During the course of its lifecycle, a plant may experience extremes of temperature and moisture, excesses and deficiencies of minerals, and challenges from herbivores and pathogens, all of which provide specific signals for altered plant growth and development. Ca2+ has been widely implicated as an intracellular messenger of physiologically and environmentally induced signaling pathways in plants (Trewavas and Malho, 1998). Because cellular Ca2+ levels are tightly regulated, small changes in intracellular Ca2+ can provide information for the modification of enzyme activity and gene expression needed for subsequent responses. Signals may trigger changes in the cellular Ca2+ signature (for example, an oscillation in the cytosolic free Ca2+ concentration), which is then perceived by various intracellular Ca2+ sensors/binding proteins to regulate a series of signaling cascades (Snedden and Fromm, 1998; Zielinski, 1998). Ca2+ sensors can be classified into sensor responders and sensor relays (Sanders et al., 2002). Upon binding Ca2+, sensor responders change their conformation and modulate their own activity or function through intramolecular interactions. In plants, the best-characterized sensor responders are the Ca2+-dependent protein kinases (Harmon et al., 2001). Ca2+-dependent protein kinases have protein kinase and calmodulin-like Ca2+- binding domains in a single protein, which allows their direct activation by Ca2+ (for review, see Cheng et al., 2002; Hrabak et al., 2003). In contrast, sensor relays such as calmodulin communicate the changed conformation to interacting partner(s) such as protein kinases, resulting in a change in kinase activity.
A number of mechanisms have been postulated to account for the wide-ranging role of Ca2+ in signaling in plants as well as for the specificity resulting from the Ca2+ signal. These include qualitative and quantitative information resident in the Ca2+ signal itself and in the Ca2+ sensors that perceive and translate the Ca2+ signal. The potential for different sensors to contribute to functional specificity is further magnified by the presence of families of interacting proteins such as protein kinases. This review focuses on recent progress in the characterization of the salt overly sensitive (SOS) 3 family of Ca2+ sensors and their associated SOS2 family of protein kinases. Recent genetic, molecular, and biochemical studies provide evidence that these two families of proteins interact differentially to form complexes that function in signaling pathways in Arabidopsis during growth and development and in response to abiotic stresses.
THE ARABIDOPSIS SOS3/SOS2 KINASE COMPLEX AND SOS SIGNALING PATHWAY
In a genetic screen designed to identify components of the mechanisms controlling salt tolerance in Arabidopsis, several SOS genes were identified. One of these genes, SOS3, encodes a novel EF-hand Ca2+ sensor (Liu and Zhu, 1998). SOS3 shares significant sequence similarity with the regulatory calcineurin B subunit from yeast (Saccharomyces cerevisiae) and neuronal Ca2+ sensors from animals (Klee et al., 1988). In spite of this similarity with calcineurin B at the primary sequence level, it is now clear that Arabidopsis does not have calcineurin and that the SOS3 Ca2+- sensing protein activates a protein kinase and not a protein phosphatase. SOS3 is a small myristoylated protein that appears to have no enzymatic activity by itself; Ca2+ binding and myristoylation are required for SOS3 function in salt tolerance (Ishitani et al., 2000). The SOS2 gene encodes a novel Ser/Thr protein kinase that also functions in salt tolerance in Arabidopsis (Liu et al., 2000). SOS2 contains an N-terminal kinase catalytic domain similar to that found in the Suc nonfermenting1 (SNF1) and AMP-activated (AMPK) kinases (Hardie, 1999) and a novel C-terminal regulatory domain. SOS3 has been shown to interact physically with SOS2 in yeast two-hybrid assays as well as in vitro (Halfter et al., 2000). Moreover, SOS3 activates SOS2 kinase activity in a Ca2+- dependent manner and sos3/sos2 double-mutant analysis also indicates that SOS3 and SOS2 function in the same pathway (Halfter et al., 2000). The first target of the SOS3-SOS2 regulatory pathway to be identified is the plasma membrane Na+/H+ exchanger (antiporter) encoded by the SOS1 gene. SOS1 gene expression during salt stress is partially controlled by SOS3 and SOS2 (Shi et al., 2000), and activation of SOS1 Na+/H+ antiport activity requires SOS3 and SOS2 (Qiu et al., 2002). Recent studies using yeast have provided additional evidence for the interaction between SOS3 and SOS2 and their regulation of SOS1. Coexpression of SOS1, 2, and 3 dramatically enhanced the salt tolerance of a yeast mutant in which all endogenous Na+ transporters had been removed (Quintero et al., 2002). The SOS3-SOS2 kinase complex phosphorylates and activates SOS1 expressed in yeast, enhancing Na+ exclusion and increasing NaCl tolerance (Quintero et al., 2002). Expression of a constitutively activated SOS2 mutant also increased salt tolerance in yeast expressing SOS1, implying that SOS2 kinase activity is partially sufficient for SOS1 activation. These results provided functional evidence that the SOS proteins function in the same signaling pathway that mediates ion homeostasis and salt tolerance in Arabidopsis (Fig. 1; Zhu, 2002). In addition, recent studies demonstrating that overexpression of SOS1 or of constitutively active SOS2 mutant kinases improves the salt tolerance of Arabidopsis (Guo et al., 2003; Shi et al., 2003) suggest that co-overexpression of SOS1, SOS2, and SOS3 may dramatically increase salt tolerance in plants.
