95% eluent A (0.1% [v/v] HCOOH/water) and 5% eluent B (0.1% [v/v] HCOOH/acetonitrile) to 95% eluent B in 45 min at a flow rate of 5.5 μL min–1 (sample injection volume: 1 μL). Formic acid was used because of its compatibility with MS analysis.
Data Analysis
Peak finding and quantification of selective ion traces was accomplished using the instrument's AnalystQS software. Automatic raw mass data deconvolution was performed using the MetaboliteID software (Applied Biosystems). Peak lists generated by MetaboliteID were further analyzed using Excel and Visual Basic.
CID-MS
2-Phenylethyl glucosinolate (gluconasturtiin): tR, 26.69 min; collision energy (CE), 20 eV; declustering potential (DP), 50 V; m/z (relative intensity, in percentages), 446.0556 (calculated for 446.0555: C15H21NO9S2Na, [M+Na]+, 10); 284.0124 (calculated for 284.0021: C9H11NO4S2Na, [M+Na-162(glucosyl)]+, 100); 266.0101 (calculated for 265.9922: C9H9NO3S2Na, [m/z 284-H2O]+, 45); 186.0300 (calculated for 186.0353: C9H9NSNa [2-phenylethyl ITC+Na]+, 8). 8-Methylsulphinyloctyl isothiocyanate (hirsutin, 8-MSOO-ITC): tR, 26.54 min; CE, 30 eV; DP, 50 V; m/z, 234.0993 (calculated for 234.0986: C10H20NOS2, [M+H]+, 80); 170.1044 (calculated for 170.0997: C9H16NS, [M+H-CH4SO]+, 100); 161.1029 (calculated for 161.0994: C8H17OS [M+H-CH3NCS]+, 18); 137.1201 (calculated for 137.1198: C9H15N, [m/z 170-SH]+, 20); 114.0407 (calculated for 114.0371: C5H8NS, 18); 69.0711 (calculated for 69.0698: C5H9, 19). Kaempferol-3-O-α-l-rhamnopyranoside-7-O-α-l-rhamnopyranoside: tR, 23.54 min; CE, 20 eV; DP, 50 V; m/z (relative intensity, in percentages), 579.1697 (calculated for 579.1708: C27H31O14, [M+H]+, 3); 433.1197 (calculated for 433.1129: C21H21O10, [M+H-146(rhamnosyl)]+, 100); 287.0538 (calculated for 287.0550: C15H11O6, [m/z 433 - 146]+, 31). Kaempferol (reference compound): CE, 40 eV; DP, 75 V; m/z (relative intensity, in percentages), 287.0557 (calculated for 287.0550: C15H11O6, [M+H]+, 66); 258.0514 (calculated for 258.0528: C14H10O5, [M+H-CHO]+, 14); 241.0503 (calculated for 241.0500: C14H9O4, [M+H-H2O-CO]+, 8); 213.0584 (calculated for 213.0547: C13 H9O3, [M+H-H2O-2CO]+, 21); 185.0602 (calculated for 185.0597: C12H9O2, [M+H-H2O-3CO]+, 10); 165.0206 (calculated for 165.0176: C8H5O4, [rings A&C-CO]+, 56) 157.0653 (calculated for 157.0653: C11H9O, [M+H-H2O-4CO]+, 14); 153.0188 (calculated for C7H5O4, [retro-Diels-Alder fragment from rings A & C]+, 100); 147.0439 (calculated for 147.0446: C9H7O2, 10); 137.0269 (calculated for 137.0238: C7H5O3, 14); 121.0339 (calculated for 121.0289: C7H5O2, +OC-C6H4OH (ring B), 13).
Acknowledgments
We thank Bernd Weisshaar and Joachim Kopka for valuable discussion, and Bernd Weisshaar for providing tt4 seeds. We also thank Claudia Horn and Kerstin Körber-Ferl for the excellent technical assistance.
This project has been part of the German plant genome project, GABI, and was supported by the Ministry of Research and Education. This work was additionally supported by the Deutsche Forschungsgemeinschaft (grant no. SCHE 235/11–1) and by the Fonds der Chemischen Industrie.
