Abstract
Bile ductular proliferation is markedly upregulated in biliary fibrosis and cirrhosis. However, the mechanisms regulating this upregulation in bile ductular proliferation have not been defined. Recently, we demonstrated that expression of the ectonucleotidase nucleoside triphosphate diphosphohydrolase-2 (NTPDase2/Entpd2) by portal fibroblasts (PF) is a critical regulator of bile ductular proliferation. Since interleukin 6 (IL-6) is markedly upregulated in biliary cirrhosis, our aims were to determine the role and mechanism of IL-6 in the regulation of NTPDase2 by PF. We found that IL-6 downregulated NTPDase2 protein expression in a concentration-dependent and time-dependent fashion but did not alter PF α-smooth muscle actin expression. IL-6 markedly downregulated NTPDase2 mRNA expression. Expression of the IL-6 receptor gp130 but not the IL-6 receptor gp80 was detected in PF. Two transcription start sites were identified in rat Entpd2 by the method of RNA ligase-mediated rapid amplification of 5′ cDNA ends. The minimal promoter construct, but not shorter constructs, was downregulated by IL-6. Three putative IL-6 response elements were identified in silico and mutated. Mutation of all three response elements, but not fewer elements, completely abolished the IL-6 response. Thus IL-6 transcriptionally downregulates NTPDase2 expression by PF via actions at specific promoter elements independently of myofibroblastic differentiation. This effect may represent a novel signaling pathway by which bile ductular proliferation is dys-regulated in biliary cirrhosis and thus provides a potential therapeutic approach for the regulation of bile ductular growth.
Keywords: bile duct epithelia, ecto-adenosine triphosphatase, P2Y, biliary cirrhosis, interleukin-6, nucleoside triphosphate diphosphohydrolase-2
Extracellular nucleotides such as ATP regulate a variety of important cellular functions via activation of specific P2X and P2Y receptors (1, 8, 18, 52). In the liver, P2Y receptors are of particular importance in such diverse processes as bile secretion (12), cell volume autoregulation (53), and cell growth (21, 49). Since P2Y receptor activation has important downstream consequences, the regulation of P2Y receptors is of great interest.
One of the primary ways in which P2X and P2Y receptors are regulated is via enzymatic catalysis of extracellular nucleotides, which then limit agonist availability (4). The chief enzymes that mediate this catalytic activity are enzymes of the ectonucleoside triphosphate diphosphohydrolase (E-NTPDase) family (28, 38). NTPDase2/CD39L1 is expressed in a variety of tissues, but there are limited observations about its potential physiologic function(s) (31, 44). In the healthy liver, NTPDase2 is expressed only in portal fibroblasts (PF), fibrogenic cells that surround intrahepatic bile ducts (11). NTPDase2 is specifically downregulated in biliary cirrhosis in both rats and humans (10). Recently, we demonstrated that NTPDase2 critically regulates bile duct epithelial growth via catalysis of extracellular nucleotides and that this regulation is lost in biliary cirrhosis (21). However, the molecular mechanism by which NTPDase2 is downregulated in biliary cirrhosis is unknown.
Bile duct epithelia upregulate production of several inflammatory mediators in biliary cirrhosis, including monocyte chemoattractant protein-1 (MCP-1/CCL2) (30), stromal cell-derived factor-1 (48), and interleukin-6 (IL-6) (58). We have shown in previous work that MCP-1 has multiple effects on PF, including induction of α-smooth muscle actin (ASMA) expression and stress filament reorganization, upregulation of collagen synthesis, and downregulation of NTPDase2 protein (27). This finding suggested that NTPDase2 expression was linked to myofibroblastic differentiation of PF.
Since bile duct epithelia markedly upregulate IL-6 expression in biliary cirrhosis, we investigated the effect of IL-6 on PF NTPDase2 expression and myofibroblastic differentiation. Presently we report that IL-6 transcriptionally downregulates NTPDase2 expression in PF without inducing myofibroblastic differentiation. To determine the mechanism by which IL-6 downregulates NTPDase2 expression in PF, we cloned and characterized the rat NTPDase2 promoter and identified specific IL-6 response elements (IL-6 REs). These findings imply that NTPDase2 can be regulated independently of myofibroblastic differentiation and provide potential new therapeutic approaches to the regulation of bile ductular proliferation.
MATERIALS AND METHODS
Confocal immunofluorescence
Expression of gp80 and gp130 by PF was determined by indirect confocal immunofluorescence. Liver sections (7 μm) were obtained using methods previously described (11) and were affixed to glass slides. Slides were washed twice in PBS and fixed with 3.7% formaldehyde. Slides were incubated with either rabbit polyclonal anti-gp80 (Santa Cruz Biotechnology, Santa Cruz, CA) or rabbit polyclonal anti-gp130 (Santa Cruz Biotechnology) (1:100 each) overnight at 4°C. Slides were then washed with cold PBS for 5 min three times and incubated with AlexaFluor 488-conjugated anti-rabbit (Molecular Probes, Eugene, OR) for 45 min at 37°C. Slides were then washed with rhodamine phalloidin to identify filamentous actin (11) and washed again with ice-cold PBS before use. Slides incubated with secondary antibody alone were used as a control for specificity of fluorescence detection. Confocal imaging of fixed liver sections was performed using a Zeiss LSM 510 confocal imaging system. Sections were excited using a krypton-argon laser at 488 nm and observed at >515 nm to detect Alexa 488 fluorescence, and they were excited at 568 nm and observed at >585 nm to detect rhodamine fluorescence.
Isolation of primary rat PF
Male adult Sprague-Dawley rats (180–250 g; Harlan Sprague Dawley, Indianapolis, IN) were used for isolation of PF in all experiments. PF were isolated as described previously (26). Briefly, rat nonparenchymal cell (NPC) fractions were obtained by collagenase and Pronase digestion of rat livers. Cell suspensions were separated using serial mesh filtration. The resulting suspension of NPC was plated in medium containing DMEM/F-12 containing 2% penicillin-streptomycin, 10% fetal calf serum, 0.3% gentamicin, and 0.1% fungizone. Cells were used 96 h after isolation, at which time cell purity approaches 100%.
Immunoblot analysis
Alterations in expression of NTPDase2 were determined by immunoblot using a rabbit polyclonal antibody directed against NTPDase2/CD39L1 (11, 44), and alterations in ASMA expression were determined by a mouse monoclonal antibody directed against rat ASMA (Sigma, St. Louis, MO) (13). Changes in expression were compared with β-tubulin expression (protein loading control). PF were treated overnight with IL-6 (1, 5, 10, or 20 ng/ml) or control (buffer alone), and protein homogenates were isolated. In separate experiments, PF were treated with or without IL-6 (10 ng/ml) for 0–24 h. Equal amounts of protein for each group were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) under non-reducing conditions and transferred onto a polyvinylidene difluoride membrane (Immobilon; Millipore, Bedford, MA). The membrane was blocked with nonfat milk (5% in PBS), hybridized to rabbit polyclonal anti-NTPDase2/CD39L1 (1:500), mouse monoclonal anti-ASMA (1: 2,000; Sigma), or mouse monoclonal anti-β-tubulin (1:1,000; Cell Signaling, Danvers, MA) and then anti-rabbit or anti-mouse secondary antibody, respectively, and developed using enhanced chemiluminescence.
In separate experiments, expression of the IL-6 receptors gp80 and gp130 was determined in untreated PF. PF were isolated, electrophoresed, transferred, and blotted as described above. Exposure to either gp80 or gp130 (1:10,000 each) was performed as described above, and detection was achieved using enhanced chemiluminescence.
Real-time RT-PCR
Changes in NTPDase2 mRNA were determined using real-time RT-PCR with an ABI PRISM 7500 sequence detection system (Applied Biosystems, Foster City, CA). Total RNA was extracted from PF treated overnight with either IL-6 (10 ng/ml) or buffer alone, and cDNA was synthesized by reverse transcriptase. The level of NTPDase2 was determined using sequence-specific oligonucleotide primers and probes [upstream, 5′-CAGCGGACAAGGAGAATGACA-3′; downstream, 5′-TGCCACCACCTTGAACATCA-3′; TaqMan (Applied Biosystems) MGB probe 6FAM-CAT CGT GGC CAG CAC AGC TCT-MGBNFQ]. Rat GAPDH primers were used as endogenous controls for mRNA concentration. PCR was performed under the following protocol: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s.