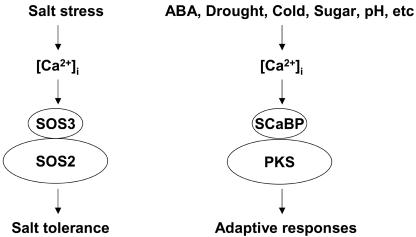
Figure 1.
Diagram showing that the SOS3-SOS2 signaling module functions in a salt stress-elicited Ca2+ signaling pathway, which mediates salt tolerance. Similarly, various SCaBP-PKS complexes have been implicated in Ca2+ signaling pathways in response to abscisic acid (ABA), sugar, high pH, or drought and cold stresses.
THE ARABIDOPSIS SOS3-LIKE Ca2+ BINDING PROTEIN (SCaBP Ca2+ SENSORS) AND SOS2-LIKE PROTEIN KINASE (PKS PROTEIN KINASE) FAMILIES
Since the initial isolation and characterization of the SOS3 Ca2+ sensor and its interacting protein kinase SOS2, many genes encoding SCaBPs and SOS2-like protein kinases (PKS) have been identified in Arabidopsis through the Arabidopsis Genome Initiative (2000; Hrabak et al., 2003). Summarized here is recent progress in our understanding of the functional domains, biochemical properties, interaction specificities, gene expression patterns, and physiological functions of these Ca2+ sensors and their interacting protein kinases.
Domain Structure of the SCaBP Calcium Sensors and PKS Protein Kinases
Six SOS3-like Ca2+ sensor/binding proteins (designated SCaBP1-6) were identified in Arabidopsis from a search of the GenBank database, and the corresponding cDNAs were isolated by reverse transcriptase-PCR (Guo et al., 2001). SCaBP1, SCaBP5, and SCaBP6 had been previously referred to as the calcineurin B-like proteins calcineurin B-like (CBL)2, CBL1, and CBL3, respectively (Kudla et al., 1999). Three additional SCaBPs have recently been identified from Arabidopsis (Luan et al., 2002). Like SOS3, these SCaBPs have no apparent enzymatic activity by themselves and thus are sensor relays (Sanders et al., 2002). These SCaBP proteins are predicted to possess three typical EF-hand Ca2+-binding motifs with the Ca2+-binding loops flanked by E and F helices (Guo et al., 2001; Luan et al., 2002). The tripeptide LYD at the junction of the E helix and the Ca2+-binding loop in the second EF-hand is deleted in the sos3-1 mutant allele (Liu and Zhu, 1998), and this sequence is conserved in the SCaBPs. A recent study of the three-dimensional structure of SCaBP1/CBL2 has shown that this protein contains two pairs of EF-hand motifs; one pair (the first and fourth) binds two Ca2+ ions, whereas the other pair (the second and third) remains in an open conformation (Nagae et al., 2003a, 2003b). Although there is currently no experimental evidence for SOS3 binding to Na+, the possibility cannot be excluded that SOS3 might serve as a Na+ sensor based on the ability of Na+ to bind within the EF-hand motifs of other proteins (Henzl et al., 2000; Ward et al., 2003).
Several SCaBPs, including SOS3 and SCaBP5, are associated with membrane fractions (Ishitani et al., 2000; Luan et al., 2002). This membrane localization is consistent with the idea that many Ca2+-signaling events are initiated by Ca2+ fluxes across membranes (Rudd and Franklin-Tong, 2001). At least three SCaBPs, SOS3, SCaBP4, and SCaBP5, contain a conserved N-myristoylation motif, MGxxxS/T[K] (Towler et al., 1988). This motif has been shown to be functional and is necessary for SOS3 function in salt tolerance (Ishitani et al., 2000) and for SOS3 to recruit SOS2 to the plasma membrane in yeast (Quintero et al., 2002; J.-K. Zhu and J.M. Pardo, unpublished data). It is possible that this cotranslational modification may help tether these Ca2+ sensors and their interacting protein kinases to specific membrane patches where Ca2+ flux takes place and/or where target proteins are localized or may contribute to pathway regulation by a Ca2+-myristoyl switch (Ishitani et al., 2000).
Twenty-three novel Arabidopsis SOS2-like protein kinases (PKS2-24, also known as CBL-interacting protein kinases) have also been identified using yeast two-hybrid assays and database searches (Guo et al., 2001; Hrabak et al., 2003). All of these proteins contain a conserved FISL motif (also known as an NAF domain), a 21-amino acid sequence in the regulatory domain (Fig. 2; Halfter et al., 2000; Albrecht et al., 2001; Guo et al., 2001). cDNAs were isolated for all PKS proteins and their sequences, and domain structures were compared. As had been found for SOS2, the deduced amino acid sequences of all PKS kinases contain an N-terminal kinase catalytic domain and a C-terminal regulatory domain. Analysis of the N-terminal kinase domain indicated that the PKS kinases are related to the SNF1 kinase from yeast and the AMPK protein kinases in animals (Hardie et al., 1998). Based on sequence similarity and domain structure, three subgroups of SNF1-related kinases (SnRK1-3) have been recognized in plants (Hardie, 2000; Hrabak et al., 2003); the PKS proteins are members of the plant-specific SnRK3 group (Halford et al., 2000; Hrabak et al., 2003).
