Abstract
Cytochrome P450 1A1 (CYP1A1) enzyme is one of the most important metabolizing enzymes responsible for the metabolism of numerous xenobiotics. Numerous individual case-control studies have investigated the associations between the CYP1A1 rs1048943 A > G and rs4646903 T > C genetic variations and colorectal cancer (CRC) risk, but the conclusions were controversial. To obtain a scientific conclusion, we performed a meta-analysis based on a total of 26 publications, including 20 studies with 8665 cases and 9953 controls on rs1048943 A > G and 19 studies with 6416 cases and 7551 controls on rs4646903 T > C, respectively. The pooled analysis indicated that rs1048943 A > G was associated with an increased risk of CRC (G vs. A: OR = 1.28, 95% CI = 1.08−1.52; GG vs. AA: OR = 1.54, 95% CI = 1.25−1.91; GA vs. AA: OR = 1.26, 95% CI = 1.00−1.60; GG/GA vs. AA: OR = 1.31, 95% CI = 1.05−1.64; GG vs. GA/AA: OR = 1.56, 95% CI = 1.26−1.91). Stratification analysis showed the association between rs1048943 A > G and CRC risk was more obvious in studies with the population-based (PB) design or high quality score. The association between rs4646903 T > C and CRC risk did not reach statistical significance in the pooled analysis as well as stratification analysis. This meta-analysis demonstrated CYP1A1 rs1048943 A > G may increase the susceptibility to CRC instead of rs4646903 T > C. This conclusion suggested CYP1A1 may contribute to the pathogenesis of CRC.
Keywords: CYP1A1, polymorphism, colorectal cancer, meta-analysis
INTRODUCTION
Although the incidence rate of CRC decreased by approximately 3% per year during the past decade, CRC is still one of the most common cancers and the third leading cause of cancer-related death worldwide [1]. Despite the fact of great improvement in chemotherapy and surgical operations, the prognosis of advanced CRC is still much worse than early-stage CRC [2]. Therefore, for cancer prevention and early diagnosis, it is important to identify risk factors and biomarkers that are associated with disease susceptibility to screen high risk population.
The development of CRC is widely considered as a multi-step, multi-factorial process involving gene-gene and gene-environment interactions [3]. Previous studies have shown that lifestyle factors, such as cigarette smoking and alcohol consumption may contribute to sporadic CRC risk [4]. In vivo, these xenobiotics (e.g. nicotine and alcohol) are metabolized by xenobiotic-metabolizing enzymes including CYP1A1 superfamily, glutathione S-transferases, N-acetyltransferase, etc [5]. Furthermore, genetic variations of these enzymes may lead to the occurrence of CRC by metabolizing environmental insults [3]. In recent years, an increasing number of individual case-control studies have investigated the association of genetic variations within cytochrome P450s (CYPs) with CRC risk.
The Human Genome Project has identified 57 human cytochrome P450 enzymes, and ordered the minto 18 families and 43 subfamilies by sequence similarities [6]. Although most chemical carcinogens are inactive in vivo, they can become bio-active via CYPs. For example, benzo[a] pyrene can be metabolized and transformed to mutagenic benzo[a] pyrene diol epoxide [7, 8]. CYPs, the key of phase I enzymes, are the main enzymes in the metabolism of carcinogenic polycyclic aromatic hydrocarbons (PAHs) [9]. Among all researches of CYP involvement in procarcinogen activation, CYP1A1s polymorphisms of rs1048943 A > G and rs4646903 T > C were the most widely studied [10]. CYP1A1 rs1048943 A > G leads to amino acid change in exon 7 of CYP1A1 from Ile to Val (nucleotides A to G) at codon 462. CYP1A1 rs4646903 T > C is characterized by the T to C mutation at nucleotide 3801 in the 3′-flanking region of the gene [11].
To date, numerous case-control studies have investigated the associations between CYP1A1 rs1048943 A > G, rs4646903 T > C and CRC, but the conclusions were inconsistent. Hence, we conducted a meta-analysis to obtain a scientific conclusion.
RESULTS
Characteristics of eligible publications
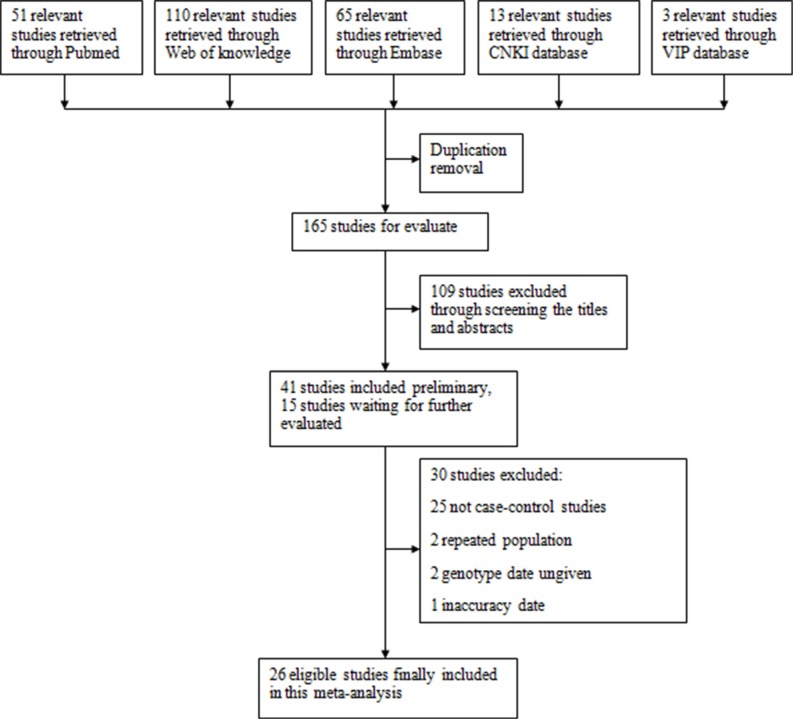
A total of 26 articles reporting the associations between CYP1A1 polymorphisms and CRC risk were included in the meta-analysis. There were 20 studies (8665 cases and 9953 controls) for rs1046943 A > G and 19 studies (6416 cases and 7551 controls) for rs4646903 T > C. The study selection process was shown in Figure 1 [5, 11–35].
