Abstract
Dengue is a viral pandemic caused by four dengue virus serotypes (DENV-1, 2, 3, and 4) transmitted by Aedes mosquitoes. Reportedly, there has been a 2-fold increase in dengue cases every decade. An efficacious tetravalent vaccine, which can provide long-term immunity against all four serotypes in all target populations, is still unavailable. Despite the progress being made in the live virus-based dengue vaccines, the World Health Organization strongly recommends the development of alternative approaches for safe, affordable, and efficacious dengue vaccine candidates. We have explored virus-like particles (VLPs)-based nonreplicating subunit vaccine approach and have developed recombinant envelope ectodomains of DENV-1, 2, and 3 expressed in Pichia pastoris. These self-assembled into VLPs without pre-membrane (prM) protein, which limits the generation of enhancing antibodies, and elicited type-specific neutralizing antibodies against the respective serotype. Encouraged by these results, we have extended this work further by developing P. pastoris–expressed DENV-4 ectodomain (DENV-4 E) in this study, which was found to be glycosylated and assembled into spherical VLPs without prM, and displayed critical neutralizing epitopes on its surface. These VLPs were found to be immunogenic in mice and elicited DENV-4-specific neutralizing antibodies, which were predominantly directed against envelope domain III, implicated in host-receptor recognition and virus entry. These observations underscore the potential of VLP-based nonreplicative vaccine approach as a means to develop a safe, efficacious, and tetravalent dengue subunit vaccine. This work paves the way for the evaluation of a DENV E-based tetravalent dengue vaccine candidate, as an alternative to live virus-based dengue vaccines.
Introduction
Dengue is an expanding global public health concern today with an estimated 390 million infected individuals per year.1,2 It is caused by any of the four antigenically distinct and closely related dengue viruses (DENV-1, 2, 3, and 4), transmitted primarily by Aedes aegypti mosquito. Dengue infection causes flu-like symptoms and these can range from mild to severe (dengue hemorrhagic fever) to fatal (dengue shock syndrome) outcomes.3–5 Long-term homotypic immunity is elicited by infection with one serotype of DENV, which provides only short-term heterotypic immunity. Therefore, subsequent secondary infection by any other serotype predisposes an individual to severe disease through antibody-dependent enhancement (ADE).6
Dengue tetravalent live-attenuated vaccines (LAVs), purified inactivated virus vaccines, plasmid DNA vaccines, recombinant subunit vaccines, etc.7–10 are at various stages of clinical development. Dengvaxia (CYD-TDV), developed by Sanofi Pasteur, which is a chimeric tetravalent LAV,11 is the first licensed dengue vaccine (licensed in Mexico, Brazil, Philippines and El Salvador). World Health Organization (WHO)'s Strategic Advisory Group of Experts has recommended limited use of Dengvaxia in regions of high dengue endemicity.5 Although Dengvaxia is a much-needed accomplishment, it is not an ideal solution to dengue. It was found to be poorly efficacious in dengue-naive individuals and resulted in increase in vaccine-induced risk of hospitalization in children under 9 years of age.12 These shortcomings of Dengvaxia are speculated to be due to vaccine-induced enhancement of DENV infection.13 The WHO recommends development of next-generation dengue vaccine candidates that could reduce concerns of viral enhancement13 and interference,14–16 associated with the current vaccine strategies.
Virus-like particles (VLPs) could be used as alternative nonreplicative vaccine candidates against dengue as they lack the viral genome. It has been reported earlier that structural proteins of many viruses self-assemble into VLPs, when expressed and purified from heterologous hosts.17–21 A critical attribute of VLPs is their ability to display epitopes in a repetitive manner on their surface like a virion particle, but lacking the infectious genome. So, these possess high immunogenicity with reduced potential of causing infection.22,23 Success of VLPs as a vaccine candidate is supported by the approved VLP-based vaccines for human papilloma and hepatitis B viral infections,17 and several others that are in clinical trials like those against influenza A virus and chikungunya virus.24,25
There are several DENV antigens that could be used to develop vaccine, but envelope glycoprotein, which is a structural protein, is the most promising of them all. The attachment of DENV to host cell receptors and fusion with endosomal membrane is mediated by envelope glycoprotein. Moreover, it is highly immunogenic, displays critical tertiary and quaternary virion surface epitopes against which natural circulating serotype-specific antibodies are directed.3,26 Because of these vital features, the envelope glycoprotein is a component of all dengue vaccine candidates, which are undergoing clinical trials. It has also been documented in the literature that co-expression of envelope glycoprotein along with pre-membrane protein (prM) produces VLPs when expressed in a heterologous system.18–21 The envelope glycoprotein has ∼500 amino acid (aa) residues of which ∼100 aa at the C-terminus are anchored in the host membrane on the mature virion surface.26 The N-terminal ∼400 aa residues ectodomain (E) contains envelope domain I (EDI), EDII, and EDIII. Among these, EDIII is a key component that possesses several effective and type-specific neutralizing epitopes.27,28 On the other hand, the prM protein has a role in virus maturation.26 Large proportions of cross-reactive antibodies that have the ability to mediate ADE have been reported to be elicited by prM.29,30
We reported that DENV-2 ectodomain (DENV-2 E), expressed in yeast Pichia pastoris, assembles into VLPs without prM. These VLPs, which showed high immunogenicity, generated DENV-2 neutralizing antibodies that were competent to provide significant protection when evaluated in challenge experiments in AG129 mice.31 The VLPs without prM have an added advantage of reducing the generation of cross-reactive antibodies, which may limit the risk of ADE. Similarly, we have expressed and purified DENV-1 E and 3 E,32,33 which also assembled into VLPs without prM and were able to generate serotype-specific DENV-neutralizing antibodies. Use of P. pastoris as expression host further makes the production process cost effective due to its characteristics of requiring inexpensive media, growth to high cell densities, high production rate, post-translational modifications, and the possibility of tightly regulating the expression through methanol-inducible alcohol oxidase 1 (AOX1) promoter.34 These factors make the development and production of a safe, affordable, and efficacious tetravalent subunit based dengue vaccine candidate feasible. Exploring this approach, we have specifically evaluated in this study the potential of DENV-4 E with respect to its assembly into VLPs, conformational display of neutralizing epitopes, and the capability of these VLPs to elicit homotypic DENV4-neutralizing antibodies.