Analysis of 5′ cDNA end by RNA ligase-mediated rapid amplification of cDNA ends
The transcription start site of the rat Entpd2 gene was determined using a modified method of 5′ rapid amplification of cDNA ends (RACE), the RNA ligase-mediated rapid amplification of 5′ cDNA ends (5′ RLM-RACE), using FirstChoice RLM-RACE kit following the manufacturer’s instructions (Ambion, Austin, TX). Total rat heart RNA was isolated with Trizol reagent (Invitrogen, Burlington, ON, Canada) and used as template for the replacement of the mRNA 5′ CAP structure by a 5′ RLM-RACE adaptor. The full-length RNA ligated to the adaptor was then reverse transcribed with SuperScript II (Invitrogen) and an oligo(dT)18. The Entpd2 transcription start sites were analyzed by nested PCR amplification. The first amplification was done using 5′ RLM-RACE and gene-specific outer primers (gsrN2-R2) at annealing temperatures of both 60 and 55°C (Table 1). An aliquot of each of these first PCR products was amplified by another PCR with the 5′ RLM-RACE and gene-specific inner primers (gsrN2-R1) at an annealing temperature of 55°C. PCR amplifications were performed using the Expand high-fidelity PCR system (Roche, Laval, QC, Canada) under the following conditions: 94°C for 3 min, 35 cycles of 94°C for 30 s, 55 or 60°C (as indicated in Table 1) for 30 s, and 72°C for 30 s, completed by a 7-min incubation at 72°C. Samples from each of these two nested amplifications were ligated into pCRII TOPO vector (Invitrogen) following the manufacturer’s instructions. Five individual colonies from each transformation were analyzed by PCR amplification following the pCRII TOPO vector kit instructions. Three clones originating from 55°C outer PCR and two from 60°C outer PCR were analyzed by sequencing on a DNA analyzer (ABI 3730xl; Applied Biosystems).
Table 1.
Primers used for these studies
| Primers | Sequence (5′-3′) | Location | Annealing Temperature, °C | Experiment |
|---|---|---|---|---|
| gsrN2-R2 | TAGAGGGGTCATTTGCATAGCTGG | +2024/+2047 (exon3) | 55 and 60 | 5′ RLM-RACE |
| gsrN2-R1 | TGTCATTCTCCTTGTCCGCTGGCC | +1477/+1500 (exon2) | 55 | 5′ RLM-RACE |
| pRN2-R1 | GAACGCGTGGGAGAACACAGC | −23/−3 | 5′ Flanking region cloning | |
| pRN2-F1 | TCTGCCTGAGCCAAGCCACAC | −1688/−1668 | 63 | 5′ Flanking region cloning |
| pRN2-F2 | CCATGAGTGTCTCCCCAGGGC | −1530/−1510 | 63 | 5′ Flanking region cloning |
| pRN2-F3 | GGAAGCATGGGATGGGAGAGC | −1302/−1282 | 63 | 5′ Flanking region cloning |
| pRN2-F4 | TGTCCCTGCCTGTTCAGCAGC | −1081/−1061 | 63 | 5′ Flanking region cloning |
| pRN2-F5 | TGTGTGCCTGGTCGGTCTAGC | −889/−869 | 63 | 55′ Flanking region cloning |
| pRN2-F6 | GGGAGGCAAAAAGAGAGCGGC | −680/−660 | 63 | 5′ Flanking region cloning |
| pRN2-F7 | CAGATGCCGAACCCCTGTGCC | −489/−469 | 63 | 5′ Flanking region cloning |
| pRN2-F8 | CAAACCCCAACACCGGGTATC | −335/−315 | 60 | 5′ Flanking region cloning |
| pRN2-F9 | ACCGCTCCACTCACCTGAGTC | −188/−168 | 60 | 5′ Flanking region cloning |
| pRN2-F10 | GTCTAACTCTGGGGCCCCGGC | −81/−61 | 63 | 5′ Flanking region cloning |
| IL-6 RE A sense | CTTGCCTTTCCCTGGCCAGA | −407/−388 | Mobility shift assays | |
| IL-6 RE A antisense | TCTGGCCAGGGAAAGGCAAG | −407/−388 | Mobility shift assays | |
| IL-6 RE B sense | GGCTGCCTAGGAATCAACCT | −358/−339 | Mobility shift assays | |
| IL-6 RE B antisense | AGGTTGATTCCTAGGCTGCC | −358/−339 | Mobility shift assays | |
| IL-6 RE C sense | CTCAGGACTCCCAGGGCAGA | −312/−293 | Mobility shift assays | |
| IL-6 RE C antisense | TCTGCCCTGGGAGTCCTGAG | −312/−293 | Mobility shift assays | |
| α2IL-6 RE sense | GATCCTTCTGGGAATTCCTA | Mobility shift assays | ||
| α2IL-6 RE antisense | TAGGAATTCCCAGAAGGATC | Mobility shift assays |
All primers are numbered with reference to the translation of the first codon (ATG) of rat Entpd2. 5′ RLM-RACE, RNA ligase-mediated rapid amplification of 5′; cDNA ends; IL-6 RE, interleukin-6 response element.
Genomic DNA isolation
Rat heart genomic DNA was rapidly isolated with the method described by Chapdelaine et al (9). Briefly, liquid nitrogen frozen tissue was ground into fine powder, resuspended in a proteinase K solution [50 μg/ml proteinase K, 0.5% sarcosyl (CinnaGen; Ottawa, ON, Canada), and 0.5 M EDTA], and incubated for 15 min at 56°C. The digested tissue was diluted 10 times in 50 mM Tris · HCl buffer, pH 8.0, and treated with 10 μg/ml RNaseA. After phenol-chloroform extraction, genomic DNA was obtained by ethanol precipitation with 0.2 volumes of 5 M NaCl and resuspended in water.
PCR amplification of Entpd2 putative promoter and construction of reporter vectors
To analyze the promoter activity of the 5′ untranslated region of rat Entpd2 gene, series of 10 fragments of the promoter region with length between 1,686 and 79 bp were generated. An extra restriction enzyme site was added to each forward (KpnI) and reverse primer (BglII). All primer sequences and positions are listed in Table 1. For PCR amplification of the longest fragment (1,686 bp) that was expected to contain the complete promoter, 0.1 μg of genomic DNA isolated from rat heart was used as template with the primers prN2-F1 and prN2-R1. The first plasmid (~10 ng) was then used as template for PCR amplification of all other promoter fragment (prN2-F2 to prN2-F10) with the same reverse primer (prN2-R1). The amplifications were done with Phusion high-fidelity DNA polymerase (Finnzyme, Ipswich, MA) for maximum of fidelity of the elongation. All PCR amplifications were done as follows: 98°C for 1 min, 30 cycles of 98°C for 10 s, annealing temperature as indicated in Table 1 for 30 s, and 72°C for 1 min, and ended by a 7-min incubation at 72°C. Amplification products were ligated into the KpnI/BglII cloning site in the luciferase reporter gene pGL2-Basic vector (Promega, Nepean, ON, Canada). The nucleotide sequence of each promoter fragment was confirmed by sequencing as described above.
Site-directed mutagenesis of putative IL-6 response promoter elements
Putative IL-6 REs were analyzed by site-directed mutagenesis. Site-directed mutants were generated using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The following sets of oligonucleotide primers were used: site A: forward, 5′-GGGTCTGGCTTGCCTTTGGCCAGAAAAAAATTTGGC-3′; reverse, 5′-GCCAAATTTTTTTCTGGCCAAAGGCAAGCCAGACCC-3′; site B: forward, 5′-CGCAAGCAAGGCTGCGGAATCAACCTAGTCAAACCC-3′; reverse, 5′-GGGTTTGACTAGGTTGATTCCGCAGCCTTGCTTGCG-3′; and site C: forward, 5′-GGGTATCCTCTCAGGACCAGGGCAGAGCTCCC-3′; reverse, 5′-GGGAGCTCTGCCCTGGTCCTGAGAGGATACCC-3′. PCR amplification of mutants was performed using Pfu Turbo DNA polymerase (Stratagene) under the following parameters: 95°C for 1 min, followed by 18 cycles of 95°C for 50 s, 60°C for 50 s, and 68°C for 6 min, and then 68°C for 7 min. Each mutation was confirmed by automated sequencing before use.