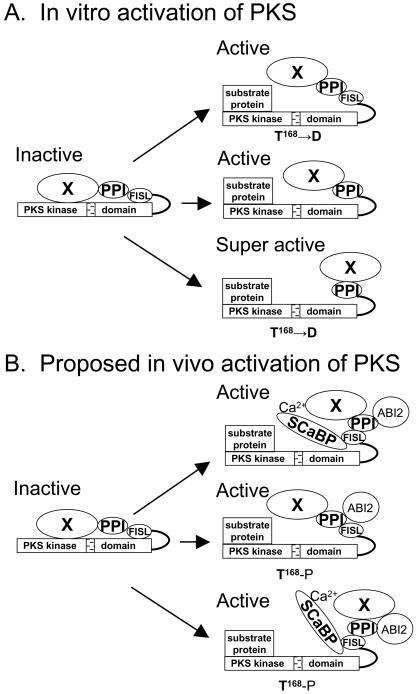
Figure 2.
Mechanisms of activation of PKS kinases in vitro and in vivo. PKS proteins consist of an N-terminal kinase catalytic domain (PKS kinase) and a C-terminal regulatory domain. An activation loop (stippled bar) is shown in the kinase catalytic domain, whereas the regulatory domain contains a FISL motif (for interaction with SCaBPs), a PPI motif (for interaction with protein phosphatase 2Cs), and an unknown functional motif(s) at the C-terminal end (indicated by X). The kinase is proposed to be kept inactive by an intramolecular interaction between the catalytic and regulatory domains. In vitro, the kinase can be activated by a T to D mutation in amino acid 168 to mimic phosphorylation of the activation loop by a putative upstream kinase (A, top panel), by deletion of the autoinhibitory FISL motif (A, middle panel), or by a combination of the two mutations (A, bottom panel). In vivo, the kinase is proposed to be activated by the binding of Ca2+-SCaBP to the FISL motif (B, to1p panel), by phosphorylation of T168 in the activation loop (B, middle panel), or by a combination of both (B, bottom panel).
An alignment of the deduced amino acid sequences of all 24 PKS kinases shows that the N-terminal kinase domains of these proteins are highly conserved and contain a putative “activation loop” (Guo et al., 2001; Gong et al., 2002b; Fig. 2A). The conserved FISL motif is found in the regulatory domain and is adjacent to the kinase catalytic domain (Guo et al., 2001; Fig. 2A). The FISL motif is responsible for binding to SCaBPs and has a consensus sequence XL (or M, I, F, Y) N (or T)AFD (or E, Q) I (or L, F, M)IS (or A, T, G, L, I) L (or M, F, Y, S, T, G)SXG (or F, D, E, S) F (or L, S) D (or N, S, G)LS (or A, E, G) G (or N, S, T, P) L (or F)FE (or D, G, A) (where X is any amino acid residue and residues in bold are absolutely conserved; Guo et al., 2001). Next to the FISL sequence is a protein phosphatase interaction (PPI) motif that mediates interaction with type 2C protein phosphatases (Ohta et al., 2003; Fig. 2A). The catalytic and regulatory domains of SOS2 have been shown to interact with each other, and this interaction requires the FISL motif (Guo et al., 2001; Fig. 2A). Intramolecular interaction in SOS2, and presumably other PKS proteins, is probably responsible for keeping the kinases inactive, as removal of the regulatory domain of SOS2 led to its constitutive activation (Guo et al., 2001; Fig. 2). It is likely that upon the binding of SCaBPs to the FISL motif, this intramolecular interaction is relieved, leading to the activation of the kinases (Fig. 2B).
Constitutive Activation of the PKS Protein Kinases
Regulation of kinase activity generally results from protein phosphorylation by another kinase(s), autophosphorylation, and/or control by regulatory domains or subunits (Sato et al., 1996; Elion, 1998). As with other protein kinases, a key feature for regulation of the PKS kinases may be phosphorylation of one or more residues within the activation loop (Vertommen et al., 2000; McCartney and Schmidt, 2001). In the absence of specific SCaBP protein(s), PKS kinases have little kinase activity when expressed and purified from bacteria as glutathione S-transferase fusion proteins. The activation loop in the kinase catalytic domain of the PKS proteins is located between the conserved DFG and APE sequences (conserved subdomain VIII; Hrabak et al., 2003). A Thr residue (T168 in SOS2) in this activation loop is completely conserved in all the PKS kinases. With site-directed mutagenesis, constitutively active forms of the kinases were generated through a substitution of the T residue with D to partially mimic phosphorylation by an upstream kinase(s) (Guo et al., 2001; Gong et al., 2002d; Fig. 2A). These results suggest that activation loop phosphorylation/dephosphorylation by an upstream kinase/phosphatase may play an important role in the regulation of PKS kinase activity in vivo (Fig. 2B). Although the upstream kinase is unknown, it is possible that the protein phosphatase 2Cs that bind to PKS may function to dephosphorylate the activation loop (Ohta et al., 2003).