Materials and Methods
Ethics statement.
The entire animal experiments were performed with prior approval from Institutional Animal Ethics Committee of International Center for Genetic Engineering and Biotechnology and Syngene International Limited, Bangalore (IAEC No. Syngene/IAEC/520/06-2014). The guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals, Government of India, were meticulously followed.
DENV-4 E gene, cell lines, vectors, viruses, antibodies, and other reagents.
The DENV-4 E gene (1.4 kb, GenBank accession no: JX292267.1) was synthesized and made available by GenScript (Piscataway, NJ) after codon optimization for expression in P. pastoris. Plasmid vector (pPICZ-A) and P. pastoris strain (KM71H) used for cloning and expression, respectively, were purchased from Invitrogen Life Technologies (Carlsbad, CA). DENV-4 E gene was cloned in pPICZ-A to permit the selection of clone by utilizing its zeocin marker that confers resistance to Zeocin antibiotic, and to enable high gene expression through methanol-inducible AOX1 promoter. DENV-1, 2, 3, and 4 were WHO reference strains as reported earlier.35 Cell lines C6/36 (for DENV production), baby hamster kidney cells (for immunofluorescence), and Vero (for neutralizing antibody titration) were obtained from American Type Culture Collection (ATCC), Manassas, VA. Ni-NTA Superflow resin, Ni-NTA His Sorb plates, and anti-Histidine monoclonal antibody (mAb) were procured from Qiagen (Hilden, Germany). For electron microscopy, carbon-coated formvar grids were obtained from TAAB Laboratories Equipment Ltd (Reading, United Kingdom). Concanavalin A-horseradish peroxidase (Con A-HRPO) conjugate, HRPO substrate 3, 3′, 5, 5′-tetramethylbenzidine, and glass beads (425–600 μm) were procured from Sigma–Aldrich (St. Louis, MO). Type-specific and cross-reactive human and murine mAbs used for characterizing purified protein have been described previously.28,36–43 4G2 mAb used in immunofluorescence assay (IFA) was procured from ATCC. Anti-mouse IgG antibody-HRPO and IgG fluorescence isothiocyanate (FITC) conjugates used in enzyme-linked immunosorbent assay (ELISA) and IFA, respectively, were purchased from Calbiochem (La Jolla, CA). Alhydrogel, used as adjuvant, was obtained from Brenntag (Frederikssund, Denmark). Escherichia coli expressing maltose-binding protein (MBP) and MBP-fused EDIII-4 (EDIII-4-MBP) were obtained from University of North Carolina at Chapel Hill.
Cloning and expression of DENV-4 E in P. pastoris.
DENV-4 E gene was cloned in pPICZ-A vector under methanol-inducible AOX1 promoter and integrated into the genome of P. pastoris strain KM71H, as accomplished hitherto for DENV-1 E, 2 E, and 3 E.31–33 The transformed clones were cultured, induced with methanol, and processed for expression evaluation as described for DENV-2 by Western blot and ELISA with serotype 4 EDIII-specific mAb, DV4 E88.31,39
Purification and characterization of DENV-4 E.