Figure 1. Flow chart of studies selection.
The ethnicities of the 20 publications about rs1048943 A > G, included Asian (6 studies), Caucasian (13 studies) and mixed populations (1 study). The distribution of genotype among controls fulfilled the criteria of HWE in14 studies. There were 18 studies with high quality scores. For rs4646903 T > C, the ethnicities of the 19 publications included Asian (7 studies) and Caucasian (12 studies). The distribution of genotype among controls fulfilled the criteria of HWE in 17 studies. There were 17 studies with high quality scores. The main characteristics of the 26 eligible studies were listed in Table 1.
Table 1. Characteristics of the 26 articles included in the meta-analysis.
| Study | Country | Ethnicity | Source | Polymorphism | Sample size | HWE | HWE | Score |
|---|---|---|---|---|---|---|---|---|
| of control | type | (case/control) | (cases) | (control) | ||||
| Hayashi,1992 | Japan | Asian | PB | rs1048943 | 85/358 | 0.114 | 0.332 | 12 |
| Sivaraman,1994 | Mixed | Caucasian | PB | rs1048943 | 43/47 | 0.227 | 0.230 | 11 |
| rs4646903 | 43/47 | 0.001 | 0.245 | 11 | ||||
| Frische,1999 | Germany | Caucasian | PB | rs1048943 | 187/101 | 0.622 | 0.839 | 13 |
| rs4646903 | 187/101 | 0.496 | 0.563 | 13 | ||||
| Ishibe,2000 | America | Caucasian | PB | rs1048943 | 212/221 | 0.018 | 0.057 | 13 |
| rs4646903 | 212/221 | 0.247 | 0.428 | 13 | ||||
| Sachse,2002 | Britain | Caucasian | PB | rs1048943 | 490/592 | 0.035 | 0.002 | 11 |
| rs4646903 | 490/592 | 0.040 | 0.596 | 14 | ||||
| Ye,2002 | Britain | Caucasian | PB | rs4646903 | 41/82 | 0.613 | 0.599 | 11 |
| Slattery,2004 | America | Caucasian | PB | rs1048943 | 1791/2180 | 0.000 | 0.000 | 12 |
| rs4646903 | 1805/2164 | 0.080 | 0.118 | 15 | ||||
| Hou,2005 | Mixed | Mixed | HB | rs1048943 | 675/679 | 0.117 | 0.478 | 12 |
| Landi,2005 | Spain | Caucasian | HB | rs1048943 | 362/323 | 0.617 | 0.469 | 11 |
| rs4646903 | 358/305 | 0.717 | 0.793 | 11 | ||||
| Chen,2005 | China | Asian | PB | rs4646903 | 139/340 | 0.978 | 0.821 | 13 |
| Little,2006 | Britain | Caucasian | PB | rs1048943 | 251/396 | 0.602 | 0.534 | 14 |
| rs4646903 | 232/378 | 0.206 | 0.448 | 14 | ||||
| Bente,2006 | Australia+ Poland | Caucasian | HB | rs4646903 | 118/100 | 0.039 | 0.637 | 10 |
| Youshida,2007 | Japan | Asian | PB | rs1048943 | 66/121 | 0.910 | 0.800 | 11 |
| rs4646903 | 66/121 | 0.244 | 0.623 | 11 | ||||
| Yeh,2007 | Taiwan, China | Asian | HB | rs1048943 | 717/729 | 0.000 | 0.280 | 12 |
| Kiss,2007 | Hungary | Caucasian | HB | rs1048943 | 500/500 | 0.172 | 0.315 | 12 |
| Pereira, 2008 | Brazil | Caucasian | PB | rs1048943 | 114/114 | 0.000 | 0.071 | 12 |
| Pande,2008 | America | Caucasian | PB | rs1048943 | 120/137 | 0.294 | 0.000 | 10 |
| rs4646903 | 120/137 | 0.046 | 0.272 | 13 | ||||
| Zheng,2009 | China | Asian | PB | rs1048943 | 79/110 | 0.056 | 0.031 | 8 |
| rs4646903 | 79/110 | 0.479 | 0.272 | 11 | ||||
| Liu,2009 | China | Asian | HB | rs4646903 | 75/100 | 0.000 | 0.000 | 6 |
| Kobayashi,2009 | Japan | Asian | HB | rs1048943 | 117/289 | 0.164 | 0.000 | 7 |
| Nisa,2010 | Japan | Asian | PB | rs1048943 | 685/778 | 0.580 | 0.970 | 14 |
| rs4646903 | 685/778 | 0.423 | 0.718 | 14 | ||||
| Cleary,2010 | Canada | Caucasian | PB | rs1048943 | 1174/1293 | 0.000 | 0.006 | 11 |
| rs4646903 | 1174/1293 | 0.281 | 0.062 | 14 | ||||
| Houlle,2011 | France | Caucasian | HB | rs1048943 | 329/419 | 0.651 | 0.546 | 11 |
| rs4646903 | 329/419 | 0.804 | 0.362 | 11 | ||||
| Darazy,2011 | Lebanon | Asian | PB | rs4646903 | 70/70 | 0.000 | 0.000 | 5 |
| Saeed,2013 | Saudi Arabia | Asian | HB | rs4646903 | 100/79 | 0.374 | 0.726 | 10 |
| Gil,2014 | Poland | Caucasian | HB | rs1048943 | 478/404 | 0.574 | 0.095 | 11 |
PB, population-based; HB, hospital-based; HWE, Hardy-Weinberg equilibrium.