Culture and transfection of HepG2 cells
HepG2 cells were cultured in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. On the day before transfection, cells were split into 12-well plates. Plasmid DNA (4 μg) was diluted into 100 μl of Opti-MEM I medium (Invitrogen), and then 6 μl of FuGENE 6 reagent (Roche Biosciences, Palo Alto, CA) was added. The mixture was incubated at room temperature for 15 min and then added to cells in a stepwise fashion.
Detection of changes in luciferase activity
Transfected HepG2 cells were treated overnight with IL-6 (10 ng/ml) or buffer alone. Changes in firefly luciferase activity were normalized to Renilla luciferase activity and detected using a Synergy HT multidetection microplate reader (BioTek, Winooski, VT).
Electromobility shift assay
Nuclear extracts from HepG2 cells after 1 h of stimulation with IL-6 (20 ng/ml) or buffer alone were prepared following the procedure outlined by Schreiber et al. (41). DNA binding assays were performed with 10 μg of nuclear extracts as described previously (51). The 32P-labeled double-stranded oligonucleotides used as probes are described in Table 1. For competition experiment, a 25-fold molar excess of unlabeled double-stranded oligonucleotides was added to the binding reaction. Complex bound to the IL-6 REs was separated by electrophoresis under nondenaturing conditions in a 4% polyacrylamide gel containing 5% glycerol that was run at 200 V for 3 h at 4°C in 0.5× Tris-borate-EDTA buffer and visualized by autoradiography.
Statistical analysis
Data are means ± SD where appropriate. Comparisons between individual groups were made with two-tailed t-tests.
RESULTS
PF express the IL-6 receptor gp130
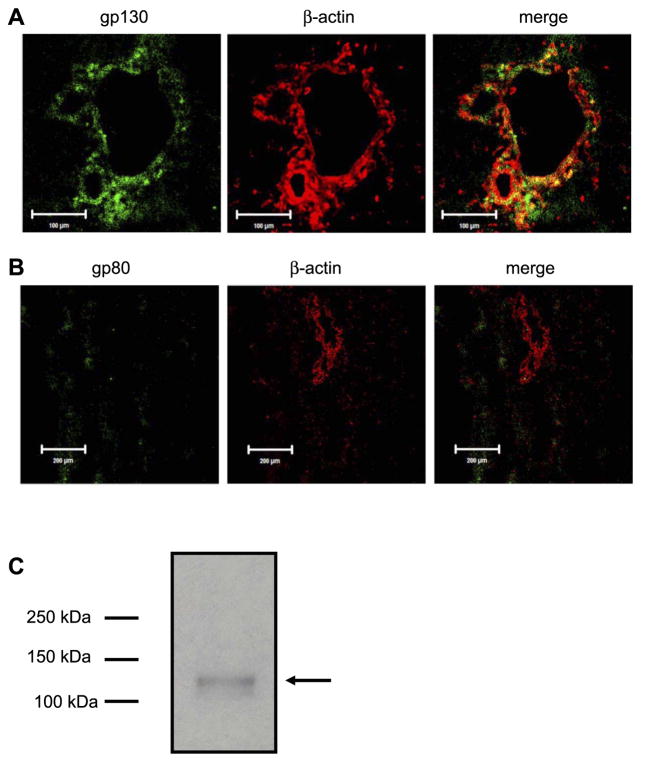
There are two IL-6 receptors that have been identified in mammalian cells: gp80 and gp130. The gp130 receptor is expressed by a variety of cell types, whereas the distribution of gp80 is more limited (40). The expression of gp130 and gp80 by PF was determined by confocal immunofluorescence in normal rat liver. As shown in Fig. 1, expression of gp130 is limited to the region in which PF are expressed. In contrast, gp80 is not expressed in this region (Fig. 1B). The expression of gp130 by PF was confirmed by immunoblot (Fig. 1C). These data demonstrate that PF express the IL-6 receptor gp130 but not gp80.
Fig. 1.
Portal fibroblasts (PF) express the IL-6 receptor gp130. Expression of IL-6 receptors gp80 and gp130 was determined by confocal immunofluorescence in normal rat liver sections. A: expression of gp130 was detected in stromal tissue surrounding intrahepatic bile ducts, portal veins, and hepatic arteries. Structures can be identified by colabeling with rhodamine phalloidin to identify β-actin. No staining outside of the portal area was noted (not shown). B: expression of gp80 was not noted within the portal area. Occasional positive cells were noted within the parenchyma. Again, portal structures were identified by rhodamine phalloidin staining. C: immunoblot analysis using the gp130 antibody in A identified a single 130-kDa band (arrow) in isolated PF. No band was detected using gp80 antibody in PF (not shown).
IL-6 downregulates NTPDase2 protein expression in a time-and concentration-dependent fashion independently of ASMA expression
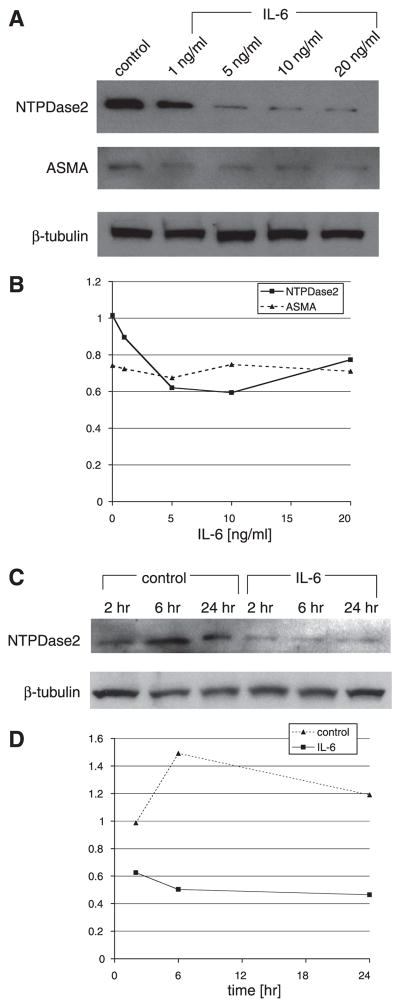
The effect of IL-6 (1–20 ng/ml) on PF NTPDase2 protein expression was determined by immunoblot (Fig. 2, A and B). Primary rat PF were treated overnight with IL-6, and the expression of NTPDase2, ASMA, and β-tubulin (loading control) was analyzed. IL-6 downregulated NTPDase2 expression in a concentration-dependent fashion, with maximal effect at 5–10 ng/ml. Importantly, IL-6 had no effect on expression of ASMA, suggesting that the regulation of NTPDase2 was independent of myofibroblastic differentiation. The concentrations of IL-6 that downregulated NTPDase2 expression were comparable to those known to be secreted by bile duct epithelia and to regulate bile duct epithelial activity (34).
Fig. 2.
IL-6 downregulates nucleoside triphosphate diphosphohydrolase (NTPDase) protein expression in PF. A: concentration downregulation of NTPDase2 protein by IL-6. Day 1 primary rat PF were treated overnight with either buffer alone or buffer containing IL-6 (1–20 ng/ml), and protein was extracted. Relative expression of NTPDase2, α-smooth muscle actin (ASMA), and β-tubulin (loading control) were determined by immunoblot. IL-6 induced downregulation of NTPDase2 in a concentration-dependent fashion but had no effect on ASMA expression. A representative of 3 blots is shown. B: quantification of changes in band intensity. Changes in band intensity were determined using ImageJ software on scanned blots. Analysis confirmed that IL-6 downregulated NTPDase2 expression (solid line) but had no effect on ASMA expression (dashed line). C: time-sensitive downregulation of NTPDase2 protein by IL-6. PF were treated at various time points (2, 6, or 24 h) with either control buffer or buffer containing IL-6 (10 ng/ml). Relative expression of NTPDase2 and β-tubulin was determined by immunoblot. Downregulation of NTPDase2 was noted at 2 h and sustained up to 24 h. D: quantification of changes in band intensity. Changes in band intensity were assessed as described in B. Analysis confirmed that IL-6 downregulated NTPDase2 expression at 2, 6, and 24 h.