Expressed protein was purified by immobilized metal (Ni) affinity chromatography resorting to its polyhistidine tag, under denaturing conditions.31 Purified DENV-4 E was characterized for its purity by SDS-PAGE, and glycosylation by Western blot and ELISA with Con A-HRPO conjugate.31 Dynamic light scattering (DLS) and electron microscopy (EM) were performed for evaluation of assembly of DENV-4 E into VLPs.33,44 Appropriate display of antigenic epitopes on VLPs was analyzed with human and murine serotype-specific and cross-reactive mAbs. Briefly, all the four VLPs, DENV-1 E, 2 E, 3 E, and 4 E, and hepatitis B surface antigen (used as negative control) were coated on high binding polystyrene plates and indirect ELISA was performed with the mAbs,28,36–43 as described earlier.33
Evaluation of immunogenicity in mice.
BALB/c mice (6-week old, in group of six) were immunized with 20 μg of purified recombinant DENV-4 E, adsorbed on alhydrogel, on days 0, 30, and 90 (intraperitoneally). Sera were collected 10 days after the third immunization, as reported earlier.31
Indirect ELISA and IFA were performed for the assessment of antibodies generated by DENV-4 E antigen, as described previously.31 Briefly, presence of antibodies against the four DENVs, DENV-Es, and EDIIIs (all eight recombinant proteins developed in-house) was evaluated by indirect ELISA and their ability to recognize virus-infected BHK-21 cells was evaluated by IFA. Fluorescence-activated cell sorting (FACS)-based neutralization test (FNT)35 was used to determine the neutralizing antibody titer against each DENV serotype, as reported previously for DENV-1 E, 2 E, and 3 E.31–33
To evaluate the antibodies that are generated against EDIII of DENV-4 E, DENV-4 E immune serum was depleted of anti-EDIII antibodies and residual neutralizing antibodies were scored by FNT, according to optimized protocol.33 Briefly, DENV-4 E immune serum was preincubated separately with EDIII-4-MBP and MBP proteins immobilized on amylose resin and the depleted sera were evaluated for residual neutralization titer against DENV-4 by FNT on Vero cells.
Results
Evaluation of DENV-4 E gene expression in P. pastoris.
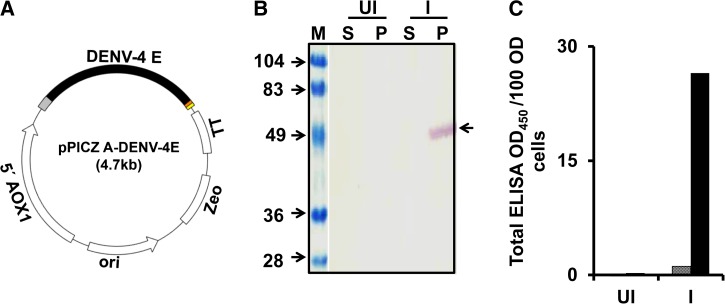
A DENV-4 E gene (Supplemental Figure 1), codon-optimized and custom-synthesized for expression in P. pastoris, was cloned in pPICZ-A expression vector (Figure 1A ) under AOX1 promoter. This gene was integrated into the genome of Pichia strain KM71H by electroporation and the transformed clone was identified by expression analysis after induction with methanol by Western blot (Figure 1B) and ELISA (Figure 1C), using serotype 4-specific mAb DENV4 E88.39 DENV4 E88 mAb recognized the DENV-4 E in the pellet fraction only (Figure 1B and C) and displayed the expected mobility (Figure 1B). This was further corroborated with Western blot and ELISA with mAb specific to 6x-His affinity tag engineered into the C-terminal end of the recombinant protein (data not shown). Methanol percentage for induction and time course optimization experiments for the expression of DENV-4 E protein in P. pastoris revealed that 1.5% methanol induction per 24 hours for 3 days is suitable for optimal expression of DENV-4 E (Supplemental Figure 2). Thus, DENV-4 E expressing biomass was prepared by inducing the cells with 0.75% methanol every 12 hours (which makes it 1.5% methanol per 24 hours), for a period of 3 days (Supplemental Figure 2). These observations of association of DENV-4 E in pellet fraction and optimal expression at 1.5% methanol for 3 days were in consonance with that observed for DENV-1 E, 2 E, and 3 E.31–33
Figure 1.
Expression of DENV-4 E in Pichia pastoris. (A) Schematic representation of DENV-4 E gene cloned in pPICZ-A expression vector with AOX1 promoter (5′ AOX1) upstream and transcription terminator sequence (TT) downstream of it. An Escherichia coli origin of replication (ori) and zeocin resistance (Zeo) marker is also present for efficient replication and transformed clone selection, respectively. (B and C) Evaluation of expression and localization of DENV-4 E in induced P. pastoris. Aliquots of un-induced (UI) and induced (I) cell cultures were subjected to lysis and separated into soluble (S) and membrane-enriched pellet (P) fractions by centrifugation. S and P fractions were evaluated through (B) Western blot and (C) ELISA on Ni-NTA His Sorb plate using DENV-4 E-specific monoclonal antibody (mAb), DENV-4 E88. The blot in panel B is a composite figure, where lane “M” denotes pre-stained markers, the sizes (in kDa) of which are indicated on the left. An arrow on the right of panel B indicates the position of DENV-4 E in the blot. In panel C, the S and P fractions are represented by the grey and black bars, respectively. DENV-4 E = dengue virus serotype 4 ectodomain; ELISA = enzyme-linked immunosorbent assay.