Meta-analysis
CYP1A1 rs1048943 A > G polymorphism
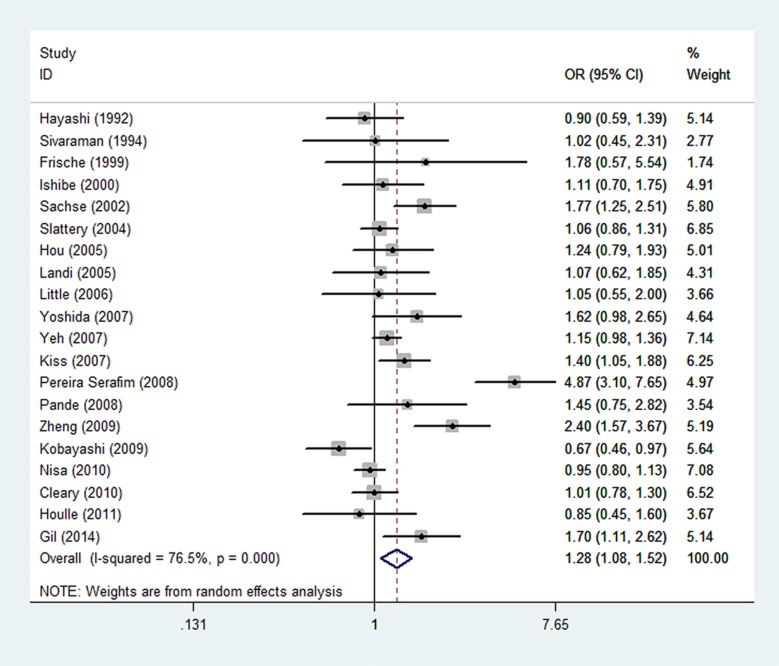
The association between CYP1A1 rs1048943 A > G and the risk of CRC was shown in Table 2 and Figure 2. To sum up, the associations under all genetic models were statistically significant (G vs. A: OR = 1.28, 95% CI = 1.08–1.52; GG vs. AA: OR = 1.54, 95% CI = 1.25–1.91; GA vs. AA: OR = 1.26,95% CI = 1.00–1.60; GG/GA vs. AA: OR = 1.31, 95% CI = 1.05–1.64; GG vs. GA/AA: OR = 1.56, 95% CI = 1.26-1.91) (Supplementary Figure 1). Further stratified analysis showed the association was especially obvious in studies with population-based (PB) designs and of high quality scores. In studies of PB designs, the association was more obvious in the homozygous model (GG vs. AA: OR = 1.61, 95% CI = 1.03–2.53). In studies of high quality score, the association was more obvious using recessive model (GG vs. GA/AA: OR = 1.45, 95% CI = 1.16–1.82). Stratification analysis by ethnicity showed the association of rs1048943 A > G and CRC were more obvious in homozygous (GG vs. AA: OR = 1.61, 95% CI = 1.04–2.47) and recessive (GG vs. GA/AA: OR = 1.59, 95% CI = 1.25-2.02) models in Asian. In Caucasian, the associations were obvious in dominant model (GA/GG vs. AA: OR=1.43, 95% CI = 1.05–1.95), heterozygous model (GA vs. AA: OR = 1.42, 95% CI = 1.04–1.96) and G allelic (G vs. A: OR = 1.33, 95% CI = 1.06–1.68). After excluding studies whose distribution of genotype in controls deviated from HWE, the association remained statistically significant (G vs. A: OR = 1.30, 95% CI = 1.05–1.61; GG vs. AA: OR = 1.51, 95% CI = 1.17–1.94; GG/GA vs. AA: OR = 1.35, 95% CI = 1.01–1.81; GG vs. GA/AA: OR = 1.52, 95% CI = 1.19–1.95). All of the results were listed in Table 2.
Table 2. Meta-analysis of the association between CYP1A1 polymorphisms and CRC risk.
| G vs. A | GG vs. AA | GA vs. AA | GA/GG vs. AA | GG vs. GA/AA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | OR(95%CI) | PORa | Phetb | OR (95% CI) | PORa | Phetb | OR (95% CI) | PORa | Phetb | OR (95% CI) | PORa | Phetb | OR (95% CI) | PORa | Phetb |
| rs1048943 | 1.28 (1.08–1.52) | 0.004 | 0.000 | 1.54 (1.25–1.91) | 0.000 | 0.228 | 1.26 (1.00–1.60) | 0.05 | 0.000 | 1.31 (1.05–1.64) | 0.018 | 0.000 | 1.56 (1.26–1.91) | 0.000 | 0.664 |
| Ethnicity | |||||||||||||||
| Asian | 1.14 (0.87–1.50) | 0.346 | 0.000 | 1.61 (1.04–2.47) | 0.031 | 0.049 | 0.93 (0.67–1.29) | 0.661 | 0.001 | 1.04 (0.75–1.48) | 0.809 | 0.000 | 1.59 (1.25–2.02) | 0.000 | 0.184 |
| Caucasian | 1.33 (1.06–1.68) | 0.015 | 0.000 | 1.48 (0.95–2.32) | 0.083 | 0.526 | 1.42 (1.04–1.96) | 0.030 | 0.000 | 1.43 (1.05–1.95) | 0.022 | 0.000 | 1.41 (0.92–2.17) | 0.113 | 0.849 |
| Source of control | |||||||||||||||
| PB | 1.40 (1.09–1.80) | 0.009 | 0.000 | 1.61 (1.03–2.53) | 0.038 | 0.054 | 1.48 (1.06–2.06) | 0.021 | 0.000 | 1.52 (1.10–2.11) | 0.011 | 0.000 | 1.37 (1.04–1.81) | 0.024 | 0.373 |
| HB | 1.13 (0.91–1.40) | 0.255 | 0.026 | 1.68 (1.22–2.33) | 0.002 | 0.935 | 0.99 (0.72–1.38) | 0.972 | 0.000 | 1.05 (0.79–1.41) | 0.727 | 0.001 | 1.82 (1.33–2.50) | 0.000 | 0.985 |
| Score | |||||||||||||||
| High | 1.28 (1.08–1.52) | 0.004 | 0.000 | 1.48 (1.18–1.86) | 0.001 | 0.591 | 1.28 (1.01–1.62) | 0.043 | 0.000 | 1.32 (1.05–1.65) | 0.017 | 0.000 | 1.49 (1.19–1.86) | 0.001 | 0.869 |
| Low | 1.32 (0.56–3.11) | 0.530 | 0.000 | 1.57 (0.42–5.93) | 0.507 | 0.030 | 1.24 (0.36–4.24) | 0.735 | 0.000 | 1.31 (0.41–4.22) | 0.652 | 0.000 | 2.02 (1.19–3.44) | 0.009 | 0.141 |
| HWE | |||||||||||||||