We next determined whether IL-6 downregulated PF expression of NTPDase2 in a time-sensitive fashion. IL-6 downregulated NTPDase2 within 2 h, and the effect was sustained to 24 h (Fig. 2, C and D). IL-6 had no effect on NTPDase2 expression in 30 or 60 min (not shown). Thus downregulation of NTPDase2 in PF by IL-6 is time- and concentration-dependent and independent of myofibroblastic differentiation.
IL-6 downregulates NTPDase2 transcription
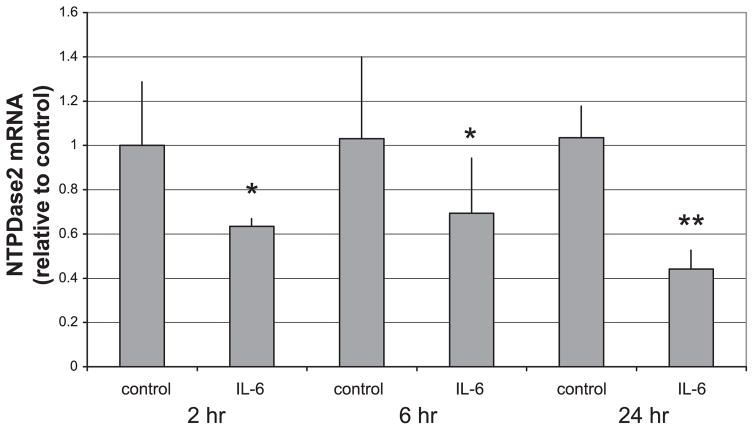
The effect of IL-6 on PF NTPDase2 mRNA expression over time was determined by real-time RT-PCR. As shown in Fig. 3. IL-6 downregulated NTPDase2 mRNA to ~40–70% of control, with maximal downregulation at 24 h. This experiment demonstrates that IL-6-dependent downregulation of NTPDase2 by PF occurs at the transcriptional level.
Fig. 3.
IL-6 downregulates NTPDase2 mRNA expression in PF. Day 1 primary rat PF were treated at various time points with either buffer alone or buffer containing IL-6 (10 ng/ml), and total RNA was extracted. Relative mRNA expression of NTPDase2 was determined using real-time RT-PCR and calculated relative to GAPDH. IL-6 decreased NTPDase2/GAPDH mRNA ratio at 2, 6, and 24 h (n = 4 for each group). *P < 0.05; **P < 0.005.
Determination of the transcription start site and promoter characterization
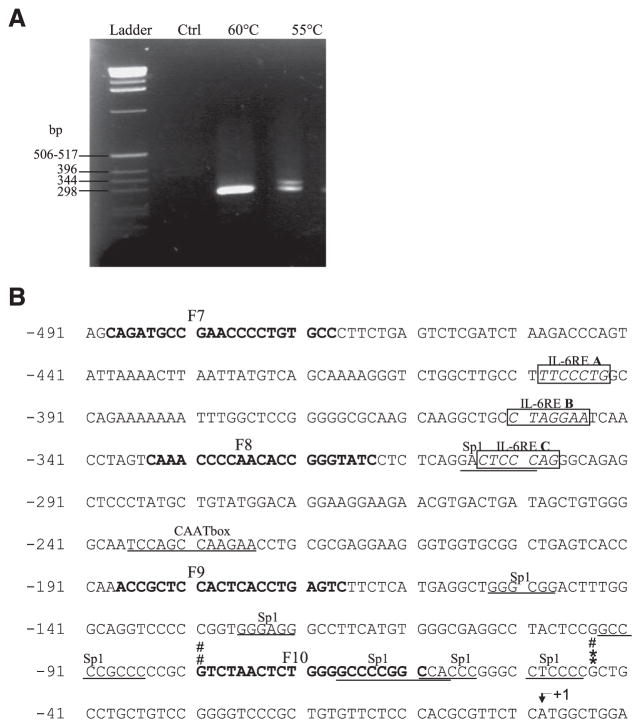
To assess the mechanism by which NTPDase2 transcription is regulated by IL-6, we located and cloned the rat Entpd2 promoter. We therefore determined the transcription start site(s) of the gene by performing 5′ RLM-RACE with total RNA purified from rat heart as template. A nested PCR amplification was done on 5′ RACE adaptor-ligated cDNAs. The PCR products obtained were separated by electrophoresis on an agarose gel as shown in Fig. 4A. The first step of the nested PCR was done at two different annealing temperatures. The annealing temperature of 55°C (Fig. 4A, 4th lane) revealed two bands at ~300 and 350 bp, whereas only the shorter band was amplified at the annealing temperature of 60°C (Fig. 4A, 3rd lane).
Fig. 4.
Rat Entpd2 transcription start sites and promoter analysis. A: electrophoretic analysis on 2% agarose gel of the nested PCR amplification products was performed on total rat heart modified cDNAs. The first round of PCR amplification was performed at 2 different annealing temperatures, 60 and 55°C, as indicated. The second lane shows the negative control (Ctrl) where 5′ rapid amplification of cDNA ends (RACE) adaptor-ligated cDNAs were omitted. B: rat Entpd2 minimal promoter nucleotide sequence. The adenine of the translation start site (ATG) is indicated by an arrow at the nucleotide +1. Transcription start sites, deduced from the sequences of 5 individual clones originating from the outer PCR at 60 and 55°C annealing temperatures, are indicated by asterisks (*) or pound signs (#), respectively. The IL-6 response elements (IL-6 REs A, B, and C) are boxed, and potential Sp-1 binding sites as well as the CAAT box-like elements are underlined. The prN2-F7 to prN2-F10 primer sequences (F7–F10) are indicated in bold type.
After the cloning of each nested PCR product separately (i.e., those from 55 and 60°C annealing temperatures), five individual colonies from each cloning experiment were analyzed by PCR, and we found that the smaller fragment was predominant (data not shown). All tested clones from the annealing temperature of 60°C corresponded to the lower fragments, whereas three lower fragments and two upper fragments were found with the annealing temperature of 55°C. Five clones were selected, and the fragments of interest were sequenced, with two corresponding to the longest fragment of ~350 bp and three to the shortest fragment of ~300 bp. The analysis of these sequences showed the presence of two transcription start sites for rat Entpd2: a major one at −45 bp and a second one at −81 bp with respect to the ATG (Fig. 4B). None of these two transcription start sites corresponds to an initiator consensus sequence (Inr) (20).
We also performed computational analysis of the GenBank database of expressed sequence tags (ESTs), looking for 5′-end sequences of rat NTPDase2 cDNAs using the sequence with the GenBank accession number BC086558 for comparison. Twelve homologous EST sequences were found with a 5′ end located between −8 and −61 bp upstream of the ATG (data not shown). Even if none of them corresponded exactly to our 5′ RLM-RACE results, it confirms that this region is the most highly probable target to contain the transcriptions starts sites.
In silico analysis of sequences upstream of transcription start site, expected to correspond to the promoter, showed a GC-rich sequence with seven putative binding sites for Sp-1 and one CAAT box-like element (Fig. 4B) but no TATA box. TATA-less promoters possess GC-rich sequences, and Sp-1 is expected to play an important role in polymerase recruitment for such promoters (36). Our analysis shows that the rat NTPDase2 promoter is among the 46% other promoters that do not possess a TATA or an Inr element (56).
The NTPDase2 promoter is IL-6 sensitive
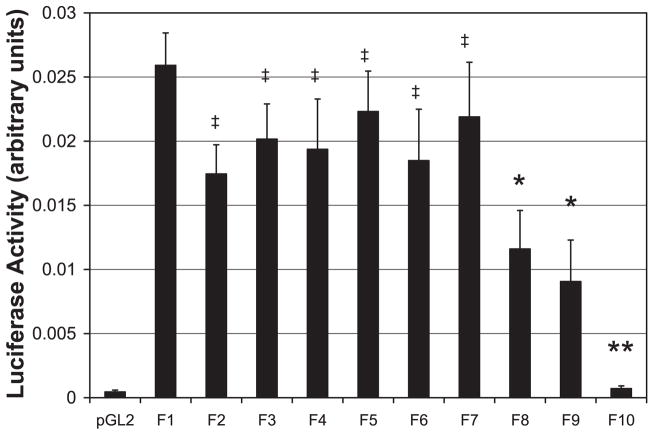
A 1,689-bp sequence upstream of the start codon, corresponding to the rat NTPDase2 promoter, was cloned. To assess the regulation of NTPDase2 promoter activity, serial truncation constructs of the promoter were made and subcloned into the luciferase mammalian expression vector pGL2-Basic. Constructs were expressed in HepG2 hepatoma cells that endogenously express NTPDase2 (17), and relative luciferase activity was assessed (Fig. 5). The shortest construct with residual luciferase activity greater than empty vector was the F9 construct extending 188 bp upstream of the NTPDase2 start codon, suggesting that this is the NTPDase2 minimal promoter (Fig. 4B).
