Sugars are central to a plant's raison d'etre as products of photosynthesis. They are the ultimate source of energy and carbon skeletons for all biomolecules, and they provide the material out of which a plant builds its cell walls, fibers, and wood. Thus, regulation of any activity involved in biosynthesis of sugars, especially Suc (the major transport form of carbon in plants), is of utmost interest in understanding the growth and development strategies of a plant. Sugars are also potent regulators of gene expression, via e.g. a hexokinase (HXK) transduction mechanism that senses hexoses, or via Suc-specific or osmoticum transduction pathways, further underlying the importance of sugars in plant homeostasis.
WHY STUDY UDP-GLC (UDPG) PYROPHOSPHORYLASE (UGPASE)?
UGPase represents an important activity in carbohydrate metabolism, catalyzing a reversible production of UDPG and pyrophosphate (PPi) from Glc-1-P and UTP. In young and mature leaves, UGPase is primarily involved in the Suc biosynthesis pathway, providing UDPG for Suc phosphate synthetase (SPS), whereas in other tissues, including immature apical leaves, which to some extent depend on imported carbon, UGPase may take part in Suc breakdown, using UDPG produced by Suc synthase (SuSy; ap Rees, 1992; Winter and Huber, 2000). In certain sink tissues, e.g. cereal seed endosperm, the reverse reaction (use of UDPG) is coupled to the activity of cytosolic ADP-Glc pyrophosphorylase, resulting in an equimolar production of ADP-Glc from UDPG (Kleczkowski, 1994), the former being the sole immediate precursor of starch in all plants. This assures close interaction of the reactions of Suc metabolism and starch synthesis in this tissue.
Despite its critical positioning at the crossroads of Suc synthesis/breakdown, UGPase has received less attention than other enzymes of Suc metabolism. One of the reasons is that the enzyme is much more active than other activities in the Suc pathways and thus, as the argument goes, it is probably not regulating the flow of carbon to/from Suc. Moreover, antisense study of potato (Solanum tuberosum) plants found that even a 96% decrease of UGPase activity in tubers had no effect on sugar levels (Zrenner et al., 1993). However, recently, several lines of evidence point toward regulation of UGPase at gene and protein levels, and questions have been raised whether, under certain conditions, the enzyme is rate limiting. Also, crystallization of human UDP-N-acetylglucosamine pyrophosphorylase (AGX; Peneff et al., 2001), which is related to UGPase, has provided a basis for interpreting earlier function/structure studies on UGPase. The present review will cover recent advances in our knowledge on UGPase. More detailed information on earlier work can be found in Kleczkowski (1994).
HOW MANY UGPASE GENES IN PLANTS?
It has been assumed that UGPase is represented by a single UGP gene in plants. A systematic screening for UGP cDNAs in barley (Hordeum vulgare) has yielded only one type of transcript in leaves, embryos, and endosperm (Eimert et al., 1996). Also, small differences in cDNA sequence of UGPases in potato have previously been explained as resulting from allelic polymorphism (Sowokinos et al., 1997). However, from the Arabidopsis genomic database, it is now evident that there are two highly homologous UGP genes in this species (At3g03250 and At5g17310, located on chromosomes 3 and 5, respectively). Both genes are expressed, and the corresponding cDNAs are found in expressed sequence tag libraries (e.g. accession nos. T44798 and T43261, respectively). Two homologous UGP cDNAs were also found in poplar (Populus spp.) and rice (Oryza sativa), and in the slime mold Dictyostelium discoideum; in the latter, the genes were proposed to have different roles under different developmental stages, based on analyses of viable mutants (Bishop et al., 2002).
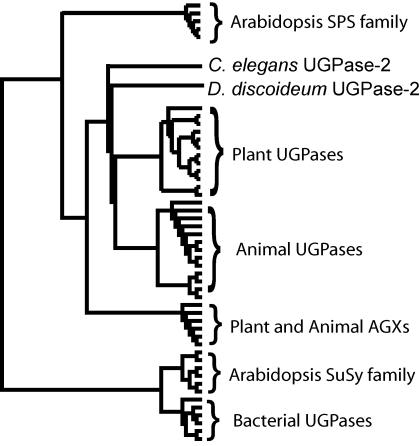
When analyzed at the derived amino acid level, there is 59% to 96% identity among plant UGPases (Katsube et al., 1990; Eimert et al., 1996; Pua et al., 2000; Abe et al., 2002) and approximately 55% identity with the corresponding proteins from slime mold (Bishop et al. 2002), mammalian liver (Konishi et al., 1993; Chang et al., 1996), and yeast (Saccharomyces cerevisiae; Daran et al., 1995). Eukaryotic UGPases are significantly divergent from those of bacterial origin, with very little or no amino acid sequence identity (approximately 8%; Fig. 1). Plant UGPases share 13% to 15% identity with AGXs from plants and animals, but have over 40% similarity when allowing for functional aa substitutions. Similarity is a better measure of a common function, whereas percentage of identity is a more useful measure of evolutionary relatedness (randomly aligned Arabidopsis proteins have approximately 5% identity and 10% similarity). There is also some relatedness (10%–16% identity and 20%–30% similarity) between plant UGPase and SPS (Eimert et al., 1996), suggesting a common early ancestor for those proteins. Interestingly, bacterial UGPase appears to be more related to SuSy (approximately 15% identity) than to plant UGPase (approximately 8% identity) or SPS (approximately 10% identity; Fig. 1), further underlying a complex evolutionary path for those enzymes.