| Yes | 1.30 (1.05–1.61) | 0.015 | 0.000 | 1.51 (1.17–1.94) | 0.001 | 0.382 | 1.31 (0.96–1.77) | 0.088 | 0.000 | 1.35 (1.01–1.81) | 0.041 | 0.000 | 1.52 (1.19–1.95) | 0.001 | 0.719 |
| No | 1.25 (0.91–1.73) | 0.173 | 0.000 | 1.63 (1.11–2.41) | 0.013 | 0.113 | 1.19 (0.79–1.79) | 0.417 | 0.000 | 1.23 (0.83–1.83) | 0.297 | 0.000 | 1.63 (1.12–2.39) | 0.011 | 0.322 |
| C vs. T | CC vs. TT | CT vs. TT | CT/CC vs. TT | CC vs. CT/TT | |||||||||||
| rs4646903 | 1.09 (0.96–1.23) | 0.186 | 0.001 | 1.11 (0.81–1.53) | 0.507 | 0.032 | 1.04 (0.91–1.18) | 0.594 | 0.034 | 1.07 (0.93–1.22) | 0.342 | 0.009 | 1.10 (0.82–1.47) | 0.530 | 0.053 |
| Ethnicity | |||||||||||||||
| Asian | 1.27 (0.92–1.75) | 0.140 | 0.000 | 1.17 (0.75–1.82) | 0.502 | 0.077 | 1.23 (0.85–1.77) | 0.267 | 0.016 | 1.32 (0.89–1.96) | 0.168 | 0.001 | 1.05 (0.84–1.32) | 0.666 | 0.279 |
| Caucasian | 1.09 (0.92–1.28) | 0.331 | 0.075 | 1.23 (0.89–1.72) | 0.213 | 0.136 | 0.98 (0.88–1.10) | 0.710 | 0.114 | 1.00 (0.90–1.12) | 0.943 | 0.150 | 1.52 (0.81–2.85) | 0.192 | 0.085 |
| Source of control | |||||||||||||||
| PB | 0.99 (0.92–1.06) | 0.774 | 0.193 | 1.04 (0.75–1.45) | 0.826 | 0.058 | 0.98 (0.90–1.08) | 0.726 | 0.175 | 0.99 (0.91–1.07) | 0.745 | 0.235 | 1.05 (0.76–1.44) | 0.783 | 0.064 |
| HB | 1.55 (0.92–2.61) | 0.100 | 0.000 | 1.61 (0.92–2.82) | 0.096 | 0.141 | 1.36 (0.86–2.14) | 0.190 | 0.017 | 1.51 (0.90–2.54) | 0.122 | 0.001 | 1.51 (0.87–2.62) | 0.144 | 0.206 |
| Score | |||||||||||||||
| High | 1.05 (0.93–1.18) | 0.465 | 0.005 | 1.02 (0.74–1.41) | 0.886 | 0.052 | 1.02 (0.90–1.17) | 0.741 | 0.029 | 1.04 (0.91–1.18) | 0.604 | 0.016 | 1.03 (0.76–1.40) | 0.859 | 0.064 |
| Low | 1.85 (1.19–2.87) | 0.006 | 0.644 | 2.24 (1.04–4.80) | 0.039 | 0.905 | 1.62 (0.80–3.28) | 0.180 | 0.696 | 1.88 (1.06–3.35) | 0.031 | 0.709 | 1.99 (0.95–4.16) | 0.068 | 0.846 |
| HWE | |||||||||||||||
| Yes | 1.05 (0.93–1.18) | 0.465 | 0.005 | 1.02 (0.74–1.41) | 0.886 | 0.052 | 1.02 (0.90–1.17) | 0.741 | 0.029 | 1.04 (0.91–1.18) | 0.604 | 0.016 | 1.03 (0.76–1.40) | 0.859 | 0.064 |
| No | 1.85 (1.19–2.87) | 0.006 | 0.644 | 2.24 (1.04–4.80) | 0.039 | 0.905 | 1.62 (0.80–3.28) | 0.180 | 0.696 | 1.88 (1.06–3.35) | 0.031 | 0.709 | 1.99 (0.95–4.16) | 0.068 | 0.846 |
P value of the Z-test for odds ratio test;
P value of the Q-test for heterogeneity test; PB, population based; HB, hospital based; HWE, Hardy-Weinberg equilibrium.
Figure 2. Forest plot of colorectal cancer risk associated with CYP1A1 rs1048943 A>G polymorphism (G vs. A).
CYP1A1 rs4646903 T > C polymorphism
In the meta-analysis, the pooled analysis showed no significant association between rs4646903 T > C and CRC risk. Further stratification analyses also found no obvious association except the subgroups of deviating from HWE and low quality score which included the same two studies showed statistical significances (C vs. T: OR = 1.85, 95% CI = 1.19–2.87; CC vs. TT: OR = 2.24, 95% CI = 1.04–4.80; CT/CC vs. TT: OR = 1.88, 95% CI = 1.06–3.35) (Table 2).
Heterogeneity and sensitivity analysis
The Chi-squared-based Q-test showed substantial heterogeneities among publications for the two polymorphisms (Table 2). Meta-regression analysis for both polymorphisms yielded no significant difference between subgroups, except subgroups of HWE and study quality score for rs464903 T > C. After excluding the inferior quality studies of deviate from HWE and low score, the conclusions of both polymorphisms were not change. Furthermore, we estimated the influence of single individual data on the combined ORs by consecutively omitting each study from the meta-analysis, no obvious differences were observed for both variations (Supplementary Figure 2).