Purification and characterization of recombinant DENV-4 E protein.
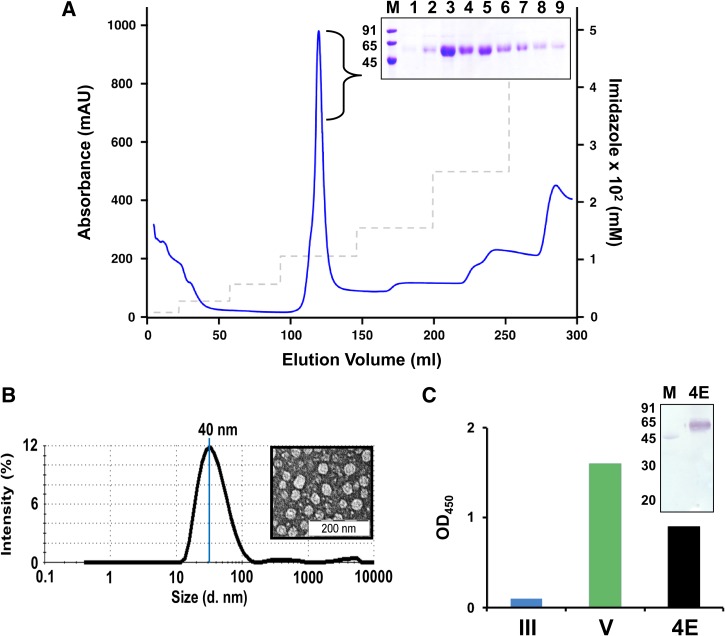
Since P. pastoris–expressed DENV-4 E protein was also found to be associated with the pellet fraction of the induced and lysed biomass, it was purified using the immobilized metal ion affinity chromatographic protocol optimized for purification of the three previously reported DENV-Es.31–33 DENV-4 E was found to elute at ∼120 mM imidazole (Figure 2A ). Different fractions of the peak were analyzed by SDS PAGE (Figure 2A inset) and were found to be > 90% pure by densitometric scan, with an overall yield of 200 mg/L methanol-induced culture. These fractions were pooled, dialyzed, and used for further characterizations. Ability of DENV-4 E to assemble into VLPs was evaluated by DLS and EM, and was found to assemble into particles of ∼40 nm (Figure 2B), which is consistent with our previous results for other recombinant DENV-Es.31–33 Like the other three DENV-Es, DENV-4 E VLPs were also found to be glycosylated as assessed by protein blot and ELISA using Con A-HRPO (Figure 2C). Similar to our previous reports for E proteins of DENV-1, 2, and 3 serotypes, the N-terminal amino acid sequence analysis of DENV-4 E protein confirmed that the N-terminal 34 aa residues of prM signal sequence had been efficiently cleaved off (data not shown).
Figure 2.
Purification and characterization of DENV-4 E. (A) DENV-4 E was purified from pellet fraction of lysate of induced Pichia pastoris biomass via Ni2+ affinity chromatography. Blue curve and dashed grey line represent the UV-absorbance profile and imidazole step gradient, respectively. The inset represents Coomassie-stained SDS-PAGE analysis of the peak fractions (lanes 1–9) of purified DENV-4 E. Lane M represents low-molecular-weight protein markers, the sizes (in kDa) of which are written on the left of the inset. (B) Formation of virus-like particles by purified DENV-4 E as assessed by DLS representing particle size distribution by intensity. The average particle size is also indicated. EM image of the same is provided in the inset. (C) Glycosylation of DENV-4 E (4E, black bar) is evaluated by ELISA using Con A-HRPO conjugate on Ni-NTA His-Sorb plate. EDIII (III, blue bar) and DENV-4 (V, green bar) is used as negative control and positive control, respectively. In the inset, a protein blot using Con A-HRPO conjugate is shown in which lane M represents low-molecular-weight protein markers, which contain ovalbumin. Low-molecular-weight protein markers serve as positive (through its only glycosylated protein, ovalbumin) and negative (proteins other than ovalbumin) controls in the assay, and the sizes (in kDa) of the protein markers are indicated on the left. Con A-HRPO = Concanavalin A-horseradish peroxidase; DENV-4 E = dengue virus serotype 4 ectodomain; ED = envelope domain; ELISA = enzyme-linked immunosorbent assay.
DENV-4 E VLPs have conformational display of critical epitopes on its surface.