Within the putative activation loop of these PKS kinases, two additional residues, S156 and Y175 (in SOS2), are also completely conserved (Gong et al., 2002c; J.-K. Zhu, unpublished data). Changing any of these residues to D in SOS2 activates the kinase in vitro (Gong et al., 2002c). These results suggest that multiple site phosphorylations may be important for activation of SOS2 (or PKS) kinase activity. Future studies will determine if these three residues are phosphorylation sites in SOS2 or other PKS kinases in vivo.
Mutational analysis has shown that the FISL motif in the regulatory domain of the SOS2 protein is autoinhibitory to the substrate phosphorylation of the PKS kinases. FISL motif deletion mutants resulted in SOS2 and several other PKS proteins that were also constitutively active (Guo et al., 2001; Gong et al., 2002b; Fig. 2A). When the T168 to D mutation was combined with the FISL deletion, superactive SOS2 and PKS kinases were generated (Guo et al., 2001; Gong et al., 2002b; Qiu et al., 2002; Fig. 2A). It is unclear whether PKS activation by Ca2+-SCaBP and by activation loop phosphorylation take place in vivo. Conceivably, both activation mechanisms may function simultaneously and synergistically (Fig. 2B).
Biochemical Properties of the PKS Protein Kinases
The biochemical properties of the PKS kinases are not well understood, due, in part, to the difficulty in isolating and purifying native PKS kinases from plants. Most recombinant PKS proteins produced in Escherichia coli are only active in autophosphorylation, but not in substrate phosphorylation. However, the availability of constitutively active recombinant PKS mutants has allowed characterization of some of the biochemical properties of these proteins. These PKS kinases require a divalent cation for autophosphorylation and substrate phosphorylation. They do not phosphorylate commonly used protein substrates, including casein, myelin basic protein, histone H1, and histone IIIS; however, they phosphorylate three peptide substrates (p1–p3) derived from the recognition sequences of protein kinase C or SNF1/AMPK (Halfter et al., 2000; Gong et al., 2002b, 2002c). Apparent Km and Kcat values for ATP with Mg2+ or Mn2+ have been determined, and the PKS kinases, including SOS2, PKS6, PKS11, and PKS18, prefer Mn2+ to Mg2+ for substrate phosphorylation (peptide substrate p3, ALARAASAAALARRR) and autophosphorylation (Gong et al., 2002b, 2002c). This cofactor preference may reflect the involvement of these kinases in a complex during full activation (Su et al., 1996). The apparent affinity of some of the constitutively activated PKS kinases for ATP and peptide substrate p3 has also been determined. The dependence of kinase activity on ATP or p3 exhibited typical Michaelis-Menten kinetics. The apparent Km values ranged from 29 to 138 μm and from 0.83 to 0.88 μm for p3 and ATP, respectively.
Active and inactive SOS2 mutants that have different molecular structures have been used to determine the mechanism underlying PKS kinase autophosphorylation. The superactive, truncated protein SOS2T168DΔ308 (in which the FISL domain and C-terminal 117 amino acids have been removed) failed to transphosphorylate the kinase-dead mutant SOS2K40 N (in which K40, a conserved amino acid in the catalytic site required for phosphotransfer activity, was changed to N; Gong et al., 2002c). This strongly suggests that unlike some other S/T kinases (Horn and Walker, 1994; Oh et al., 2000; Shah et al., 2001), SOS2 autophosphorylation takes place via an intramolecular reaction mechanism. Whether other PKS kinases autophosphorylate using a similar mechanism remains to be determined.
In most cases, the physiological substrates(s) of the PKS kinases are largely unknown. However, several lines of evidence indicate that SOS1, a Na+/H+ antiporter, is one physiological substrate of SOS2 in Arabidopsis. First, the C-terminal portion of the SOS1 protein contains several putative consensus recognition motifs found in the preferred peptide substrate p3 of the SOS2 mutants. Second, a Histagged SOS1 protein purified from yeast membranes could be phosphorylated in vitro by the SOS2T168DΔ308 mutant (Quintero et al., 2002). Third, SOS3-SOS2 activates the Na+/H+ antiport activity of SOS1 (Qiu et al., 2002).