Figure 1.
Cladogram of UGPase and related proteins. Plant and animal UGPases form separate clades, related to AGXs and, to a lesser degree, to SPS. There is little or no homology between eukaryotic and prokaryotic UGPases, but the latter share significant homology with the Arabidopsis SuSy family. UGPase-2 is the second divergent UGPase protein in C. elegans and slime mold, whereas UGPases-1 from both organisms fall within the animal UGPase clade. This figure was generated with Clustal-X (v 1.83) alignment and Tree-view (v 1.66).
SUC-SPECIFIC REGULATION OF UGP EXPRESSION
The expression of UGP from Arabidopsis leaves was strongly up-regulated by Suc, but not by Glc or osmoticum, and the Suc effect was, to a large extent, mimicked by exposing leaves to the light; the light effect was probably at least partly mediated by Suc formed during photosynthesis (Ciereszko et al., 2001b). Suc up-regulation was also observed for UGP from potato tubers (Spychalla et al., 1994). Suc-specific signaling may have different components from HXK-mediated Glc (hexose) signaling or osmoticum signal pathways (Rook et al. 1998, Loreti et al., 2001). Glc is likely to be a cell autonomous signal as it is not usually exported to other cells, whereas Suc is readily transported via the phloem.
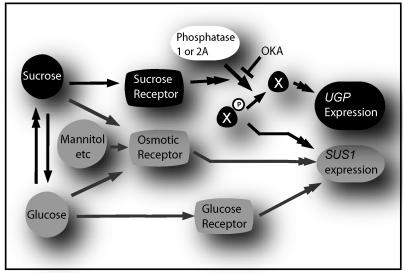
In Arabidopsis, the Suc effect on UGP was independent of HXK status, and was completely blocked by okadaic acid (OKA), a potent inhibitor of protein phosphatases (PP) 1 and 2A (Ciereszko et al., 2001b). This was accompanied by a marked OKA-dependent up-regulation of a SuSy (SUS1) gene that is prone to hexose (via HXK) and osmoticum regulation (Déjardin et al., 1999; Ciereszko and Kleczkowski, 2002). The OKA effects indicate that a phosphoprotein (X-P) that serves as a substrate for PP1 and/or PP2A is mediating the up-regulating signal for SUS1, whereas up-regulation of UGP requires dephosphorylation of X-P or some other phosphoprotein that acts as a substrate for PP1 and/or PP2A (Fig. 2). Thus, hexose (and/or osmoticum) and Suc-signaling pathways may intersect antagonistically, at least for SUS1 and UGP. The presence of distinct signaling pathways for UGP and SUS1 genes may represent a mechanism where UDPG is assured to be produced even if one of the pathways is inactive or blocked.
Figure 2.
OKA-sensitive regulation of UGP and SUS1 genes. The boxes and arrows represent hypothetical pathways for Suc, osmoticum, and Glc signaling. Double arrowheads represent multiple or unknown steps. An unknown phosphorylated intermediate (X) antagonistically regulates SUS1 or UGP gene expression depending on phosphorylation status. OKA inhibits PP1 and 2A, and prevents dephosphorylation of X. This probably causes accumulation of X-P and depletion of X, which in turn strongly up-regulates SUS1, and suppresses expression of UGP. The enzymatic interconvertability of Glc and Suc is indicated, both contributing also to osmotic status, similarly to some unmetabolizable compounds (e.g. mannitol). For simplicity, the same PP is indicated as important for the expression of UGP and SUS1; in fact, we cannot exclude that the two genes are regulated by distinct OKA-sensitive PPs.