Publication bias
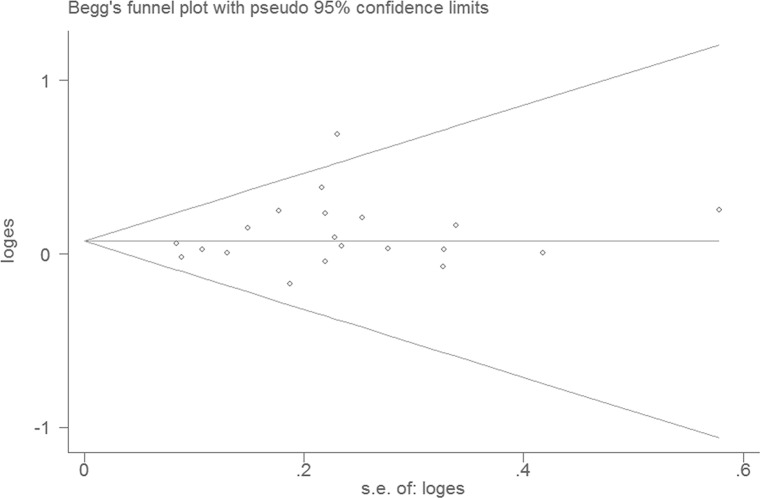
For rs1048943 A > G, the shapes of funnel plots were symmetric (Figure 3) and Egger's linear regression test provided no evidence of publication bias (G vs. A: P = 0.185, t = 1.38; GG vs. AA: P = 0.325, t = 1.01; GA vs. AA: P = 0.132, t = 1.58; GG/GA vs. AA: P = 0.116, t = 1.65; GG vs. GA/AA: P = 0.587, t = 0.56). For rs4646903 T > C the funnel plots were also symmetric and Egger's linear regression test provided no evidence of publication bias (CC vs. TT: P = 0.174, t = 1.42; CT vs. TT: P = 0.056, t = 2.05; CC vs. CT/TT: P = 0.234, t = 1.24), except two models of G allelic (C vs. T: P = 0.005, t = 3.27) and dominant model (CT/CC vs. TT: P = 0.013, t = 2.78).
Figure 3. Begg's funnel plot of colorectal cancer risk associated with CYP1A1 rs1048943 A >G polymorphism (G vs. A).
DISCUSSION
In this meta-analyses, we comprehensively evaluated the associations between two polymorphisms (rs1048943 A > G and rs4646903 T > C) and CRC risk through 26 studies. We observed that rs1048943 A > G genetic variation was associated with increased risk of CRC. The association remained statistically significant in subgroups (Asians, Caucasians, PB, high quality score and the studies consistent with HWE). On the contrary, the association between rs4646903 T > C and CRC risk did not reach the significance level.
CYP1A1 protein is a member of CYP superfamily and widely distributes in lung, kidney, intestine, skin, larynx, placenta, lymphocyte, brain tissues [36]. Because the main role of this enzyme is to metabolize environmental carcinogens, such as PAHs, heterocyclic amines, aflatoxin B1 and estrogen [37], variations within CYP1A1 gene may induce the occurrence of CRC. Currently, a widely accepted paradigm for CYP1A1 enzyme mediated carcinogens activation is that CYP1A1 metabolizes polycyclic aromatic hydrocarbons to reactive epoxide intermediates, which could covalently bind to DNA and then induce tumors [38].
Due to the high minor allele frequency (MAF) as well as their possible involvement in an increased risk of several carcinomas [39], including lung cancer, colorectal cancer, breast cancer, leukemia, esophageal carcinoma and prostate cancer [40], CYP1A1 rs1048943 A > G and rs4646903 T > C polymorphisms are more widely studied. In addition to inducing the occurrence of cancers, the polymorphisms of CYP1A1 may also lead to other diseases, such as ulcerative colitis, colorectal adenoma, atherosclerosis, myocardial infarction and so on [41–43].
Partial meta-analyses on the association of CYP1A1 polymorphisms and colorectal cancer risk showed that CYP1A1 rs1048943 A > G might be associated with increased risk of CRC [44, 45]. In the present study, we combined the studies to increase sample size and further validated this results. However, contradictory results were observed in a previous meta-analysis which concluded that there was no association between rs1048943 A > G and CRC risk [46]. This discrepancy mainly results from sample size. We analyzed 20 studies with 8665 cases and 9953 controls while they only included two studies with 238 cases and 280 controls. For rs4646903 T > C, previous studies showed no association with CRC risk which was consistent with us.
Compared to previous meta-analyses, the present analysis has some advantages. Firstly, we have the largest sample size with the statistic power of 92% to evaluate the associations. Secondly, 90% studies included in our meta-analyses were of high quality score. Thirdly, we conducted sensitivity analysis and found no obvious influence of a single study on the pooled ORs and 95% CIs for rs1048943 A > G. In addition, negative result of publication bias evaluation indicated that our conclusions were unbiased. However, between-study heterogeneity existed, we should draw the conclusion with caution, subgroups analysis of rs1048943 A > G indicated that heterogeneities may be from ethnicity, source of control or controls deviated from HWE. For rs4646903 T > C polymorphism, substantial between-study heterogeneities may originate from ethnicity, source of control and controls deviated from HWE. Besides, for unavailable original data, we failed to analyze clinical features, such as tumor stage, age and sex, etc.
In conclusion, CYP1A1 rs1048943 A > G polymorphism may increase the CRC risk. However, our study still existed in some limitations, further studies with higher quality and larger sample size are necessary.
MATERIALS AND METHODS
Search strategy
We searched publications from PubMed, Web of knowledge, Embase and Chinese database of China National Knowledge Infrastructure (CNKI) and VIP database with the following search items: “CYP1A1” or “Cytochrome P450 1A1” and “polymorphism” or “variant” or “SNP” and “colorectal cancer” or “colon cancer” or “rectal cancer”. The languages were limited to English and Chinese. We updated the search results on January 2016 and confirmed potential relevant studies through the titles and abstracts.