Recent evidence indicates that H+-coupled antiporters on the vacuolar membrane (tonoplast) may also be substrates or targets for SOS2. Tonoplast Na+/H+-antiport activity originating from AtNHX proteins was compared in vesicles isolated from wild-type and sos2 cultures of Arabidopsis (Qiu et al., 2003). Antiport activity was greatly reduced in sos2, and this activity could be restored when activated SOS2 protein was added in vitro. Coexpression of SOS2 and the vacuolar Ca2+/H+ antiporter, CAX1, in yeast restored growth on high levels of Ca2+ and activated CAX1 activity (Cheng et al., 2003). SOS2 regulation of the activity of these transporters did not depend on the presence or activity of SOS3 and appears to involve mechanisms that do not involve changes in phosphorylation of the transporters (Qiu et al., 2003; Cheng et al., 2003, J.-K. Zhu, unpublished data). SOS2 regulation of Na+/H+ antiporters on the tonoplast and plasma membrane provides additional support that this kinase plays an important role in the maintenance of cellular Na+ homeostasis and is a critical component of the salt tolerance machinery in Arabidopsis. In addition to identifying substrates for SOS2, these studies suggest links between the regulation of Na+ and Ca2+ homeostasis in Arabidopsis.
Interaction Specificity of the SCaBP Calcium Sensors and PKS Protein Kinases
In vivo and in vitro assays have been used to investigate the interaction specificity of individual members of the SCaBP and PKS families (Halfter et al., 2000; Kim et al., 2000; Guo et al., 2001, 2002). Some SCaBPs can interact with multiple PKS kinases and certain PKS kinases interact with several SCaBP proteins. However, some SCaBP/PKS proteins can only interact specifically with one PKS/SCaBP protein, and several SCaBP members do not interact with any of the PKS kinases investigated (Albrecht et al., 2001; Guo et al., 2001), suggesting that these members interact with other PKS proteins or have distinct protein targets. As examples of these varied specificities, SOS3 interacts strongly with SOS2 (Halfter et al., 2000), whereas SCaBP1 only weakly interacts with SOS2 and shows very strong interaction with PKS6 (Guo et al., 2001). An in vivo interaction between SCaBP5 and PKS3 has also been observed (Guo et al., 2002). These interactions are mediated via the highly conserved FISL motif in the kinase regulatory domain (Guo et al., 2001). Sequence variations within the FISL motif and/or in neighboring structures likely determine which PKS interacts with which SCaBP, and the interaction specificity between the SCaBP and PKS proteins may provide the structural basis for some of the functional specificity of these proteins in plant cells.
Interactions Between PKS Protein Kinases and Protein Phosphatases
SOS2, as well as several other PKS proteins, has been shown to interact with the type 2C protein phosphatases, Abscisic Acid Insensitive (ABI)1 and ABI2 (Guo et al., 2002; Ohta et al., 2003). A novel protein domain of 37 amino acid residues in the SOS2 kinase C-terminal regulatory region, designated as the PPI motif, is necessary and sufficient for interaction with the ABI protein phosphatases. The PPI motif is not only conserved in the PKS protein kinases, but also in the DNA damage repair and replication block checkpoint kinase, Chk1, from various organisms, including humans (Ohta et al., 2003). Mutations in conserved amino acid residues in the PPI motif abolish the interaction of SOS2 with ABI2. In contrast, a mutation in the FISL motif disrupts the interaction between SOS2 and SOS3, but has no effect on the interaction between SOS2 and ABI2. A protein kinase-interaction (PKI) domain exists in the ABI protein phosphatase subfamily (Ohta et al., 2003). The interaction between SOS2 and ABI2 is disrupted by the abi2-1 mutation in the PKI domain, and the mutation causes increased tolerance to salt shock as well as ABA insensitivity in plants (Ohta et al., 2003).
When the interaction specificity of several PKS and ABI proteins was compared, some PKS members were found to interact specifically with ABI2, whereas others interacted preferentially with ABI1 (Ohta et al., 2003). Sequence variations in the PPI motif of PKS may determine whether a particular PKS interacts with ABI2, ABI1, or other related protein phosphatases. For example, residues R340 and S343 of SOS2 are conserved in PKS3, PKS11, and PKS24, which all interact strongly with ABI2 but not ABI1. In contrast, these two residues are changed to K and T in PKS24, which likely explains why PKS24 interacts strongly with ABI1 but only weakly with ABI2. Variations in the PKI domain of the protein phosphatase may determine its interaction with a specific PKS. Although the wild-type ABI1 protein interacts only weakly with SOS2, substitutions of A197 and A201 within the PKI domain with corresponding residues (T and V, respectively) in ABI2 led to an interaction with SOS2 nearly as strong as that of ABI2 (Ohta et al., 2003).