A comprehensive immunological characterization of the purified DENV-4 E protein was performed to assess the presentation of critical epitopes, using a panel of conformational, type-specific, and cross-reactive murine and human mAbs against E, EDI/II, EDIII, and fusion loop epitopes. The reactivity of these mAbs was determined in an indirect ELISA (Table 1), using DENV-4 E VLPs as the coating antigen and compared head-to-head with the VLPs corresponding to the remaining three DENV serotypes.31–33 DENV-4 E42 and E43, mAbs specific to the E protein of DENV serotype 4,39 which recognized purified DENV-4 E VLPs efficiently, did not react to any discernible extent with DENV E VLPs corresponding to the remaining three serotypes. Further, the recognition of the DENV-4 E VLPs by DENV-4 mAbs E29, E76, and E88 substantiate the conclusion that EDIII epitopes are intact and accessible. DENV4-E29 mAb binds to Y377 and H390 in the F and G strand of the EDIII, respectively; DENV4-E76 mAb binds to N-terminal linker (M301), B-strand (E338, V347, and V348), CC' loop (R350), D-strand (P356), and G-strand (L387) of EDIII; DENV4 E88 binds to A331 in BC loop and T361 in DE loop of EDIII.39 Of note, DENV-4 mAb E88 is specific to the EDIII lateral ridge (LR) epitope, which is implicated in the induction of virus-neutralizing antibodies.27,28 EDIII displayed on DENV-4 E VLPs retained the antigenic structure characteristic of DENV-4 serotype, which is corroborated by the lack of reactivity of mAbs (such as DENV-1 E103, DENV-2 3H5, DENV-3 8A1, and many others in Table 1) specific to the EDIII epitopes of the remaining three DENV serotypes. Consistent with the data so far, the DENV-4 E VLP-displayed EDIII was recognized by mAb 12 C1, an antibody that recognizes EDIII of all four DENV serotypes. However, mAb h-2J20, a human EDIII-specific mAb, recognized DENV-4 VLPs albeit with lesser efficiency when compared with the ELISA reactivity toward DENV E VLPs of the remaining three serotypes. This may presumably reflect subtle differences in the antigenic structure of DENV-4 EDIII. Aside from EDIII epitopes, the DENV-4 E VLPs also displayed the fusion loop epitope (evidenced by noticeable reactivity manifested by mAbs 4G2, h-1M7, and h-1N5) and EDI/II (based on mAb h-23.13 reactivity). Consistent with its design, the DENV-4 E VLPs, which do not contain prM, failed to be recognized by mAb h-2K2.
Table 1.
Evaluation of epitope integrity of DENV-1, 2, 3, and 4 VLPs*
| Serotype-specific murine mAbs | ||||||
|---|---|---|---|---|---|---|
| mAb† | Epitope specificity | Absorbance at 450 nm | ||||
| 1E | 2E | 3E | 4E | HBsAg | ||
| DENV1 E103 | Anti-EDIII LR | 3.48 | 0.06 | 0.06 | 0.05 | 0.03 |
| DENV1 E24 | Anti-EDIII | 1.23 | 0.03 | 0.02 | 0.03 | 0.03 |
| DENV1 E29 | Anti-EDIII | 1.87 | 0.03 | 0.09 | 0.14 | 0.03 |
| DENV1 E37 | Anti-EDIII | 0.71 | 0.02 | 0.03 | 0.01 | 0.04 |
| DENV2 3H5 | Anti-EDIII LR | 0.14 | 3.46 | 0.06 | 0.11 | 0.03 |
| DENV2 104 | Anti-EDIII CC loop | 0.08 | 3.45 | 0.1 | 0.05 | 0.04 |
| DENV2 70 | Anti-EDIII AS/LR | 0.02 | 0.92 | 0.03 | 0.03 | 0.02 |
| DENV2 106 | Anti-EDIII LR | 0.05 | 2.5 | 0.03 | 0.03 | 0.05 |
| DENV3 8A1 | TS | 0.08 | 0.05 | 1.35 | 0.03 | 0.03 |
| DENV3 E3 | Anti-EDIII | 0.13 | 0.03 | 2.90 | 0.02 | 0.02 |
| DENV3 E1 | Anti-EDIII | 0.04 | 0.03 | 3.43 | 0.02 | 0.03 |
| DENV3 E4 | Anti-EDI/II | 0.12 | 0.04 | 3.18 | 0.03 | 0.03 |
| DENV4 E29 | Anti-EDIII | 0.04 | 0.04 | 0.03 | 2.80 | 0.02 |
| DENV4 E42 | Anti-E | 0.03 | 0.02 | 0.02 | 2.90 | 0.02 |
| DENV4 E43 | Anti-E | 0.02 | 0.02 | 0.02 | 2.15 | 0.03 |
| DENV4 E76 | Anti-EDIII | 0.05 | 0.07 | 0.03 | 2.32 | 0.04 |
| DENV4 E88 | Anti-EDIII LR | 0.02 | 0.03 | 0.02 | 2.94 | 0.03 |
| Cross-reactive murine and human mAbs | ||||||
| 4G2‡ | FL | 0.23 | 0.2 | 0.83 | 0.41 | 0.04 |
| 12 C1‡ | Anti-EDIII, not LR | 3.76 | 3.86 | 3.72 | 3.68 | 0.04 |
| h-1M7‡ | FL | 2.97 | 3.03 | 2.48 | 2.73 | 0.03 |
| h-23.13 | EDI/II | 0.23 | 0.1 | 0.52 | 0.26 | 0.03 |
| h-1N5‡ | FL | 0.95 | 0.45 | 1.47 | 1.06 | 0.05 |
| h-2J20 | Anti-EDIII | 3.67 | 3.46 | 3.58 | 0.27 | 0.02 |
| h-2K2 | prM | 0.07 | 0.06 | 0.06 | 0.06 | 0.05 |
AS = A strand; DENV = dengue virus; ED = envelope domain; ELISA = enzyme-linked immunosorbent assay; FL = fusion loop; LR = lateral ridge; mAb = monoclonal antibody; prM = pre-membrane; TS = type specific; VLP = virus-like particle.