STRESS REGULATION OF UGP EXPRESSION
In Arabidopsis, UGP expression and UGPase activity/protein content were strongly up-regulated by conditions resulting in phosphorus deficiency. The approaches included the use of mutants affected in inorganic phosphate (Pi) status, growth of wild-type plants on liquid media lacking Pi, and the feeding of Man to leaves to decrease internal content of Pi (Ciereszko et al., 2001a). The Pi stress did not result in significant increase in Suc in pho1 mutant (Pi deficient), suggesting distinct signaling mechanisms for transmitting P deficiency and Suc effects on UGP expression in this species. In pea (Pisum sativum) roots, UGPase protein increased after cadmium-excess stress, possibly reflecting Cd-induced Pi deficiency effects at the UGP expression level (Repetto et al., 2003). The UGPase step may constitute an important mechanism increasing Pi availability during P stress, at least in source tissues where UGPase is involved in Suc synthesis. The PPi released in the synthesis of UDPG by UGPase is hydrolyzed to Pi by pyrophosphatase(s), thereby increasing the availability of Pi for P-deprived plants.
Low temperature had a strong up-regulating effect on UGP from potato tubers (Spychalla et al., 1994; Borovkov et al., 1996) and leaves of Arabidopsis (Ciereszko et al., 2001b). In Arabidopsis, the cold signal was transmitted via an ABA-independent pathway, as found using aba mutants. Cold treatment frequently leads to sugar accumulation and, in potato, UGPase has been the main marker for processes leading to “cold sweetening”, i.e. accumulation of sugars when tubers are stored in the cold (Sowokinos et al., 1997). With UGPase being intrinsically involved in Suc metabolism, low temperature up-regulation of UGP may have direct effects on the “cold-sweetening” phenomenon. The UGP was also down-regulated by drought and flooding conditions (Ciereszko et al., 2001b). The latter strongly induced SUS2 gene from Arabidopsis leaves, with the concomitant increase in SuSy protein (Déjardin et al., 1999). Thus, it appears that flooding (O2 deprivation) may exert differential effects on UGPase and SuSy, suggesting that the two enzymes are not metabolically linked via UDPG under these conditions.
THE PREDICTED THREE-DIMENSIONAL STRUCTURE OF UGPASE AGREES WITH MUTANT FUNCTION DATA
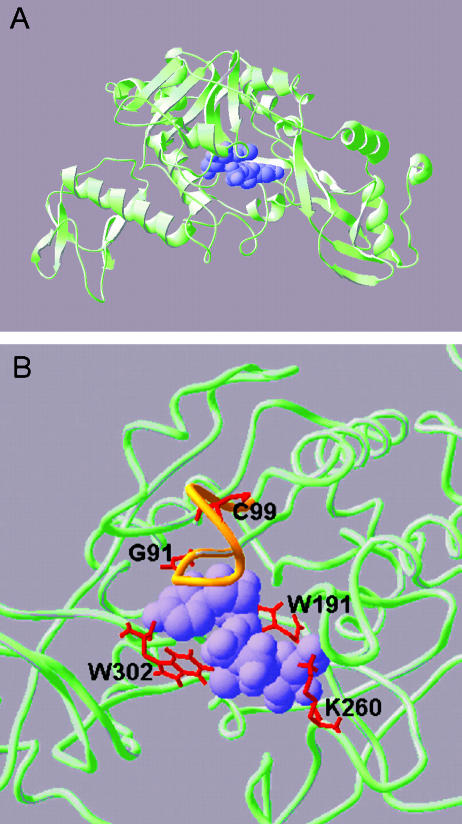
A major recent advance in studies on pyrophosphorylases has been the crystallization of human AGX1 (Peneff et al. 2001). This protein is functionally related and has approximately 40% protein sequence similarity to UGPases. A predicted structure for the UGPase consensus sequence (reference no. KOG2638) can be found in the National Center for Biotechnology Information conserved domain database (http://www3.ncbi.nlm.nih.gov/) and visualized with the cn3d program (available at www.ncbi.nlm.nih.gov/Structure/cn3d/). Swissmodel (http://swissmodel.expasy.org/) enables easy template-homology modeling and visualization using their DeepView program (http://swissmodel.expasy.org/spdbv/). The primary sequence for barley UGPase (GenBank accession no. X91347) was homology modeled using the crystal structure for AGX1 (Expasy file name IJV1.pdb) to produce a predicted three-dimensional structure in Figure 3. The visualization programs are very user friendly, and we recommend down-loading and viewing the key mutant residues in three-dimensional images rather than relying on the two-dimensional images we present here.
Figure 3.