Expression Patterns of the SCaBP Calcium Sensors and PKS Protein Kinases
The functional specificity of the SCaBPs and PKS kinases is not only determined by their biochemical properties, subcellular localization, and their interacting proteins, but also by the spatial, temporal, and environmental responsive expression patterns of their genes. Expression patterns of a number of the SCaBPs and PKS kinases have been determined. Different SCaBP genes are expressed differentially in various tissues and in response to stresses (Kudla et al., 1999; Guo et al., 2002). For example, SCaBP1 is preferentially expressed in roots, whereas SCaBP6 is constitutively expressed in leaves, stems, roots, and flowers. Expression of SCaBP1 or SCaBP6 is not induced by drought, cold, or wounding treatment, whereas SCaBP5 expression is up-regulated by these stresses and by the phytohormone ABA (Luan et al., 2002), indicating a potential involvement of the Ca2+ sensor in ABA and abiotic stress responses. Promoter-glucuronidase (GUS) reporter fusions have been used to analyze the tissue distribution of SCaBP gene expression in Arabidopsis seedlings (Guo et al., 2002). SCaBP5 promoter-GUS was detected in imbibing seeds, cotyledons, hypocotyls, and roots of young seedlings, and also in the leaves, roots, stems, and floral organ of adult plants (Guo et al., 2002). These results suggest that different members of the SCaBP family may function in distinct tissues, at specific developmental stages, and in response to particular environmental conditions.
Different PKS members also exhibit specific tissue expression patterns. For example, PKS6 is expressed in leaves, stems, flowers, and siliques, but is not detectable in roots of mature plants (Gong et al., 2002b). In striking contrast, PKS11 expression in roots is substantially higher than that in leaves, stems, flowers, or siliques (Gong et al., 2002a). PKS11 promoter-GUS staining is also readily detected in roots, but the staining in other tissues is below the detection limit. These PKS genes also display differential expression in response to a variety of stresses. For example, PKS6 expression is up-regulated by ABA, salt, and drought, but is not affected by cold treatment (Gong et al., 2002b). However, no significant induction or repression of PKS11 expression is observed after any of these treatments. The differential expression patterns of the SCaBPs and PKS kinases strongly indicate diverse functions for their gene products in plant growth and development, as well as in the response of the plant to stress.
In Planta Functions of the SCaBP Calcium Sensors and PKS Protein Kinases
The differential expression patterns of SCaBP and PKS genes and the range of interactions among these proteins indicate that they may have varied physiological functions in the plant. The in vivo functions of several SCaBP and PKS proteins have been uncovered recently using forward and reverse genetic approaches. Via changes in protein activation, as well as regulation of gene expression, these proteins have been implicated in the plant's response to ABA and sugar signaling and in cellular pH regulation (Fig. 1).
ABA plays an important role in plant growth and development, as well as in the response of the plant to environmental stress. There is accumulating evidence that changes in protein phosphorylation may be an important part of ABA signaling. Because ABA is known to activate Ca2+ signaling, it seemed likely that PKS kinases and SCaBPs also play a role in the response of the plant to ABA. To test this, transgenic Arabidopsis plants with altered levels of the leaf-specific PKS18 were evaluated for their response to ABA. Transgenic plants overexpressing a constitutively active form of PKS18 (PKS18T/D) were hypersensitive to ABA during seed germination and seedling growth, whereas silencing the kinase gene by RNA interference conferred ABA insensitivity (Gong et al., 2002d). Recently, Arabidopsis mutants in which SCaBP5 or its interacting PKS, PKS3, had been silenced were found to be hypersensitive to ABA during seed germination, seedling growth, and stomatal closure. In addition, ABA induction of a number of genes (Leung and Giraudat, 1998), including COR47, COR15A, RD29A, and RAB18, was enhanced in the scabp5 and pks3 mutants (Guo et al., 2002). These results indicate that SCaBP5 and closely related Ca2+ sensors and interacting protein kinases may be global negative regulators of ABA signaling in plants (Guo et al., 2002). Because ABI2 has been shown to control the influx of Ca2+ (a second messenger in ABA signaling), we have speculated that SCaBP5-PKS3 may sense intracellular Ca2+ levels in response to ABA and feedback regulate Ca2+ influx (Guo et al., 2002). Thus, SCaBP5-PKS3, together with ABI2, may be important for the generation of ABA-specific Ca2+ oscillations. It is possible that the SCaBP5-PKS3-ABI2 complex directly modulates Ca2+ oscillation “generators” (Ca2+ channels, Ca2+- ATPases, and Ca2+/H+ exchangers). More recently, Cheong et al. (2003) showed that overexpression of SCaBP5/CBL1 enhanced drought induction, but reduced cold induction of stress genes. Conversely, the scabp5/cbl1 null mutant in Arabidopsis showed increased cold induction and decreased drought induction of stress genes. Using a T-DNA insertional mutant of CBL-interacting protein kinase 3/PKS12, Kim et al. (2003) showed that this kinase is involved in mediating ABA signaling. The mutant plants were more sensitive than wild type to ABA during seed germination and seedling growth, and expression of a number of ABA- and stress-responsive genes was altered in the mutant.
In a manner similar to plant hormones, sugars can act as signaling molecules that control gene expression and developmental processes in plants (Finkelstein and Gibson, 2002). Recent evidence indicates that the PKS proteins may also be involved in sugar signaling. When a constitutively active PKS11 mutant, PKS11T161D, was expressed in transgenic Arabidopsis plants, the transgenic plants were more resistant to high levels of Glc. Based on observations with Glc analogs (no difference was seen between the control and transgenic plants in their responses to the Glc analogs 2-deoxy-Glc or 3-O-methyl-Glc; Gong et al., 2002a), it is likely that PKS11 functions in mediating Ca2+ signaling in response to a sugar signal, and that its function may be independent of the hexokinase pathway (Sheen et al., 1999). Identification of PKS11-interacting partners and substrate proteins will help to clarify the precise role of this kinase in sugar responses.