Evaluated by indirect ELISA using Pichia Pastoris–expressed purified DENV E VLPs as coating antigens.
Serotype-specific mAbs used in ELISA: DENV-1 mAbs E103, E24, E29, and E3728; DENV-2 mAb 3H536; DENV-2 mAbs 104, 70, and 10638; DENV-3 mAb 8A140; DENV-3 mAbs E3, E1, and E437; DENV-4 mAbs E29, E42, E43, E76, and E88.39 Cross-reactive mAbs: 4G236; 12C140; h-1N5 and h-1M7,43 h-23.1341; h-2J2044; h-2k2. All mAbs are murine, except those that are prefixed with h-, which indicates human.
Complex-specific mAbs; 4G2, h-1N5, and h-1M7 bind the fusion loop of all four DENV-Es; 12 C1 binds recombinant EDIII (outside the LR epitope) of all four DENV serotypes.
Finally, the DENV-4 E VLPs were not recognized by any of the DENV-1, 2, or 3 type-specific mAbs (Table 1). Since the antibodies that were unable to recognize DENV-4 E VLPs identified their cognate type-specific antigens with strong reactivity, it ruled out any possibility of mAbs being nonfunctional. HBsAg, used as negative control, was not recognized by any antibody used in the assay. Overall, this mAb analysis suggests that DENV-4 E VLPs display the critical neutralizing epitopes of E protein, particularly those on EDIII.
DENV-4 E VLPs are immunogenic in mice and elicit EDIII-focused serotype-specific neutralizing antibody response.
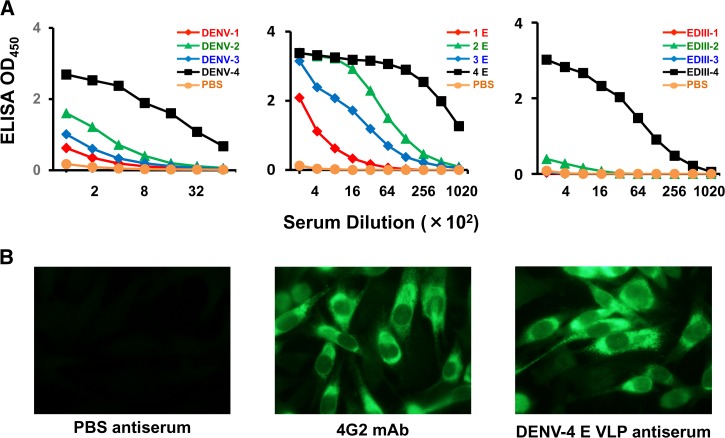
DENV-4 E adsorbed on alhydrogel was injected into BALB/c mice on days 0, 30, and 90. Sera obtained from the bleeds collected on day 100 were pooled and analyzed for the presence of antibodies by ELISA (Figure 3A ), for the presence of DENV-4-recognizing antibodies by IFA (Figure 3B) and for the presence of DENV-neutralizing antibodies by FNT (Figure 4A ).
Figure 3.
DENV-4 E VLPs are immunogenic in mice. (A) BALB/c mice were immunized with DENV-4 E VLPs adsorbed on alhydrogel on days 0, 30, and 90. On day 100, sera were collected and evaluated (at various dilutions) for the presence of antibodies against the four DENVs (first panel), four DENV-Es (second panel), and four EDIIIs (third panel) by ELISA. In each of the three cases, curves in red, green, blue, and black represent DENV serotype 1, 2, 3, and 4, respectively. (B) Recognition of DENV-4-infected BHK-21 cells by PBS-immunized negative control mouse serum (first panel), 4G2 mAb (second panel), and DENV-4 E immune serum (third panel) by IFA. DENV-4 E = dengue virus serotype 4 ectodomain; ED = envelope domain; ELISA = enzyme-linked immunosorbent assay; mAb = monoclonal antibody; VLP = virus-like particle.