Predicted three-dimensional structure of barley UGPase. The overall UGPase structure (A) was predicted using Swissmodel first approach method using human AGX-1 as a template fold and barley UGPase protein sequence. The model shows segments of UGPase as α-helices (curls), β-sheets (ribbons with arrows), and coils (wire tubes). The predicted location of the substrate (UDPG) is shown as a purple space-filled model. In the enlargement of the active site, shown in wireframe view (B), the nucleotide binding (NB) loop conserved in all pyrophosphorylases is colored orange. Residues shown by mutation to be critical to activity are shown in red and labeled in amino acid unicode (e.g. W = Trp, K = Lys) along with their position in the barley UGPase protein sequence. Illustrations were generated by Swissprotein Deepview (v 3.7).
The overall putative three-dimensional structure of UGPase monomer is bowl-like, with an active site positioned in a central groove (Fig. 3A). This shape is common for AGX and UGPase, and perhaps for all pyrophosphorylase-like proteins (Peneff et al. 2001). The active site of UGPase contains several amino acid residues that were shown to be important for substrate binding and catalysis of the enzyme (Fig. 3B). For instance, Trp-191, Trp-302, and Lys-260 (based on a barley UGPase amino acid sequence) were suggested to be involved in UDPG binding for potato and liver UGPase (Kazuta et al., 1991; Chang et al., 1996). In the derived structure of barley UGPase (Fig. 3B), the positive charge of Lys-260 may well stabilize the negative charge of the two phosphates of UDPG. In other studies, Cys-99 was implicated in PPi binding for the liver and barley enzyme (Chang et al., 1996; Martz et al., 2002), whereas in hamster lung fibroblasts, a mutation in a Gly residue corresponding to Gly-91 of barley UGPase resulted in a persistently low UDPG level (Flores-Diaz et al., 1997), consistent with the positioning of Gly-91 at the active site of the protein. Gly-91 and Cys-99 lie within what Peneff et al. (2001) refer to as the NB loop (Fig. 3B, labeled in orange), a conserved domain common for pyrophosphorylases. Cys-99 sits in the middle of this loop, and lies above the two phosphates of UDPG, an ideal position for binding and stabilization of the pyrophosphate bridge. On the other hand, Gly-91 sits on the edge of the NB loop, interacting with the uracil residue of UDPG.
OLIGOMERIZATION AS GENERAL REGULATORY MECHANISM FOR PYROPHOSPHORYLASES?
Recent work with barley UGPase has established that the protein readily oligomerizes and that only monomeric form of the enzyme is active. This was based on activity staining for forward and reverse reactions of UGPase (after native PAGE). Based on mutant studies, the deoligomerization step (formation of monomers) was found as rate limiting (Martz et al., 2002). A monomer is also the only active form of human AGX; a dimer of this enzyme was proposed to dissociate to monomers under assay conditions (Peneff et al., 2001). The dimerization process apparently modifies the structural environment of the active site, which is open in the monomer and occluded at the dimer interface. Oligomerization was also demonstrated as an important regulatory mechanism for ADP-Glc pyrophosphorylase from several species (Hendriks et al., 2003). Because a change in quaternary state of those distinct but functionally related proteins is coupled with changes in enzymatic activity, it is tempting to propose that the oligomerization phenomenon is a common regulatory feature of the pyrophosphorylase family.
UGPASE AND CELL WALL BIOSYNTHESIS
Any consideration of the role of UGPase should take into account that UDPG, the substrate/product of the enzyme, is a key metabolite for carbohydrate metabolism in photosynthetic and nonphotosynthetic tissues. In addition to being used in Suc pathways, UDPG is used, directly or indirectly, in the biosynthesis of cell wall polysaccharides, reflecting the key role of UDPG as a precursor for cell wall biogenesis (Gibeaut, 2000). Among UDPG's other roles is its involvement in the synthesis of carbohydrate moiety of glycolipids and glycoproteins (Flores-Diaz et al., 1997; Bishop et al., 2002).