Selection criteria
All studies included in the meta-analysis are selected according to the following criteria: (a) case-control studies; (b) studies about the associations between CYP1A1 rs1048943 A > G or rs4646903 T > C and colorectal cancer; (c) studies that contain genotype data; (d) when studies had overlapping populations, the most recent ones with the most complete data set were included. In addition, exclusion criteria were as follows: (a) overlapped articles or studies with overlapping data; (b) review articles, conference reports and dissertations.
Data extraction
Two investigators (ZXR and ZML) extracted data independently from the eligible studies with the following items: the first author's last name, year of publication, country, ethnicity, source of controls, polymorphism type, number of cases and controls, the frequency of each genotype in cases and controls (Supplementary Table 3), minor allele frequency (MAF) and p-value of Hardy Weinberg equilibrium (HWE). To get the accurate data, two investigators discussed together to reach a consensus.
Quality assessment
We evaluated the quality of the included studies respectively, according to the quality assessment criteria (Supplementary Table 1) [47–50]. The range of quality scores are from 0 (worst) to 15 (best). Publications with quality scores < 10 were categorized as “low quality” and those with quality scores ≥ 10 were categorized as “high quality” [51]. The process of scoring was listed in Supplementary Table 2.
Statistical analysis
Stata software (version 12.0; Stata Corporation, College Station, TX) was used to perform statistical analyses. We used allelic, heterozygote, homozygote, dominant and recessive as the models. The strength of associations between the CYP1A1 rs1048943 A > G and rs4646903 T > C and the risk of colorectal cancer were evaluated by the pooled odds ratios (ORs) and 95% confidence intervals (CIs). We set the significance cutoff as a p-value of 0.05 for the pooled OR. Between-study heterogeneity was assessed using Chi-squared-based Q-test. If heterogeneity P value was lower than 0.10, we considered the heterogeneity to be significant and random-effects model was used [52]. Otherwise, the fixed effects model was used [53]. We also performed stratified analyses by ethnicity (Asian and Caucasian), control source (population-based and hospital-based), quality score of studies (low and high) and HWE. Sensitivity analyses were performed to measure the stability of the results by consecutively omitting each study from the meta-analysis (leave-one-out sensitivity analysis). Begg's funnel plot [54] and Egger's test [55] (P <0.05 was considered significant) were used to evaluate the publication bias among the literatures.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Footnotes
CONFLICTS OF INTEREST
The authors have declared that no competing interests exist.
FUNDING
This study was supported by funding from the Science and Technology Commission of Shanghai Municipality, China (Grant No.15ZR1427700) and Xin Hua Hospital affiliated to Shanghai Jiaotong University School of Medicine, China (Grant No.13YJ22).
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Morris M, Platell C, McCaul K, Millward M, van Hazel G, Bayliss E, Trotter J, Ransom D, Iacopetta B. Survival rates for stage II colon cancer patients treated with or without chemotherapy in a population-based setting. Int J Colorectal Dis. 2007;22:887–895. doi: 10.1007/s00384-006-0262-y. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed FE. Gene-gene, gene-environment & multiple interactions in colorectal cancer. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2006;24:1–101. doi: 10.1080/10590500600614295. [DOI] [PubMed] [Google Scholar]
- 4.Khan N, Afaq F, Mukhtar H. Lifestyle as risk factor for cancer: Evidence from human studies. Cancer Lett. 2010;293:133–143. doi: 10.1016/j.canlet.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gil J, Gaj P, Misiak B, Ostrowski J, Karpinski P, Jarczynska A, Kielan W, Sasiadek MM. CYP1A1 Ile462Val polymorphism and colorectal cancer risk in Polish patients. Med Oncol. 2014;31:72. doi: 10.1007/s12032-014-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preissner S, Kroll K, Dunkel M, Senger C, Goldsobel G, Kuzman D, Guenther S, Winnenburg R, Schroeder M, Preissner R. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res. 2010;38:D237–243. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guengerich FP. Roles of cytochrome P-450 enzymes in chemical carcinogenesis and cancer chemotherapy. Cancer Res. 1988;48:2946–2954. [PubMed] [Google Scholar]
- 8.Guengerich FP, Kim DH, Iwasaki M. Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem Res Toxicol. 1991;4:168–179. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- 9.Gelboin HV. Benzo[alpha]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol Rev. 1980;60:1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- 10.Wrensch MR, Miike R, Sison JD, Kelsey KT, Liu M, McMillan A, Quesenberry C, Wiencke JK. CYP1A1 variants and smoking-related lung cancer in San Francisco Bay area Latinos and African Americans. Int J Cancer. 2005;113:141–147. doi: 10.1002/ijc.20537. [DOI] [PubMed] [Google Scholar]
- 11.Sivaraman L, Leatham MP, Yee J, Wilkens LR, Lau AF, Le Marchand L. CYP1A1 genetic polymorphisms and in situ colorectal cancer. Cancer Res. 1994;54:3692–3695. [PubMed] [Google Scholar]