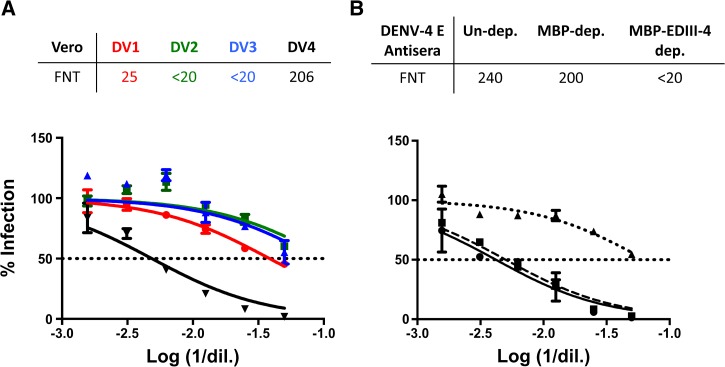
Figure 4.
DENV-4 E VLPs elicit EDIII-directed serotype-specific neutralizing antibodies. (A) Immune sera collected 10 days after the final boost from DENV-4 E VLP-immunized BALB/c mice were evaluated for existence of neutralizing antibodies against DENV-1 (DV1, red curve), DENV-2 (DV2, green curve), DENV-3 (DV3, blue curve), and DENV-4 (DV4, black curve) by FNT on Vero cells. (B) The contribution of anti-EDIII antibodies in DENV-4 E immune serum (black solid curve) in neutralizing DENV-4 was evaluated by FNT on Vero cells after depleting it of anti-EDIII antibodies by incubating it with serotype 4 EDIII-MBP (black dotted curve). DENV-4 E immune serum was also incubated with MBP (black dashed curve) to score nonspecific depletion of antibodies (if any). The percentage of Vero cells infected with the DENV is represented on y axis, while log of reciprocal of sera dilution is represented on x axis. FNT50 titers, tabulated on the top of each of the graphs, represent the sera dilution resulting in 50% neutralization of respective DENV, and are calculated from the dotted horizontal line in the graphs representing 50% infection (or 50% neutralization). DENV-4 E = dengue virus serotype 4 ectodomain; ED = envelope domain; FNT = fluorescence-activated cell sorting (FACS)-based neutralization test; MBP = maltose-binding protein; VLP = virus-like particle.
Serum pooled on day 100 was evaluated, at various dilutions, for reactivity toward three sets of coating antigens in an indirect ELISA format. The first set of coating antigens were the virus particles corresponding to the four serotypes (Figure 3A, first panel), the second set of coating antigens were purified, P. pastoris–expressed E VLPs of the four DENV serotypes (Figure 3A, second panel), and the third set of coating antigens were purified, P. pastoris–expressed EDIII proteins (Figure 3A, third panel). It was observed that DENV-4 E VLP-induced antiserum reacted the best with EDIII, E, and DENV of serotype 4 (black curve in all the three panels). The reactivity to EDIII of the other three serotypes was only marginal (Figure 3A, third panel). The order of reactivity to E and DENV was similar, with highest reactivity to serotype 4, followed by reactivity to serotypes 2, 3, and 1 (Figure 3A, first and second panels). In these ELISAs, phosphate buffered saline-immunized serum served as negative control and failed to react to EDIIIs, DENV-Es, and DENVs.
The ability of the elicited DENV-4 E antibodies to recognize DENV-4 in infected cells was evaluated by IFA. Here, DENV-4-infected BHK-21 cells were incubated with pooled anti-DENV-4 E VLP antiserum and the bound antibodies were detected with FITC-labeled anti-mouse IgG using fluorescence microscopy (Figure 3B). It was observed that anti-DENV-4 E VLP antiserum (Figure 3B, third panel) recognized DENV-4 in infected BHK-21 cells as efficiently as positive control, 4G2 mAb (Figure 3B, second panel); PBS-immunized negative control mouse serum failed to recognize DENV-4 (Figure 3B, first panel).
The DENV-neutralizing efficiency of the antibodies induced by DENV-4 E VLPs and their specificity was evaluated by FNT on Vero cells. This revealed that DENV-4 E VLPs elicited antibodies that were primarily capable of neutralizing DENV-4 (FNT50 ∼200; Figure 4A), with no discernible neutralizing activity toward the remaining three DENV serotypes (FNT50 titers of 25, < 20, and < 20 against DENV-1, 2, and 3, respectively). FNT50 titers elicited by PBS-immunized negative control mouse serum against the four DENV serotypes were found to be < 20 (data not shown). The generation of serotype-specific neutralizing antibody response is consistent with the immune response pattern generated by the DENV-E VLPs corresponding to the remaining three serotypes, reported earlier.31–33
The DENV-4 E VLP immune serum (Figure 4B, solid curve) ceased to neutralize DENV-4 once it was depleted of EDIII-4 antibodies by incubation with EDIII-4-MBP (Figure 4B, dotted curve). On the contrary, there was no significant reduction in the titers of anti-DENV-4 E VLP antiserum when subjected to depletion of antibodies on MBP (Figure 4B, dashed curve). This is in agreement with the observation made with DENV-1 E, 2 E, and 3 E VLPs,31–33 all of which were shown to elicit EDIII-focused virus-neutralizing antibodies.