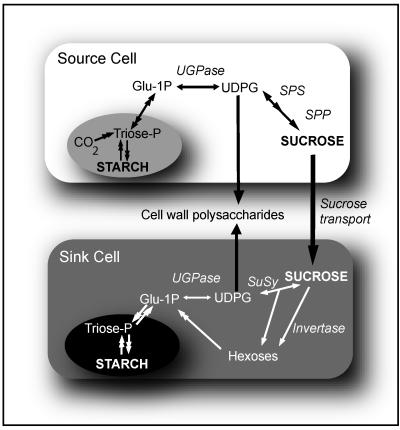
The prevailing view is that UGPase is localized mainly in the cytosol. Immunogold labeling of rice cells has revealed that most of the UGPase is located in the cytosol, but also to some extent in the amyloplast and Golgi (Kimura et al., 1992). Fractionation of rice and tobacco (Nicotiana tabacum) cells yielded some UGPase activity in the microsomes (Mikami et al., 2001), whereas in barley, high UGPase activities were found in a membrane fraction (Becker et al., 1995). The membrane association of UGPase is reminiscent of properties of SuSy, which has been proposed to bind to plasmalemma and to directly provide UDPG to cellulose synthase (Amor et al., 1995). Phosphorylation/dephosphorylation events are essential for binding of SuSy to the plasmalemma (Winter and Huber, 2000); the phosphorylated enzyme is fully soluble, and dephosphorylation promotes binding of SuSy to the membrane. Whereas there is no doubt that SuSy contributes strongly to cell wall biosynthesis (Amor et al., 1995; Ruan et al., 2003), the possible role of UGPase in this process still needs to be considered, especially in source tissues. Mature leaves under normal physiological conditions generally contain low, if any, SuSy protein (Déjardin et al., 1999), whereas UGPase activity in leaf extracts is well in excess of photosynthetic fluxes (Igamberdiev and Kleczkowski, 2000). In leaves, the overall carbon flow overwhelmingly favors production of UDPG from fixed carbon via the UGPase reaction (ap Rees, 1992; Kleczkowski, 1994; Fig. 4). On the other hand, for UGPase to be effective in making UDPG in sink tissues, there must be an effective supply of Glc-1-P. This may arise indirectly from Fru, a product of the SuSy reaction, which releases one Fru for every UDPG made during Suc hydrolysis, or from the invertase reaction (Fig. 4).
Figure 4.
Possible involvement of UGPase in cell wall synthesis in source and sink tissues. UGPase in photosynthetic (source) tissues generates UDPG, and works in conjunction with SPS and Suc-P phosphatase (SPP) to produce Suc. In a sink (e.g. roots), UGPase can work in conjunction with SuSy and/or invertases to break down Suc, but can also remobilize Glc-1-P back to UDPG. Please note that UDPG is used for cell wall polysaccharide synthesis in both source and sink tissues.
Cellulose and callose are produced from UDPG supplied directly to the plasmalemma, whereas other polysaccharides (e.g. hemicelluloses and pectins) are made in Golgi bodies and are then transported outside the plasmalemma via vesicular transport (Gibeaut, 2000). Activities providing UDPG for the latter processes must be Golgi associated or there should be some UDPG transporters present to allow transport of UDPG from cytosol through the Golgi membrane. In pea stems, UDPG was shown to be transported to Golgi vesicles in exchange for UMP and there was rapid incorporation of the Glc moiety of UDPG into endogenous acceptors (Neckelmann and Orellana, 1998). There is also a turnover of Glc in Golgi vesicles (Gibeaut, 2000) and likely a reassimilation into UDPG, which may be carried out via a Golgi-associated UGPase. The involvement of UGPase in cell wall synthesis was directly demonstrated in a yeast mutant where a 10-fold-reduced UGPase activity led to a decreased β-glucan content in cell walls (Daran et al., 1995). Also, cellulose-negative mutants of Acetobacter xylinum were deficient in UGPase (Valla et al., 1989). Those organisms do not contain SuSy activity, therefore the strong effects of UGPase deficiency on cell wall synthesis are probably not surprising. However, overexpression of UGP and SUS genes (single and double transformations) in tobacco resulted in a significant increase in height and an increased cellulose content in several single transformed lines and in all double transformants (Coleman et al., 2003). This suggested a correlation between the up-regulation of both genes and overall growth in tobacco. In DNA microarray studies on xylogenesis in the cambial region of poplar stems, a gene for UGPase was found to be up-regulated during the late cell expansion and secondary cell wall formation phases (Hertzberg et al., 2001). Also, in tissues undergoing heartwood formation in black walnut (Juglans nigra) trunks, UGPase activity was correlated with increased Suc breakdown occurring from late summer until early winter (Magel et al., 2001).
IS UGPASE A RATE-LIMITING STEP IN METABOLISM?
There is some confusion in the literature as to whether changes in UGPase content/activity bring about subsequent changes in metabolite flux. Zrenner et al. (1993) showed that UGPase activity in developing potato tubers could be reduced (by an antisense approach) by as much as 96% with no effect on carbohydrate metabolism. On the other hand, Spychalla et al. (1994) and Borovkov et al. (1996) failed to produce transgenic potatoes with an inhibition of enzyme activity greater than 30% and 50%, respectively, but this level of inhibition was sufficient to decrease sugar content in stored tubers. In Arabidopsis, a relatively small inhibition (30%) of UGPase activity in leaves of antisense plants led to a decrease in sugar content and, to some extent, to a decrease in starch level (Johansson, 2003), suggesting that the enzyme may play a rate-limiting role in carbohydrate synthesis. Given the presence of two UGP genes in Arabidopsis, the possibility cannot be excluded that the apparently conflicting results of antisense studies in potato may in fact reflect differential inhibition of UGP genes.