The SOS2-like protein, PKS5, has been shown to be involved in the regulation of intracellular pH homeostasis through its effect on the plasma membrane H+-ATPase (Y. Guo et al., unpublished data). PKS5 interacts strongly with SCaBP1, but not with SOS3. Loss-of-function mutants (pks5), generated by RNA interference or ethyl methanesulfonate, have been used to characterize the role of this kinase in the plant's response to various environmental stimuli. When compared with the growth of wild-type seedlings, pks5 seedlings were able to maintain growth at higher external pH and to maintain neutral cytoplasmic pH at a more alkaline extracellular pH. H+- transport assays using isolated plasma membrane vesicles showed that the activity of the plasma membrane H+-ATPase was 40% higher in the pks5 mutant compared with activity in vesicles isolated from wild-type plants. When activated PKS5 protein kinase was added in vitro, H+-transport activity in the mutant was reduced to wild-type levels. PKS5 directly phosphorylates the AHA2 isoform of the plasma membrane H+-ATPase, and this phosphorylation was reduced in the presence of the interacting Ca2+-binding protein, SCaBP1 (Y. Guo et al., unpublished data).
FUTURE DIRECTIONS
The novel families of SCaBP Ca2+ sensors and PKS protein kinases appear to constitute important Ca2+- decoding systems that sense and interpret Ca2+ signals during plant growth and development and in response to abiotic stresses (Fig. 1). Although significant progress has been made in our understanding of the biochemical properties and physiological roles of some of these proteins, many questions remain to be answered.
First, what mechanisms underlie Ca2+ sensing and kinase activity regulation? To answer this question, the crystal structures of the SCaBP-PKS complexes need to be solved. Second, what are the upstream kinase(s) that phosphorylate the Thr, Ser, or Tyr residues in the putative activation loop of the PKS kinases? What is the relationship between the regulation of PKS activity in vivo by Ca2+-SCaBP and by the upstream kinase? Both mechanisms may work in vivo, and may even function simultaneously and synergistically. Third, what are the in planta functions of the remaining SCaBP and PKS proteins? This information will be revealed using forward and reverse genetic approaches. Identifying the physiological inputs and outputs of each SCaBP-PKS pair will be required to fully understand their biological roles. In comparative studies of wild-type and mutant plants, it will be important to assess phenotypes at the appropriate stages of development, as well as in response to numerous biotic and abiotic stresses. Fourth, what are the in vivo substrates of the PKS kinases? Additionally, although substantial progress has been made relative to SCaBP-PKS kinase signaling pathways in Arabidopsis, as yet, little is known about these signaling modules in other plant species. These signaling modules are likely conserved among different plant species, although the exact number and combination of the SCaBPs and PKSs may differ. Future studies that combine genetic, genomic, and proteomic approaches promise to rapidly elucidate the roles of SCaBP and PKS proteins in Ca2+ signaling in plants.
This work was supported by the National Institutes of Health (grant no. R01GM59138 to J.-K.Z.), by the U.S. Department of Energy (grant no. DE–FG03–93ER20120 to K.S.S.), and by the Southwest Consortium on Plant Genetics and Water Resources (grant to K.S.S. and J.-K.Z.).
References
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20: 1051-1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Cheng N-H, Pittman JK, Zhu J-K, Hirschi KD (2004) The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem 279: 2922-2926 [DOI] [PubMed] [Google Scholar]
- Cheng S-H, Willmann MR, Chen H-C, Sheen J (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469-485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Kim K-N, Pandey GK, Gupta R, Grant JJ, Luan S (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15: 1833-1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA (1998) Routing MAP kinase cascades. Science 281: 1625-1626 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI (2002) ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol 5: 26-32 [DOI] [PubMed] [Google Scholar]
- Gong D, Gong Z, Guo Y, Chen X, Zhu J-K (2002a) Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J Biol Chem 277: 28340-28350 [DOI] [PubMed] [Google Scholar]
- Gong D, Gong Z, Guo Y, Zhu J-K (2002b) Expression, activation, and biochemical properties of a novel Arabidopsis protein kinase. Plant Physiol 129: 225-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Guo Y, Jagendorf A, Zhu J-K (2002c) Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol 130: 256-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Zhang C, Chen X, Gong Z, Zhu J-K (2002d) Constitutive activation and transgenic evaluation of the function of an Arabidopsis PKS protein kinase. J Biol Chem 277: 42088-42096 [DOI] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu J-K (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13: 1383-1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Qiu Q, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu J-K (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopis thaliana. Plant Cell 16: epub January 23, 2004 [DOI] [PMC free article] [PubMed]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu J-K (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233-244 [DOI] [PubMed] [Google Scholar]
- Halford NG, Bouly JP, Thomas M (2000) SNF1-related protein kinases (SnRKs): regulators at the heart of the control of carbon metabolism and partitioning. Adv Bot Res 32: 405-434 [Google Scholar]