References
- Bjergegaard C, Buskov S, Sørensen H, Sørensen JC, Sørensen M, Sørensen S (2000) Reactions between glucosinolate products and thiol groups in food components. Czech J Food Sci 18: 193–195 [Google Scholar]
- Chaney RL (1988) Plants can utilize iron from Fe-N, N′-di-(2-hydroxybenzoyl)-ethylenediamine-N, N′-diacetic acid, a ferric chelate with 106 greater formation constant than FE-EDDHA. J Plant Nutr 11: 1033–1050 [Google Scholar]
- Chen F, Tholl D, D'Auria JC, Farooq A, Pichersky E, Gershenzon J (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernushevich IV, Loboda AV, Thomson BA (2001) An introduction to quadrupole-time-of-flight mass spectrometry. J Mass Spectrom 36: 849–865 [DOI] [PubMed] [Google Scholar]
- De Hoffmann E (1996) Tandem mass spectrometry: a primer. J Mass Spectrom 31: 129–137 [Google Scholar]
- Dixon RA (2001) Natural products and plant disease resistance. Nature 411: 843–847 [DOI] [PubMed] [Google Scholar]
- Fiehn O (2002) Metabolomics: the link between genotypes and phenotypes. Plant Mol Biol 48: 155–171 [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey R, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Graham TL (1991) A rapid, high resolution high performance liquid chromatography profiling procedure for plant and microbial aromatic secondary metabolites. Plant Physiol 95: 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Bednarek P, Ciolkowski I, Hamberger B, Heise A, Liedgens H, Logemann E, Nürnberger T, Schmelzer E, Somssich IE et al. (2003) Non-self recognition, transcriptional reprogramming, and secondary metabolite accumulation during plant/pathogen interactions. Proc Natl Acad Sci USA 100: 14569–14576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhman DV, Sumner LW (2002) Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59: 347–360 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata J, Fukushi Y, Hayashi R, Suzuki K, Mishima Y, Yamane A, Mizutani J (1989) 8-Methylsulfinyloctyl isothiocyanate as an allelochemical candidate from Rorippa sylvestris Besser. Agric Biol Chem 53: 3361–3362 [Google Scholar]
- Kjær A, Christensen B (1958) Isothiocyanates XXX: glucohirsutin, a new naturally occurring glucoside furnishing (-)-8-methylsulfinyloctyl isothiocyanate on enzymatic hydrolysis. Acta Chem Scand 12: 833–838 [Google Scholar]
- Kjær A, Ohashi M, Wilson JM, Djerassi C (1963) Mass spectra of isothiocyanates. Acta Chem Scand 17: 2143–2154 [Google Scholar]
- Kutchan TM (2001) Ecological arsenal and developmental dispatcher: the paradigm of secondary metabolism. Plant Physiol 125: 58–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Ketchum RE, Croteau RB (2001) Isoprenoid biosynthesis: metabolite profiling of peppermint oil gland secretory cells and application to herbicide target analysis. Plant Physiol 127: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen WMA (1999). Liquid Chromatography-Mass Spectrometry, Chapter 14: Natural Products and Endogenous Compounds, Ed 2. Marcel Dekker, New York, pp 465–500
- Peters NK, Frost JW, Long SR (1986) A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233: 977–980 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HW (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T (2002) Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem 74: 2233–2239 [DOI] [PubMed] [Google Scholar]
- Spencer GF, Daxenbichler M (1980) Gas chromatography-mass spectrometry of nitriles, isothiocyanates, oxazoloidinethiones derived from cruciferous glucosinolates. J Sci Food Agric 31: 359–367 [Google Scholar]
- Sumner LW, Mendes P, Dixon RA (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62: 817–836 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Tolstikov VV, Fiehn O (2002) Analysis of highly polar compounds of plant origin: combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal Biochem 301: 298–307 [DOI] [PubMed] [Google Scholar]
- Trethewey RN, Krotzky AJ, Willmitzer L (1999) Metabolic profiling: a Rosetta Stone for genomics? Curr Opin Plant Biol 2: 83–85 [DOI] [PubMed] [Google Scholar]
- Veit M, Pauli GF (1999) Major flavonoids from Arabidopsis thaliana leaves. J Nat Prod 62: 1301–1303 [DOI] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62: 887–900 [DOI] [PubMed] [Google Scholar]