Fig. 5.
The NTPDase2 minimal promoter is located 188 bp upstream of the NTPDase2 start site. HepG2 cells were transfected with control plasmid (pGL2) or serial truncations of the putative NTPDase2 promoter (F10, −81 bp; F9, −188 bp; F8, −335 bp; F7, −489 bp; F6, −680 bp; F5, −889 bp; F4, −1,080 bp; F3, −1,302 bp; F2, −1,530 bp; F1, −1,688 bp), and luciferase activity was determined. Luciferase activity was equal to the full-length F1 construct for constructs F2–F7 but was markedly decreased in shorter constructs F8–F9 and was essentially absent in the shortest construct, F10 (n = 3). *P < 0.05; **P < 0.005; ‡P = NS (not significant) vs. F1.
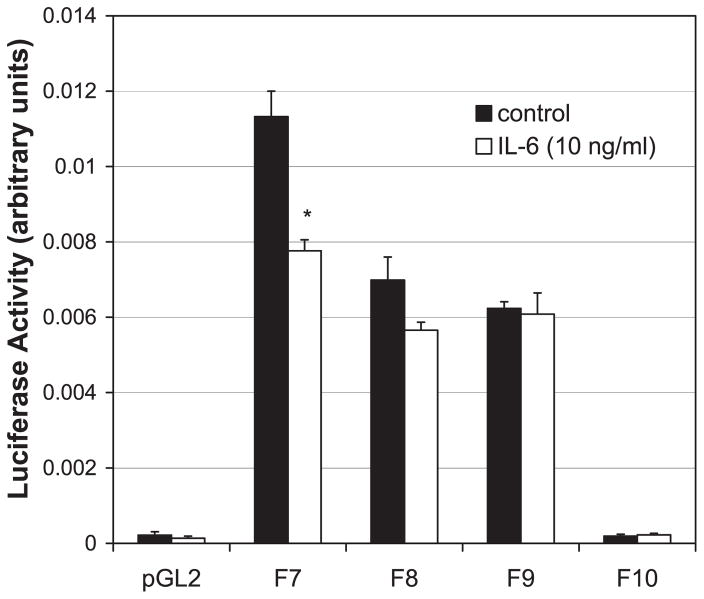
To determine whether the F7 construct was IL-6 sensitive, HepG2 cells expressing this and further truncated constructs were treated overnight with IL-6 (10 ng/ml). Cells expressing the F7 construct downregulated luciferase activity in response to IL-6 (Fig. 6), whereas, cells expressing shorter constructs did not. These data demonstrate that the IL-6 RE of the NTPDase2 promoter is located within the F7 construct.
Fig. 6.
F7 promoter construct activity is downregulated by IL-6. HepG2 cells were transfected with control plasmid (pGL2) or IL-6 promoter truncation constructs F7–F10 and treated overnight with IL-6 (10 ng/ml), and then luciferase activity was determined. Luciferase activity was downregulated by IL-6 in the F7 construct but not in shorter constructs (n = 3). *P < 0.02.
Mutation of all three putative IL-6 REs in the NTPDase2 promoter abrogates the response to IL-6
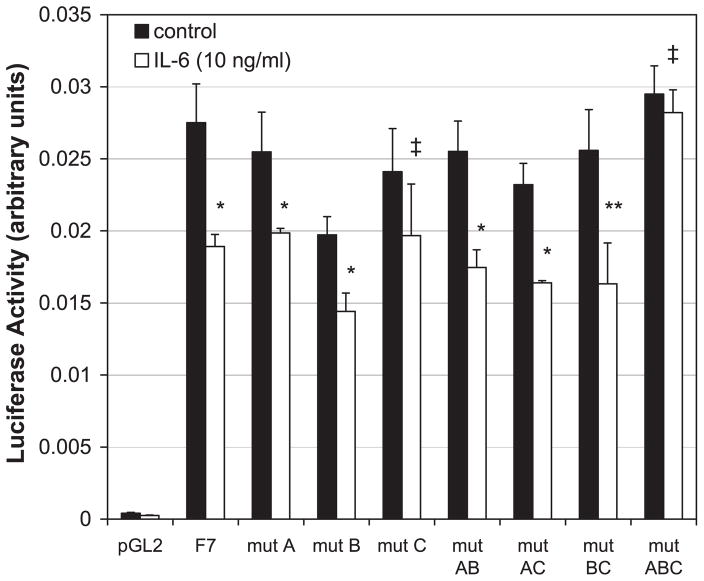
As shown in Fig. 4B, in silico analysis of the NTPDase2 promoter using the transcription element search system (TESS; University of Pennsylvania Computational Biology and Informatics Biology Laboratory, www.cbil.upenn.edu/tess) revealed three potential IL-6 REs. To determine whether these putative IL-6 REs are responsible for IL-6-mediated downregulation of the NTPDase2 promoter, the three elements were mutated either alone or in various combinations. IL-6 sensitivity was determined in each of these constructs by luciferase activity in HepG2 cells. As shown in Fig. 7, mutation of each element individually or two by two had no impact on the IL-6 repressive effects, except for the construct harboring a single mutation in the IL-6 RE C, where the IL-6-mediated repression did not reach statistical significance in this particular set of experiments. A reporter construct with all three IL-6 RE elements mutated (mut ABC), however, was no longer responsive to IL-6 (Fig. 7). These results indicate that IL-6 represses the NTPDase2 promoter through the IL-6 REs and that the three IL-6 REs appear to be redundant.
Fig. 7.
Mutation of all 3 putative IL-6 REs abrogates downregulation of IL-6 promoter activity by IL-6. HepG2 cells were transfected with control plasmid (pGL2), the F7 luciferase construct, or luciferase constructs containing mutations (mut) in various combinations of the 3 putative IL-6 REs and treated overnight with IL-6 (10 ng/ml), and then luciferase activity was assessed. As shown in Fig. 6, IL-6 downregulated NTPDase2 promoter activity in the F7 construct. IL-6 also downregulated NTPDase2 promoter activity for all of the singly or doubly mutated constructs (except for mut C, which failed to reach statistical significance). Conversely, mutation of all 3 putative IL-6 REs (mut ABC) completely blocked the downregulation of NTPDase2 promoter activity by IL-6 (n = 3). *P < 0.05; **P = 0.052; ‡P = NS.
The three IL-6 RE in the Entpd2 promoter are bound by proteins from HepG2 nuclear extracts
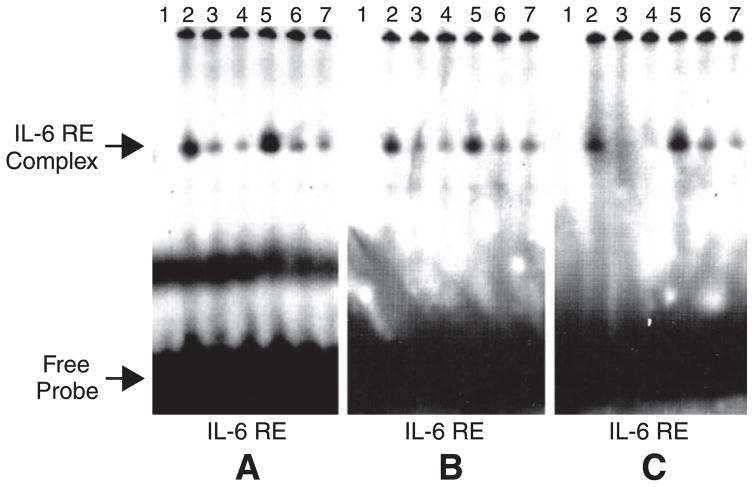
Electromobility shift assays with nuclear extracts from IL-6-stimulated and unstimulated HepG2 cells are presented in Fig. 8. We detected a single protein-DNA complex with all three probes corresponding to IL-6 REs A, B, and C found in the rat NTPDase2 promoter. The same band was also seen with the α2-macroglobulin-specific probe (data not shown). The bands obtained are similar to the data previously published with a typical IL-6 RE (19). The specificity of the binding was confirmed by competition assays. Addition of a 25-fold molar excess of unlabeled probes of IL-6 REs A, B, and C (Fig. 8, lanes 3 and 5) or rat α2-macroglobulin-specific unlabeled probe (59) (Fig. 8, lanes 4 and 7) to the reaction mixture efficiently competed the binding. Altogether, these data indicate that a protein specifically binds to each of the three IL-6 REs in the NTPDase2 promoter and that this protein can also bind the consensus IL-6 RE from the rat α2-macroglobulin.