- 12.Hayashi S, Watanabe J, Kawajiri K. High susceptibility to lung cancer analyzed in terms of combined genotypes of P450IA1 and Mu-class glutathione S-transferase genes. Jpn J Cancer Res. 1992;83:866–870. doi: 10.1111/j.1349-7006.1992.tb01992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritsche E, Bruning T, Jonkmanns C, Ko Y, Bolt HM, Abel J. Detection of cytochrome P450 1B1 Bfr I polymorphism: genotype distribution in healthy German individuals and in patients with colorectal carcinoma. Pharmacogenetics. 1999;9:405–408. [PubMed] [Google Scholar]
- 14.Ishibe N, Stampfer M, Hunter DJ, Hennekens C, Kelsey KT. A prospective study of cytochrome P450 1A1 polymorphisms and colorectal cancer risk in men. Cancer Epidemiol Biomarkers Prev. 2000;9:855–856. [PubMed] [Google Scholar]
- 15.Sachse C, Smith G, Wilkie MJ, Barrett JH, Waxman R, Sullivan F, Forman D, Bishop DT, Wolf CR. A pharmacogenetic study to investigate the role of dietary carcinogens in the etiology of colorectal cancer. Carcinogenesis. 2002;23:1839–1849. doi: 10.1093/carcin/23.11.1839. [DOI] [PubMed] [Google Scholar]
- 16.Ye Z, Parry JM. Genetic polymorphisms in the cytochrome P450 1A1, glutathione S-transferase M1 and T1, and susceptibility to colon cancer. Teratog Carcinog Mutagen. 2002;22:385–392. doi: 10.1002/tcm.10035. [DOI] [PubMed] [Google Scholar]
- 17.Slattery ML, Samowtiz W, Ma K, Murtaugh M, Sweeney C, Levin TR, Neuhausen S. CYP1A1, cigarette smoking, and colon and rectal cancer. Am J Epidemiol. 2004;160:842–852. doi: 10.1093/aje/kwh298. [DOI] [PubMed] [Google Scholar]
- 18.Chen K, Jin MJ, Fan CH, Song L, Jiang QT, Yu WP, Ma XY, Yao KY. A case-control study on the association between genetic polymorphisms of metabolic enzymes and the risk of colorectal cancer [Article in Chinese] Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26:659–664. [PubMed] [Google Scholar]
- 19.Hou L, Chatterjee N, Huang WY, Baccarelli A, Yadavalli S, Yeager M, Bresalier RS, Chanock SJ, Caporaso NE, Ji BT, Weissfeld JL, Hayes RB. CYP1A1 Val462 and NQO1 Ser187 polymorphisms, cigarette use, and risk for colorectal adenoma. Carcinogenesis. 2005;26:1122–1128. doi: 10.1093/carcin/bgi054. [DOI] [PubMed] [Google Scholar]
- 20.Landi S, Gemignani F, Moreno V, Gioia-Patricola L, Chabrier A, Guino E, Navarro M, de Oca J, Capella G, Canzian F. A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet Genomics. 2005;15:535–546. doi: 10.1097/01.fpc.0000165904.48994.3d. [DOI] [PubMed] [Google Scholar]
- 21.Little J, Sharp L, Masson LF, Brockton NT, Cotton SC, Haites NE, Cassidy J. Colorectal cancer and genetic polymorphisms of CYP1A1, GSTM1 and GSTT1: a case-control study in the Grampian region of Scotland. Int J Cancer. 2006;119:2155–2164. doi: 10.1002/ijc.22093. [DOI] [PubMed] [Google Scholar]
- 22.Talseth BA, Meldrum C, Suchy J, Kurzawski G, Lubinski J, Scott RJ. Genetic polymorphisms in xenobiotic clearance genes and their influence on disease expression in hereditary nonpolyposis colorectal cancer patients. Cancer Epidemiol Biomarkers Prev. 2006;15:2307–2310. doi: 10.1158/1055-9965.EPI-06-0040. [DOI] [PubMed] [Google Scholar]
- 23.Kiss I, Orsos Z, Gombos K, Bogner B, Csejtei A, Tibold A, Varga Z, Pazsit E, Magda I, Zolyomi A, Ember I. Association between allelic polymorphisms of metabolizing enzymes (CYP 1A1, CYP 1A2, CYP 2E1, mEH) and occurrence of colorectal cancer in Hungary. Anticancer Res. 2007;27:2931–2937. [PubMed] [Google Scholar]
- 24.Yeh CC, Sung FC, Tang R, Chang-Chieh CR, Hsieh LL. Association between polymorphisms of biotransformation and DNA-repair genes and risk of colorectal cancer in Taiwan. J Biomed Sci. 2007;14:183–193. doi: 10.1007/s11373-006-9139-x. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida K, Osawa K, Kasahara M, Miyaishi A, Nakanishi K, Hayamizu S, Osawa Y, Tsutou A, Tabuchi Y, Shimada E, Tanaka K, Yamamoto M, Takahashi J. Association of CYP1A1, CYP1A2, GSTM1 and NAT2 gene polymorphisms with colorectal cancer and smoking. Asian Pac J Cancer Prev. 2007;8:438–444. [PubMed] [Google Scholar]
- 26.Pande M, Amos CI, Osterwisch DR, Chen J, Lynch PM, Broaddus R, Frazier ML. Genetic variation in genes for the xenobiotic-metabolizing enzymes CYP1A1, EPHX1, GSTM1, GSTT1, and GSTP1 and susceptibility to colorectal cancer in Lynch syndrome. Cancer Epidemiol Biomarkers Prev. 2008;17:2393–2401. doi: 10.1158/1055-9965.EPI-08-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira Serafim PV, Cotrim Guerreiro da Silva ID, Manoukias Forones N. Relationship between genetic polymorphism of CYP1A1 at codon 462 (Ile462Val) in colorectal cancer. Int J Biol Markers. 2008;23:18–23. doi: 10.1177/172460080802300103. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi M, Otani T, Iwasaki M, Natsukawa S, Shaura K, Koizumi Y, Kasuga Y, Sakamoto H, Yoshida T, Tsugane S. Association between dietary heterocyclic amine levels, genetic polymorphisms of NAT2, CYP1A1, and CYP1A2 and risk of colorectal cancer: a hospital-based case-control study in Japan. Scand J Gastroenterol. 2009;44:952–959. doi: 10.1080/00365520902964721. [DOI] [PubMed] [Google Scholar]
- 29.Zheng LJ, Pang EG, Yang ZW, Ma J, Zhu ZY. Study on the Relationship between CYP1A1 Gene Ile-Val and Msp1 Polymorphism and Susceptibility of Colorectal Cancer. Journal of Shanxi Datong University (Natural Science) 2009;25:36–39. [Google Scholar]
- 30.Liu Y, Zhang PY, Shi CL, Qiu B, Meng XW. Realationship between cytochrome P450 1A1 gene polymorphism and susceptibility to colorectal carcinoma. Journal of Shanghai Jiaotong University (Medical Science) 2009;29:58–60. 67. [Google Scholar]