- Halfter U, Ishitani M, Zhu J-K (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735-3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG (1999) Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol 50: 97-131 [DOI] [PubMed] [Google Scholar]
- Hardie DG (2000) Plant protein-serine/threonine kinases: classification into subfamilies and overview of function. In M Kreis, JC Walker, eds, Plant Protein Kinases. Academic Press, San Diego, pp 1-44
- Hardie DG, Carling D, Carlson M (1998) The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem 67: 821-855 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Phytol 151: 175-183 [DOI] [PubMed] [Google Scholar]
- Henzl MT, Larson JD, Agah S (2000) Influence of monovalent cations on rat α- and β-parvalbumin stabilities. Biochemistry 39: 5859-5867 [DOI] [PubMed] [Google Scholar]
- Horn MA, Walker JC (1994) Biochemical properties of the autophosphorylation of RLK5, receptor-like protein kinase from Arabidopsis thaliana. Biochim Biophys Acta 1208: 65-74 [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR et al. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666-680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim C-S, Shi W, Zhu J-K (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667-1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-N, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15: 411-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-N, Cheong YH, Gupta R, Luan S (2000) Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol 124: 1844-1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee CB, Draetta GF, Hubbard MJ (1988) Calcineurin. Adv Enzymol 61: 149-200 [DOI] [PubMed] [Google Scholar]
- Kudla J, Xu Q, Harter K, Gruissem W, Luan S (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96: 4718-4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49: 199-222 [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730-3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943-1945 [DOI] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14: S389-S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney RR, Schmidt MC (2001) Regulation of Snf1 kinase: activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem 276: 36460-36466 [DOI] [PubMed] [Google Scholar]
- Nagae M, Nozawa A, Koizumi N, Sano H, Hashimoto H, Sato M, Shimizu T (2003a) Crystallization and preliminary X-ray characterization of a novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. Acta Crystallogr D Biol Crystallogr 59: 1079-1080 [DOI] [PubMed] [Google Scholar]
- Nagae M, Nozawa A, Koizumi N, Sano H, Hashimoto H, Sato M, Shimizu T (2003b) The crystal structure of the novel calcium binding protein AtCBL2 from Arabidopsis thaliana. J Biol Chem 278: 42240-42246 [DOI] [PubMed] [Google Scholar]
- Oh M-H, Ray WK, Huber SC, Asara JM, Gage DA, Clouse SD (2000) Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol 124: 751-766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu J-K (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA 100: 11771-11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q-S, Guo Y, Dietrich M, Schumaker KS, Zhu J-K (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436-8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q-S, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu J-K (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem 279: 207-215 [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu J-K, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99: 9061-9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd JJ, Franklin-Tong VE (2001) Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol 151: 7-33 [DOI] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14: S401-417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Aoto M, Mori K, Akasofu S, Tokmakov AA, Sahara S, Fukami Y (1996) Purification and characterization of a Src-related p57 protein-tyrosine kinase from Xenopus oocytes. J Biol Chem 271: 13250-13257 [DOI] [PubMed] [Google Scholar]
- Shah K, Vervoort J, Vries SC (2001) Role of threonines in the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J Biol Chem 276: 41263-41269 [DOI] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang J-C (1999) Sugars as signaling molecules. Curr Opin Plant Biol 2: 410-418 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu J-K (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896-6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wu S-J, Zhu J-K (2003) Overexpression of a plasma membrane Na+/H+ antiporter improves salt tolerance in Arabidopsis. Nat Biotechnol 21: 81-85 [DOI] [PubMed] [Google Scholar]
- Snedden WA, Fromm H (1998): Calmodulin, calmodulin-related proteins and plant response to the environment. Trends Plant Sci 3: 299-304 [Google Scholar]
- Su JY, Eriksob E, Maller JL (1996) Cloning and characterization of a novel serine/threonine protein kinase expressed in early Xenopus embryos. J Biol Chem 271: 14430-14437 [DOI] [PubMed] [Google Scholar]
- Towler DA, Adams SP, Eubanks SR, Towery DS, Jackson-Machelski E, Glaser L, Gordon JI (1988) Myristoyl CoA: protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J Biol Chem 263: 1784-1790 [PubMed] [Google Scholar]
- Trewavas AJ, Malho R (1998) Ca2+ signaling in plant cells: the big network! Curr Opin Plant Biol 1: 428-433 [DOI] [PubMed] [Google Scholar]
- Vertommen D, Rider M, Ni Y, Waelkens E, Merlevede W, Vandenheede JR, Lint JV (2000) Regulation of protein kinase D by multisite phosphorylation. J Biol Chem 275: 19567-19576 [DOI] [PubMed] [Google Scholar]
- Ward JM, Hirschi KD, Sze H (2003) Plants pass the salt. Trends Plant Sci 8: 200-201 [DOI] [PubMed] [Google Scholar]
- Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski RE (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 697-725 [DOI] [PubMed] [Google Scholar]