The extractable UGPase activity, especially in sink tissues (but also in leaves), is far in excess of carbon flow in plants (Igamberdiev and Kleczkowski, 2000), underlying the potential of UGPase for maintaining substantial fluxes, even if regulated in vivo. However, it is quite possible that under in vivo conditions, the UGPase activity is much lower and, in fact, is limiting to the metabolic pathway(s) that are connected to soluble sugar formation/degradation. For instance, the enzyme undergoes reversible oligomerization, with the deoligomerization step shown as rate limiting (Martz et al., 2002). Because only the monomer of UGPase is active, any process stimulating formation of dimers and higher order oligomers (e.g. binding to other proteins during in vivo metabolite channeling) may affect its enzymatic efficiency. Bacterial UGPase interacts with a GalF protein that modulates its activity in vivo, especially during stress conditions (Marolda and Valvano, 1996). In yeast, UGPase is regulated by phosphorylation by a Ser/Thr kinase, with the phosphorylation resulting in lower UGPase activity (Rutter et al., 2002). A similar observation was made for UGPase from spinach (Spinacia oleracea), but there the efficiency of enzyme phosphorylation was also modulated by binding to a 14:3:3 protein (Huber et al., 2000). Mammalian UGPase apparently undergoes O-glycosylation (Wells et al., 2003), but the significance of this modification is not clear at the moment. Plant UGPases contain several putative glycosylation and phosphorylation sites (Eimert et al., 1996). Although those motifs are not ultimate proofs that the posttranslational modification occurs in vivo, it seems possible that plant UGPase is prone to undergo posttranslational modifications that may affect its activation state before and after extraction, so the activity measured in leaf extracts overestimates that existing in vivo.
PROSPECTS
The UGP from Arabidopsis is one of a limited number of genes where Suc-specific regulation has been documented (Ciereszko et al., 2001b), and the transduction is likely to occur via a different pathway from those for hexose- or osmoticum-sensitive genes (Rook et al., 1998; Loreti et al., 2001; Moore et al., 2003; see also Fig. 2). The UGP genes can serve as useful tools for identifying mutants in transduction of the Suc signaling, using e.g. a negative selection marker fused to the UGP promoter. Based on observation that Suc up-regulation of UGP expression is disrupted by OKA (an inhibitor of PP1/2A; Ciereszko et al., 2001b), T-DNA mutagenized lines impaired in PP1/2A can be screened to identify plants where UGP expression is specifically abolished. DNA microarrays performed on selected plants that are impaired in Suc sensing or in specific PP1/2A involved in Suc signaling should reveal a global perspective concerning Suc sensing/signaling, i.e. allowing the identification of genes that are regulated by Suc and those involved in Suc signaling. The same concerns the possible use of UGP promoters to study P deficiency and low temperature transduction pathways.
The resolution of the crystal structure of AGX (Peneff et al., 2001) and its relatedness to plant UGPase (Fig. 3) should dramatically facilitate studies on structure/function properties of UGPase. We have now a model to test, e.g. via site-directed mutagenesis, to verify details of the plant enzyme catalysis and substrate binding, as well as oligomerization processes. Another milestone should be studies on the role of specific UGP genes, using e.g. gene-specific antisense or RNA interference technologies. This should resolve the long-standing question of the importance of UGPase for overall carbon flux in plants and whether it is rate limiting (Zrenner et al., 1993, Spychalla et al., 1994, Borovkov et al. 1996). Also, whereas the involvement of UGPase in cell wall biosynthesis in source tissues is rather clear (Fig. 4), its possible role in sink tissues (along with SuSy) needs more experimental evidence. Finally, the knowledge of posttranslational regulation of yeast and mammalian UGPases (Rutter et al., 2002; Wells et al., 2003) should allow more studies on similar regulatory mechanisms for plant UGPase.
CONCLUSIONS
Considerable progress has been made in studies on plant UGPase, one of key enzymes of Suc synthesis/breakdown. The UGPase genes are specifically up-regulated by Suc, involving an OKA-sensitive transduction component(s), and by a variety of abiotic stresses, including cold and P deficiency. The predicted protein structure of the UGPase monomer closely resembles a recently resolved crystal structure of a bowl-shaped AGX, with an active site in the central groove that includes several amino acids earlier shown as critical for catalysis/substrate binding. The UGPase is now recognized as existing as at least two isozymes, regulated by oligomerization and, possibly, phosphorylation. In source tissues, the enzyme provides UDPG for Suc and cell wall biosynthesis. Although present at high activity in plant extracts, the UGPase may represent a rate-limiting step in sugar metabolism in vivo.