Fig. 8.
IL-6 treatment decreases the binding of a protein from HepG2 cells on NTPDase2 IL-6 RE. Electromobility shift assays were performed using 10 μg of nuclear extracts from 1-h IL-6-stimulated or unstimulated HepG2 cells that were incubated with 32P-labeled double-stranded oligonucleotide probes corresponding to IL-6 RE A, B, or C found in the promoter of rat Entpd2. Lanes 1, probe alone; lanes 2–4, unstimulated nuclear extracts with the labeled probes; lanes 5–7, IL-6-stimulated nuclear extract with the labeled probe. Competition assays with a 25-fold excess of unlabeled probes from corresponding IL-6 RE or from consensus IL-6 RE of rat α2-macroglobulin correspond to lanes 3 and 6 and to lanes 4 and 7, respectively. IL-6 treatment induced a 36% increase in band intensity for IL-6 RE A, a 17% increase in band intensity for IL-6 RE B, and a 71% increase in band intensity for IL-6 RE C.
DISCUSSION
Ecto-nucleotidases of the E-NTPDase family regulate extracellular concentrations of nucleotides and their hydrolytic products (4). As such, NTPDases are critical regulators of the activity of plasma membrane receptors for extracellular nucleotides, including P2Y and P2X receptors. NTPDase2 is a 75-kDa ectonucleotidase with a well-defined organ and suborgan distribution (7, 23, 44). However, the physiological functions of NTPDase2 have only recently begun to be defined. NTPDase2 has been implicated in brain development, largely since NTPDase2 is expressed in germinal zones of developing neural tissue (6, 31). NTPDase2 has been shown to be responsible for hydrolysis of extracellular nucleotides by retinal pigment epithelium (37); however, the specific physiological function of NTPDase2 in this tissue has not been determined. Although NTPDase1 appears to be the dominant ectonucleotidase regulating platelet aggregation and vascular inflammation in solid organ transplantation (14, 39), NTPDase2 also appears to have a role in this process (4, 44).
The most clearly documented function of NTPDase2 has been demonstrated in the liver, where it appears to have an important physiological role that is disrupted in disease. Specifically, NTPDase2 regulates liver cell growth. NTPDase2 is expressed by PF that surround intrahepatic bile ducts. PF expressing NTPDase2 markedly block the proliferation of bile duct epithelia, whereas those lacking NTPDase2 expression have no effect on the proliferation of bile duct epithelia (21). Bile ductular proliferation is upregulated by biliary cirrhosis (3, 46), and NTPDase2 transcription and expression are down-regulated by biliary cirrhosis (10). Restoration of NTPDase2 into portal myofibroblasts from animals with biliary cirrhosis restores their ability to block proliferation of bile duct epithelia (21). Thus identification of factors that regulate NTPDase2 expression by portal fibroblasts/myofibroblasts is of great importance.
Although much is known about the acute regulation of NTPDase enzymatic activity (24, 42, 54), relatively little is known about the transcriptional regulation of NTPDases as a group, and about NTPDase2 in particular. An ectonucleotidase related to NTPDase2 is transcriptionally regulated by follicle-stimulating hormone in rat testicular Sertoli cells, although the signal transduction mechanism by which this occurs is unknown (29). NTPDase2 transcription is upregulated in hepatoma cells (24), but the mechanism by which this occurs is also unknown. Interestingly, NTPDase2 transcription is markedly upregulated by the environmental toxin 2,3,7,8-tetrachlorodibenzo-p-dioxin via a mechanism involving the aromatic hydrocarbon receptor (AhR)/AhR nuclear translocator system in mouse hepatoma cells (17). Finally, downregulation of transcription of both NTPDase2 (as noted in the preceding paragraph) and the hepatocyte canalicular ecto-ATPDase (now designated NTPDase8) (15, 25, 43) have both been demonstrated in animal models of biliary cirrhosis (2, 10). This is mediated in part by the inflammatory cytokine MCP-1; however, the effects of MCP-1 are likely indirect, since MCP-1 induces multiple morphological and functional changes in PF (27). At the transcriptional level, NTPDase2 appears to be the most highly regulated member of the E-NTPDase enzyme family.
In this study we have identified and cloned the rat Entpd2 promoter and report that the inflammatory cytokine IL-6 has specific direct effects on NTPDase2 transcription that are mediated by particular NTPDase2 promoter elements. We focused on IL-6 as a regulator of NTPDase2 function for several reasons. First, IL-6 is among several inflammatory mediators thought to be important in liver fibrosis (22, 33, 50). Second, IL-6 is upregulated in patients with biliary cirrhosis (32, 57). Finally, IL-6 has been shown to be secreted by bile duct epithelia (55, 58), which are the cells in closest proximity to PF. We found that IL-6 transcriptionally downregulated NTPDase2 expression in a time- and concentration-dependent fashion in PF. We cloned the NTPDase2 promoter and found three putative IL-6 REs by in silico analysis. Interestingly, combined inactivation of two of these elements was insufficient to prevent the effect of IL-6 on promoter activity; only mutation of all three elements prevented downregulation of NTPDase2 promoter activity by IL-6. This example of promoter element redundancy is somewhat uncommon but has been noted previously for a variety of genes. For example, the human 7SK encoding a small nuclear RNA contains two octamer-like elements, both of which must be mutated to block transcription (5), and the killer cell Ig-like receptor promoter contains functionally redundant AP4 sites (35). However, since single mutation of IL-6 RE C (most proximal to the NTPDase2 start site) had no effect on either IL-6-sensitive NTPDase2 promoter activity or protein/DNA interaction, it is likely that any effects mediated at this promoter site are indirect.
IL-6 is thought to induce gene regulation via an indirect signal transduction system. IL-6 binds to cells at the plasma membranes via a receptor complex consisting of two molecules: IL-6 receptor (IL-6R)/gp80 and gp130 (40). Whereas gp130 is thought to be widely expressed, the cell distribution of gp80 has been reported to be limited (47). In this study we have shown that PF express gp130, providing a molecular basis for IL-6 responsiveness. IL-6-sensitive changes in transcription are mediated via actions of specific promoter IL-6 REs (16, 19). Although the intermediate steps coupling IL-6 receptor activation and IL-6 REs are unclear, a set of IL-6 RE binding proteins have been identified, which mediate IL-6-sensitive induction of α2-macroglobulin (19). The intermediate steps coupling IL-6 and NTPDase2 transcriptional downregulation have not yet been identified.
In summary, we have identified a novel and specific regulator of NTPDase2 transcription and have characterized the signal transduction system by which this is regulated. Since NTPDase2 is of great importance in the regulation of liver cell growth, we hope that this work will lead to novel pharmacological approaches to the dysregulation of cell growth in the cirrhotic liver and will be extended to cell growth dysregulation in other organs in fibrotic conditions.
Acknowledgments
We are grateful to Pierre Chapdelaine and Maxime A. Tremblay, both from the Ontogeny-Reproduction Research Unit, Centre Hospitalier Universitaire de Québec, for technical assistance.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant DK-070849 (to J. A. Dranoff), an American Heart Association Grant-in-Aid (to J. A. Dranoff), Yale Liver Center NIDDK Grant DK-45710, and grants from the Canadian Institutes of Health Research (CIHR) (to J. Sévigny). É. G. Lavoie was a recipient of a scholarship from Fonds de recherche en santé du Québec. J. Sévigny was a recipient of a CIHR New Investigator Award.