- 31.Cleary SP, Cotterchio M, Shi E, Gallinger S, Harper P. Cigarette smoking, genetic variants in carcinogen-metabolizing enzymes, and colorectal cancer risk. Am J Epidemiol. 2010;172:1000–1014. doi: 10.1093/aje/kwq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nisa H, Kono S, Yin G, Toyomura K, Nagano J, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, Maekawa T. Cigarette smoking, genetic polymorphisms and colorectal cancer risk: the Fukuoka Colorectal Cancer Study. BMC Cancer. 2010;10:274. doi: 10.1186/1471-2407-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darazy M, Balbaa M, Mugharbil A, Saeed H, Sidani H, Abdel-Razzak Z. CYP1A1, CYP2E1, and GSTM1 gene polymorphisms and susceptibility to colorectal and gastric cancer among Lebanese. Genet Test Mol Biomarkers. 2011;15:423–429. doi: 10.1089/gtmb.2010.0206. [DOI] [PubMed] [Google Scholar]
- 34.Houlle S, Charbonnier F, Houivet E, Tinat J, Buisine MP, Caron O, Benichou J, Baert-Desurmont S, Frebourg T. Evaluation of Lynch syndrome modifier genes in 748 MMR mutation carriers. Eur J Hum Genet. 2011;19:887–892. doi: 10.1038/ejhg.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saeed HM, Alanazi MS, Nounou HA, Salaby MA, Semlali A, Azzam N, Aljebreen A, Alharby O, Parine NR, Shaik J, Arafaha M. Cytochrome P450 1A1, 2E1 and GSTM1 gene polymorphisms and susceptibility to colorectal cancer in the Saudi population. Asian Pac J Cancer Prev. 2013;14:3761–3768. doi: 10.7314/apjcp.2013.14.6.3761. [DOI] [PubMed] [Google Scholar]
- 36.Crewe HK, Notley LM, Wunsch RM, Lennard MS, Gillam EM. Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: formation of the 4-hydroxy, 4′-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos. 2002;30:869–874. doi: 10.1124/dmd.30.8.869. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez J, Marchand-Geneste N, Giraudel JL, Shimada T. Docking and QSAR comparative studies of polycyclic aromatic hydrocarbons and other procarcinogen interactions with cytochromes P450 1A1 and 1B1. SAR QSAR Environ Res. 2012;23:87–109. doi: 10.1080/1062936X.2011.636380. [DOI] [PubMed] [Google Scholar]
- 38.He X, Feng S. Role of Metabolic Enzymes P450 (CYP) on Activating Procarcinogen and their Polymorphisms on the Risk of Cancers. Curr Drug Metab. 2015;16:850–863. doi: 10.2174/138920021610151210164501. [DOI] [PubMed] [Google Scholar]
- 39.Hussein AG, Pasha HF, El-Shahat HM, Gad DM, Toam MM. CYP1A1 gene polymorphisms and smoking status as modifier factors for lung cancer risk. Gene. 2014;541:26–30. doi: 10.1016/j.gene.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Daly AK. Polymorphic Variants of Cytochrome P450:Relevance to Cancer and Other Diseases. Adv Pharmacol. 2015;74:85–111. doi: 10.1016/bs.apha.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Ma C, Lu XC, Fan L, Luo Y, Yang B, Gao Y, Liu XF. Irbesartan regulates inflammatory gene expressions related to atherosclerosis in EA. hy926 cells [Article in Chinese] Nan Fang Yi Ke Da Xue Xue Bao. 2011;31:1835–1839. [PubMed] [Google Scholar]
- 42.Buyukgoze O, Osmanoglu N, Arslan S, Sen A. Association of the CYP1A1*2A, GSTT1 null, GSTM1 null, mEPHX*3, and XRCC1-399 genetic polymorphisms with ulcerative colitis. Int J Colorectal Dis. 2013;28:593–595. doi: 10.1007/s00384-012-1507-6. [DOI] [PubMed] [Google Scholar]
- 43.Hamachi T, Tajima O, Uezono K, Tabata S, Abe H, Ohnaka K, Kono S. CYP1A1, GSTM1, GSTT1 and NQO1 polymorphisms and colorectal adenomas in Japanese men. World J Gastroenterol. 2013;19:4023–4030. doi: 10.3748/wjg.v19.i25.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, Wang JJ, Sun L, Li HL. Association between CYP1A1 polymorphism and colorectal cancer risk: a meta-analysis. Mol Biol Rep. 2012;39:3533–3540. doi: 10.1007/s11033-011-1126-2. [DOI] [PubMed] [Google Scholar]
- 45.Jin JQ, Hu YY, Niu YM, Yang GL, Wu YY, Leng WD, Xia LY. CYP1A1 Ile462Val polymorphism contributes to colorectal cancer risk: a meta-analysis. World J Gastroenterol. 2011;17:260–266. doi: 10.3748/wjg.v17.i2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen K, Jiang QT, He HQ. Relationship between metabolic enzyme polymorphism and colorectal cancer. World J Gastroenterol. 2005;11:331–335. doi: 10.3748/wjg.v11.i3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakkinstian A, McKay GJ, McEvoy M, Chakravarthy U, Chakrabarti S, Silvestri G, Kaur I, Li X, Attia J. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol. 2011;173:1365–1379. doi: 10.1093/aje/kwr025. [DOI] [PubMed] [Google Scholar]
- 48.Attia J, Thakkinstian A, D'Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003;56:297–303. doi: 10.1016/s0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]
- 49.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. Jama. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 50.Ioannidis JP, Boffetta P, Little J, O'Brien TR, Uitterlinden AG, Vineis P, Balding DJ, Chokkalingam A, Dolan SM, Flanders WD, Higgins JP, McCarthy MI, McDermott DH, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 51.He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, Chen W, Jia WH. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159. doi: 10.1038/srep06159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 53.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 54.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 55.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.