This work was supported, in part, by the Swedish Research Council and by the Umeå University biotechnology fund.
References
- Abe T, Niiyama H, Sasahara T (2002) Cloning of cDNA for UDP-glucose pyrophosphorylase and the expression of mRNA in rice endosperm. Theor Appl Genet 105: 216-221 [DOI] [PubMed] [Google Scholar]
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP (1995) A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA 92: 9353-9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ap Rees T (1992) Synthesis of storage starch. In CR Pollock, JF Farrar, AJ Gordon, eds, Carbon Partitioning Within and Between Organisms. Bios Scientific Publishers, Oxford, pp 115-131
- Becker M, Vincent C, Reid JSG (1995) Biosynthesis of (1, 3)(1, 4)-β-glucan and (1, 3)-β-glucan in barley (Hordeum vulgare L.): properties of the membrane-bound glucan synthases. Planta 195: 331-338 [DOI] [PubMed] [Google Scholar]
- Bishop JD, Moon BC, Harrow F, Gomer RH, Dottin RP, Brazill DT (2002) A second UDP-glucose pyrophosphorylase is required for differentiation and development in Dictyostelium discoideum. J Biol Chem 277: 32430-32437 [DOI] [PubMed] [Google Scholar]
- Borovkov AY, McClean PE, Sowokinos JR, Ruud SH, Secor GA (1996) Effect of expression of UDP-glucose pyrophosphorylase ribozyme and antisense RNAs on the enzyme activity and carbohydrate composition of field-grown transgenic potato plants. J Plant Physiol 147: 644-652 [Google Scholar]
- Chang HY, Peng HL, Chao YC, Duggleby RG (1996) The importance of conserved residues in human liver UDP-glucose pyrophosphorylase. Eur J Biochem 236: 723-728 [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Hurry V, Kleczkowski LA (2001a) Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 212: 598-605 [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Kleczkowski LA (2001b) Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in Arabidopsis. Biochem J 354: 67-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko I, Kleczkowski LA (2002) Glucose and mannose regulate the expression of a major sucrose synthase gene in Arabidopsis via hexokinase-dependent mechanisms. Plant Physiol Biochem 40: 907-911 [Google Scholar]
- Coleman H, Ellis D, Gilbert M, Mansfield SD (2003) Increased growth and yield by altered carbohydrate allocation. In B Sundberg, ed, Tree Biotechnology 2003. UPSC Publishers, Umeå, Sweden, pp S10.41
- Daran JM, Dallies N, Thines-Sempoux D, Francois J (1995) Genetic and biochemical characterization of the UGP1 gene encoding UDP-glucose pyrophosphorylase from Saccharomyces cerevisiae. Eur J Biochem 233: 520-530 [DOI] [PubMed] [Google Scholar]
- Déjardin A, Sokolov LN, Kleczkowski LA (1999) Sugar/osmoticum levels modulate differential abscisic acid-independent expression of two stress-responsive sucrose synthase genes in Arabidopsis. Biochem J 344: 503-509 [PMC free article] [PubMed] [Google Scholar]
- Eimert K, Villand P, Kilian A, Kleczkowski LA (1996) Cloning and characterization of several cDNAs for UDP-glucose pyrophosphorylase from barley (Hordeum vulgare) tissues. Gene 170: 227-232 [DOI] [PubMed] [Google Scholar]
- Flores-Diaz M, Alape-Girón A, Persson B, Pollesello P, Moss M, Eichel-Streiber C, Thelestam M, Florin I (1997) Cellular UDP-glucose deficiency caused by a single point mutation in the UDP-glucose pyrophosphorylase. J Biol Chem 272: 23784-23791 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM (2000) Nucleotide sugars and glucosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiol Biochem 38: 69-80 [Google Scholar]
- Hendriks JH, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838-849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlen M, Teeri TT, Lundeberg J et al. (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA 98: 14732-14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Winter H, Toroser D, Athwal GS (2000) Metabolic regulation of nitrate and sucrose metabolism. Plant Cell Physiol 41S: 8 [Google Scholar]
- Igamberdiev AU, Kleczkowski LA (2000) Capacity for NADPH/NADP turnover in the cytosol of barley seed endosperm: the role of NADPH-dependent hydroxypyruvate reductase. Plant Physiol Biochem 38: 747-753 [Google Scholar]
- Johansson H (2003) Gene regulation of UDP-glucose synthesis and metabolism in plants. PhD thesis, Umeå University, Umeå, Sweden