References
- 1.Abbracchio MP, Burnstock G, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, Knight GE, Fumagalli M, Gachet C, Jacobson KA, Weisman GA. International Union of Pharmacology LVIII: update on the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol Rev. 2006;58:281–341. doi: 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accatino L, Pizarro M, Solis N, Arrese M, Vollrath V, Ananthanarayanan M, Chianale J, Koenig CS. Differential expression of canalicular membrane Ca2+/Mg2+-ecto-ATPase in estrogen-induced and obstructive cholestasis in the rat. J Lab Clin Med. 2000;136:125–137. doi: 10.1067/mlc.2000.108151. [DOI] [PubMed] [Google Scholar]
- 3.Alpini G, Ulrich CD, 2nd, Phillips JO, Pham LD, Miller LJ, LaRusso NF. Upregulation of secretin receptor gene expression in rat cholangiocytes after bile duct ligation. Am J Physiol Gastrointest Liver Physiol. 1994;266:G922–G928. doi: 10.1152/ajpgi.1994.266.5.G922. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson B, Dwyer K, Enjyoji K, Robson SC. Ecto-nucleotidases of the CD39/NTPDase family modulate platelet activation and thrombus formation: potential as therapeutic targets. Blood Cells Mol Dis. 2006;36:217–222. doi: 10.1016/j.bcmd.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 5.Boyd DC, Turner PC, Watkins NJ, Gerster T, Murphy S. Functional redundancy of promoter elements ensures efficient transcription of the human 7SK gene in vivo. J Mol Biol. 1995;253:677–690. doi: 10.1006/jmbi.1995.0582. [DOI] [PubMed] [Google Scholar]
- 6.Braun N, Sévigny J, Mishra SK, Robson SC, Barth SW, Gerstberger R, Hammer K, Zimmermann H. Expression of the ecto-ATPase NTPDase2 in the germinal zones of the developing and adult rat brain. Eur J Neurosci. 2003;17:1355–1364. doi: 10.1046/j.1460-9568.2003.02567.x. [DOI] [PubMed] [Google Scholar]
- 7.Braun N, Sévigny J, Robson SC, Hammer K, Hanani M, Zimmermann H. Association of the ecto-ATPase NTPDase2 with glial cells of the peripheral nervous system. Glia. 2004;45:124–132. doi: 10.1002/glia.10309. [DOI] [PubMed] [Google Scholar]
- 8.Burnstock G. Purinergic signalling—an overview. Novartis Found Symp. 2006;276:26–48. discussion 48–57, 275–281. [PubMed] [Google Scholar]
- 9.Chapdelaine P, Delahaye S, Gauthier E, Tremblay RR, Dube JY. A one-hour procedure for the preparation of genomic DNA from frozen tissues. Biotechniques. 1993;14:163–164. [PubMed] [Google Scholar]
- 10.Dranoff JA, Kruglov E, Toure J, Braun N, Zimmermann H, Jain D, Knowles AF, Sévigny J. Ecto-nucleotidase NTPDase2 is selectively down-regulated in biliary cirrhosis. J Investig Med. 2004;52:475–482. doi: 10.1136/jim-52-07-42. [DOI] [PubMed] [Google Scholar]
- 11.Dranoff JA, Kruglov EA, Robson SC, Braun N, Zimmermann H, Sévigny J. The ecto-nucleoside triphosphate diphosphohydrolase NTPDase2/CD39L1 is expressed in a novel functional compartment within the liver. Hepatology. 2002;36:1135–1144. doi: 10.1053/jhep.2002.36823. [DOI] [PubMed] [Google Scholar]
- 12.Dranoff JA, Masyuk AI, Kruglov EA, LaRusso NF, Nathanson MH. Polarized expression and function of P2Y ATP receptors in rat bile duct epithelia. Am J Physiol Gastrointest Liver Physiol. 2001;281:G1059–G1067. doi: 10.1152/ajpgi.2001.281.4.G1059. [DOI] [PubMed] [Google Scholar]
- 13.Dranoff JA, Ogawa M, Kruglov EA, Gaca MD, Sévigny J, Robson SC, Wells RG. Expression of P2Y nucleotide receptors and ectonucleotidases in quiescent and activated rat hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2004;287:G417–G424. doi: 10.1152/ajpgi.00294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enjyoji K, Sévigny J, Lin Y, Frenette PS, Christie PD, Esch JS, 2nd, Imai M, Edelberg JM, Rayburn H, Lech M, Beeler DL, Csizmadia E, Wagner DD, Robson SC, Rosenberg RD. Targeted disruption of cd39/ATP diphosphohydrolase results in disordered hemostasis and thrombo-regulation. Nat Med. 1999;5:1010–1017. doi: 10.1038/12447. [DOI] [PubMed] [Google Scholar]
- 15.Fausther M, Lecka J, Kukulski F, Levesque SA, Pelletier J, Zimmermann H, Dranoff JA, Sévigny J. Cloning, purification and identification of the liver canalicular ecto-ATPase as NTPDase8. Am J Physiol Gastrointest Liver Physiol. 2007;292:G785–G795. doi: 10.1152/ajpgi.00293.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fey GH, Hattori M, Hocke G, Brechner T, Baffet G, Baumann M, Baumann H, Northemann W. Gene regulation by interleukin 6. Biochimie. 1991;73:47–50. doi: 10.1016/0300-9084(91)90073-a. [DOI] [PubMed] [Google Scholar]
- 17.Gao L, Dong L, Whitlock JP., Jr A novel response to dioxin. Induction of ecto-ATPase gene expression. J Biol Chem. 1998;273:15358–15365. doi: 10.1074/jbc.273.25.15358. [DOI] [PubMed] [Google Scholar]
- 18.Gever JR, Cockayne DA, Dillon MP, Burnstock G, Ford AP. Pharmacology of P2X channels. Pflügers Arch. 2006;452:513–537. doi: 10.1007/s00424-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 19.Hocke GM, Barry D, Fey GH. Synergistic action of interleukin-6 and glucocorticoids is mediated by the interleukin-6 response element of the rat alpha 2 macroglobulin gene. Mol Cell Biol. 1992;12:2282–2294. doi: 10.1128/mcb.12.5.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jhandier MN, Kruglov EA, Lavoie ÉG, Sévigny J, Dranoff JA. Portal fibroblasts regulate the proliferation of bile duct epithelia via expression of NTPDase2. J Biol Chem. 2005;280:22986–22992. doi: 10.1074/jbc.M412371200. [DOI] [PubMed] [Google Scholar]
- 22.Kershenobich Stalnikowitz D, Weissbrod AB. Liver fibrosis and inflammation. A review. Ann Hepatol. 2003;2:159–163. [PubMed] [Google Scholar]
- 23.Kittel A, Pelletier J, Bigonnesse F, Guckelberger O, Kordas K, Braun N, Robson SC, Sévigny J. Localization of nucleoside triphosphate diphosphohydrolase-1 (NTPDase1) and NTPDase2 in pancreas and salivary gland. J Histochem Cytochem. 2004;52:861–871. doi: 10.1369/jhc.3A6167.2004. [DOI] [PubMed] [Google Scholar]
- 24.Knowles AF, Chiang WC. Enzymatic and transcriptional regulation of human ecto-ATPase/E-NTPDase 2. Arch Biochem Biophys. 2003;418:217–227. doi: 10.1016/j.abb.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Knowles AF, Li C. Molecular cloning and characterization of expressed human ecto-nucleoside triphosphate diphosphohydrolase 8 (E-NTPDase 8) and its soluble extracellular domain. Biochemistry. 2006;45:7323–7333. doi: 10.1021/bi052268e. [DOI] [PubMed] [Google Scholar]
- 26.Kruglov EA, Jain D, Dranoff JA. Isolation of primary rat liver fibroblasts. J Investig Med. 2002;50:179–184. doi: 10.2310/6650.2002.33431. [DOI] [PubMed] [Google Scholar]
- 27.Kruglov EA, Nathanson RA, Nguyen T, Dranoff JA. Secretion of MCP-1/CCL2 by bile duct epithelia induces myofibroblastic transdifferentiation of portal fibroblasts. Am J Physiol Gastrointest Liver Physiol. 2006;290:G765–G771. doi: 10.1152/ajpgi.00308.2005. [DOI] [PubMed] [Google Scholar]
- 28.Kukulski F, Levesque SA, Lavoie ÉG, Lecka J, Bigonesse F, Knowles AF, Robson TL, Sévigny J. Comparative hydrolysis of P2 receptor antagonists by NTPDases 1, 2, 3, and 8. Purinergic Signal. 2005;1:193–204. doi: 10.1007/s11302-005-6217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Q, Porter LD, Cui X, Sanborn BM. Ecto-ATPase mRNA is regulated by FSH in Sertoli cells. J Androl. 2001;22:289–301. [PubMed] [Google Scholar]
- 30.Marra F, DeFranco R, Grappone C, Milani S, Pastacaldi S, Pinzani M, Romanelli RG, Laffi G, Gentilini P. Increased expression of monocyte chemotactic protein-1 during active hepatic fibrogenesis: correlation with monocyte infiltration. Am J Pathol. 1998;152:423–430. [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra SK, Braun N, Shukla V, Fullgrabe M, Schomerus C, Korf HW, Gachet C, Ikehara Y, Sévigny J, Robson SC, Zimmermann H. Extracellular nucleotide signaling in adult neural stem cells: synergism with growth factor-mediated cellular proliferation. Development. 2006;133:675–684. doi: 10.1242/dev.02233. [DOI] [PubMed] [Google Scholar]
- 32.Muller C, Zielinski CC. Interleukin-6 production by peripheral blood monocytes in patients with chronic liver disease and acute viral hepatitis. J Hepatol. 1992;15:372–377. doi: 10.1016/0168-8278(92)90071-v. [DOI] [PubMed] [Google Scholar]
- 33.Napoli J, Bishop GA, McCaughan GW. Increased intrahepatic messenger RNA expression of interleukins 2, 6, and 8 in human cirrhosis. Gastroenterology. 1994;107:789–798. doi: 10.1016/0016-5085(94)90128-7. [DOI] [PubMed] [Google Scholar]
- 34.Park J, Gores GJ, Patel T. Lipopolysaccharide induces cholangiocyte proliferation via an interleukin-6-mediated activation of p44/p42 mitogen-activated protein kinase. Hepatology. 1999;29:1037–1043. doi: 10.1002/hep.510290423. [DOI] [PubMed] [Google Scholar]
- 35.Presnell SR, Zhang L, Ramilo CA, Chan HW, Lutz CT. Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. Int Immunol. 2006;18:1221–1232. doi: 10.1093/intimm/dxl043. [DOI] [PubMed] [Google Scholar]
- 36.Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 37.Reigada D, Lu W, Zhang X, Friedman C, Pendrak K, McGlinn A, Stone RA, Laties AM, Mitchell CH. Degradation of extracellular ATP by the retinal pigment epithelium. Am J Physiol Cell Physiol. 2005;289:C617–C624. doi: 10.1152/ajpcell.00542.2004. [DOI] [PubMed] [Google Scholar]
- 38.Robson SC, Sévigny J, Zimmermann H. The E-NTPDase family of ectonucleotidases: structure, function, relationships, and pathophysiological significance. Purinergic Signal. 2006;2:409–430. doi: 10.1007/s11302-006-9003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robson SC, Wu Y, Sun X, Knosalla C, Dwyer K, Enjyoji K. Ecto-nucleotidases of CD39 family modulate vascular inflammation and thrombosis in transplantation. Semin Thromb Hemost. 2005;31:217–233. doi: 10.1055/s-2005-869527. [DOI] [PubMed] [Google Scholar]
- 40.Scheller J, Rose-John S. Interleukin-6 and its receptor: from bench to bedside. Med Microbiol Immunol. 2006;195:173–183. doi: 10.1007/s00430-006-0019-9. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senger MR, Rico EP, de Bem Arizi M, Frazzon AP, Dias RD, Bogo MR, Bonan CD. Exposure to Hg2+ and Pb2+ changes NTPDase and ecto-5′-nucleotidase activities in central nervous system of zebrafish (Danio rerio) Toxicology. 2006;226:229–237. doi: 10.1016/j.tox.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Sévigny J, Robson SC, Waelkens E, Csizmadia E, Smith RN, Lemmens R. Identification and characterization of a novel hepatic canalicular ATP diphosphohydrolase. J Biol Chem. 2000;275:5640–5647. doi: 10.1074/jbc.275.8.5640. [DOI] [PubMed] [Google Scholar]
- 44.Sévigny J, Sundberg C, Braun N, Guckelberger O, Csizmadia E, Qawi I, Imai M, Zimmermann H, Robson SC. Differential catalytic properties and vascular topography of murine nucleoside triphosphate diphosphohydrolase 1 (NTPDase1) and NTPDase2 have implications for thromboregulation. Blood. 2002;99:2801–2809. doi: 10.1182/blood.v99.8.2801. [DOI] [PubMed] [Google Scholar]
- 46.Strazzabosco M, Spirli C, Okolicsanyi L. Pathophysiology of the intra-hepatic biliary epithelium. J Gastroenterol Hepatol. 2000;15:244–253. doi: 10.1046/j.1440-1746.2000.02091.x. [DOI] [PubMed] [Google Scholar]
- 47.Taga T. IL6 signalling through IL6 receptor and receptor-associated signal transducer, gp130. Res Immunol. 1992;143:737–739. doi: 10.1016/0923-2494(92)80013-b. [DOI] [PubMed] [Google Scholar]
- 48.Terada R, Yamamoto K, Hakoda T, Shimada N, Okano N, Baba N, Ninomiya Y, Gershwin ME, Shiratori Y. Stromal cell-derived factor-1 from biliary epithelial cells recruits CXCR4-positive cells: implications for inflammatory liver diseases. Lab Invest. 2003;83:665–672. doi: 10.1097/01.lab.0000067498.89585.06. [DOI] [PubMed] [Google Scholar]
- 49.Thevananther S, Sun H, Li D, Arjunan V, Awad SS, Wyllie S, Zimmerman TL, Goss JA, Karpen SJ. Extracellular ATP activates c-jun N-terminal kinase signaling and cell cycle progression in hepatocytes. Hepatology. 2004;39:393–402. doi: 10.1002/hep.20075. [DOI] [PubMed] [Google Scholar]
- 50.Tsukamoto H. Cytokine regulation of hepatic stellate cells in liver fibrosis. Alcohol Clin Exp Res. 1999;23:911–916. [PubMed] [Google Scholar]
- 51.Viger RS, Mertineit C, Trasler JM, Nemer M. Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Mullerian inhibiting substance promoter. Development. 1998;125:2665–2675. doi: 10.1242/dev.125.14.2665. [DOI] [PubMed] [Google Scholar]
- 52.Volonte C, Amadio S, D’Ambrosi N, Colpi M, Burnstock G. P2 receptor web: complexity and fine-tuning. Pharmacol Ther. 2006;112:264–280. doi: 10.1016/j.pharmthera.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Roman R, Lidofsky SD, Fitz JG. Autocrine signaling through ATP release represents a novel mechanism for cell volume regulation. Proc Natl Acad Sci USA. 1996;93:12020–12025. doi: 10.1073/pnas.93.21.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Y, Sun X, Kaczmarek E, Dwyer KM, Bianchi E, Usheva A, Robson SC. RanBPM associates with CD39 and modulates ecto-nucleotidase activity. Biochem J. 2006;396:23–30. doi: 10.1042/BJ20051568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu B, Broome U, Ericzon BG, Sumitran-Holgersson S. High frequency of autoantibodies in patients with primary sclerosing cholangitis that bind biliary epithelial cells and induce expression of CD44 and production of interleukin 6. Gut. 2002;51:120–127. doi: 10.1136/gut.51.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasoshima M, Kono N, Sugawara H, Katayanagi K, Harada K, Nakanuma Y. Increased expression of interleukin-6 and tumor necrosis factor-alpha in pathologic biliary epithelial cells: in situ and culture study. Lab Invest. 1998;78:89–100. [PubMed] [Google Scholar]
- 58.Yokoyama T, Komori A, Nakamura M, Takii Y, Kamihira T, Shimoda S, Mori T, Fujiwara S, Koyabu M, Taniguchi K, Fujioka H, Migita K, Yatsuhashi H, Ishibashi H. Human intrahepatic biliary epithelial cells function in innate immunity by producing IL-6 and IL-8 via the TLR4-NF-kappaB and -MAPK signaling pathways. Liver Int. 2006;26:467–476. doi: 10.1111/j.1478-3231.2006.01254.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Z, Fuentes NL, Fuller GM. Characterization of the IL-6 responsive elements in the gamma fibrinogen gene promoter. J Biol Chem. 1995;270:24287–24291. doi: 10.1074/jbc.270.41.24287. [DOI] [PubMed] [Google Scholar]