- Katsube T, Kazuta Y, Mori H, Nakano K, Tanizawa K, Fukui T (1990) UDP-glucose pyrophosphorylase from potato tuber: cDNA cloning and sequencing. J Biochem 108: 321-326 [DOI] [PubMed] [Google Scholar]
- Kazuta Y, Omura Y, Tagaya M, Nakano K, Fukui T (1991) Identification of lysyl residues located at the substrate-binding site in UDP-glucose pyrophosphorylase from potato tuber: affinity labeling with uridine di- and triphosphopyridoxals. Biochemistry 30: 8541-8545 [DOI] [PubMed] [Google Scholar]
- Kimura S, Mitsui T, Matsuoka T, Igaue I (1992) Purification, characterization and localization of rice UDP-glucose pyrophosphorylase. Plant Physiol Biochem 30: 683-693 [Google Scholar]
- Kleczkowski LA (1994) Glucose activation and metabolism through UDP-glucose pyrophosphorylase in plants. Phytochemistry 37: 1507-1515 [Google Scholar]
- Konishi Y, Tanizawa K, Muroya S, Fukui T (1993) Molecular cloning and affinity labeling of bovine liver UDP-glucose pyrophosphorylase. J Biochem 114: 61-68 [DOI] [PubMed] [Google Scholar]
- Loreti E, De Bellis L, Alpi A, Perata P (2001) Why and how do plant cells sense sugars? Ann Bot 88: 803-812 [Google Scholar]
- Magel E, Abdel-Latif A, Hampp R (2001) Non-structural carbohydrates and catalytic activities of sucrose metabolizing enzymes in trunks of two Juglans species and their role in heartwood formation. Holzforschung 55: 135-145 [Google Scholar]
- Marolda CL, Valvano MA (1996) The GalF protein of Escherichia coli is not a UDP-glucose pyrophosphorylase but interacts with the GalU protein possibly to regulate cellular levels of UDP-glucose. Mol Microbiol 22: 827-840 [DOI] [PubMed] [Google Scholar]
- Martz F, Wilczynska M, Kleczkowski LA (2002) Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochem J 367: 295-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami S, Hori H, Mitsui T (2001) Separation of distinct compartments of rice Golgi complex by sucrose density gradient centrifugation. Plant Sci 161: 665-675 [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332-336 [DOI] [PubMed] [Google Scholar]
- Neckelmann G, Orellana A (1998) Metabolism of uridine 5′-diphosphate-glucose in Golgi vesicles from pea stems. Plant Physiol 117: 1007-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peneff C, Ferrari P, Charrier V, Taburet Y, Monnier C, Zamboni V, Winter J, Harnois M, Fassy F, Bourne Y (2001) Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J 20: 6191-6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua EC, Lim SSW, Liu P, Liu JZ (2000) Expression of UDP-glucose pyrophosphorylase cDNA during fruit ripening of banana. Aust J Plant Physiol 27: 1151-1159 [Google Scholar]
- Repetto O, Bestel-Corre G, Dumas-Gaudot E, Berta G, Gianinazzi-Pearson V, Gianinazzi S (2003) Targeted proteomics to identify cadmium-induced protein modifications in Glomus mosseae-inoculated pea roots. New Phytol 157: 555-567 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15: 253-263 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15: 952-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Probst BL, McKnight SL (2002) Coordinate regulation of sugar flux and translation by PAS kinase. Cell 111: 17-28 [DOI] [PubMed] [Google Scholar]
- Sowokinos JR, Thomas C, Burrell MM (1997) Allelic polymorphism of UDP-glucose pyrophosphorylase in potato cultivars and its association with tuber resistance to sweetening in the cold. Plant Physiol 113: 511-517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spychalla JP, Scheffler BE, Sowokinos JR, Bevan MW (1994) Cloning, antisense RNA inhibition and the coordinated expression of UDP-glucose pyrophosphorylase with starch biosynthetic genes in potato tubers. J Plant Physiol 144: 444-453 [Google Scholar]
- Valla S, Coucheron DH, Fjaervik E, Kjosbakken J, Weinhouse H, Ross P, Amikam D, Benziman M (1989) Cloning of a gene involved in cellulose biosynthesis in Acetobacter xylinum: complementation of cellulose-negative mutants by the UDPG pyrophosphorylase structural gene. Mol Gen Genet 217: 26-30 [DOI] [PubMed] [Google Scholar]
- Wells L, Whalen SA, Hart GW (2003) O-GlcNAc: a regulatory posttranslational modification. Biochem Biophys Res Commun 302: 435-441 [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Plant Sci 19: 31-67 [DOI] [PubMed] [Google Scholar]
- Zrenner R, Willmitzer L, Sonnewald U (1993) Analysis of the expression of potato uridinediphosphate-glucose pyrophosphorylase and its inhibition by antisense RNA. Planta 190: 247-252 [DOI] [PubMed] [Google Scholar]