Discussion
Dengue is a huge public health problem around the globe. A preventive vaccine is required to protect all “at risk” population from dengue. A subunit-based vaccine approach, capable of generating serotype-specific neutralizing antibodies, could be an effective strategy. In this context, the envelope glycoprotein appears to be an appropriate target since it harbors the maximum DENV-neutralizing epitopes and also contains the host-receptor-binding domain EDIII, which has been reported to possess strongly neutralizing type-specific epitopes.27,28,45 Tetravalent mixture of Merck's E glycoprotein expressed in insect cells is in clinical phase I.46 Preclinical studies of this vaccine candidate revealed encouraging data in mice and macaques, as they were found to be immunogenic.47,48 Unlike insect cell-expressed E glycoprotein, P. pastoris–expressed E glycoprotein of serotypes 1, 2, and 3 have been found to assemble into VLPs, despite the lack of prM31–33; VLPs being nanoparticles offer an advantage of improved immunogenicity, and their formation without prM could be a potential benefit, as anti-prM antibodies are known to be disease enhancing.29
With the aim to generate E VLP-based tetravalent dengue vaccine candidate, development of DENV-1, 2, and 3 E VLPs has already been reported.31–33 Encouraged by the results of such studies, we expressed DENV-4 E glycoprotein in P. pastoris, the findings of which are discussed in this study. Overall, like the E glycoprotein of serotypes 1, 2, and 3, DENV-4 E could also be expressed in P. pastoris and assembled into VLPs in the absence of prM, displaying the essential neutralizing epitopes on its surface. DENV-4 E VLPs were found to be immunogenic in mice and generated neutralizing antibodies only against DENV-4; cross-neutralization of DENV-1, 2, and 3 was not discerned. Moreover, anti-DENV-4 E antiserum ceased to effectively neutralize DENV-4 when depleted of anti-EDIII antibodies, highlighting the importance of EDIII antibodies in virus neutralization.
DENV-4 E gene was similar in its design to DENV-1, 2 and 3 E genes.31–33 The DENV-4 E expressing biomass was prepared and protein was purified according to protocols optimized for the other three E proteins. Purified DENV-4 E was found to be devoid of the engineered prM signal peptide (assessed through N-terminal sequencing) and, glycosylated on the basis of its reactivity with Con A-HRPO. It assembled into VLPs with efficient display of critical neutralizing epitopes like the LR, which has been reported to generate neutralizing antibodies. Of special significance is the recognition of DENV-4 E VLPs by serotype 4-specific anti-EDIII mAb E88. This mAb, which does not recognize isolated EDIII, has been reported to recognize a quaternary epitope on the E dimer, which involves EDIII.39 Therefore, recognition of DENV-4 E VLPs by mAb E88 strongly suggests conformational display of critical EDIII epitopes on its surface. This indicates that the VLPs may be potentially immunogenic. These VLPs were indeed found to be immunogenic in BALB/c mice. The anti-DENV-4 E VLP antiserum collected 10 days post-final boost was found to possess high ELISA titer antibodies against EDIIIs, DENV-Es, and DENVs. ELISA reactivity of the DENV-4 E immune serum against EDIII was found to be largely homotypic (Figure 3A, third panel). However, moderate extent of cross-reactivity in ELISA was observed with E and DENV of the other three serotypes (Figure 3A, first and second panels). Presumably, the EDIII and non-EDIII epitopes on the DENV-4 E VLPs, respectively, induce homotypic and cross-reactive antibodies. Unlike the moderate cross-reactivity observed in ELISA against DENVs and the four DENV E VLPs, the anti-DENV-4 E VLP antiserum exhibited homotypic neutralization. This capacity of DENV-4 E VLPs to elicit homotypic neutralizing immune response is consistent with the other three DENV E VLPs, which have also been found to induce cognate homotypic neutralizing immune responses.31–33 Absence of cross-reactive neutralization and presence of homotypic neutralization might be a potential advantage, as cross-reactive neutralizing antibodies have been found to enhance DENV infection.49 Watanabe and others reported in AG129 mouse lethal model that DENV-2 immune complexes (ICs) formed with 3H5 mAb (specific toward DENV-2 EDIII) protected the mice at all mAb concentrations, whereas ICs with cross-reactive neutralizing mAbs 4G2 or 6B6C-1 (directed against fusion loop of EDII) induced lethal disease even at highly neutralizing concentrations.49 Moreover, the serotype-specific neutralizing antibodies of anti-DENV-4 E VLP antiserum were found to be directed against EDIII, offering support to the argument that EDIII generates strongly neutralizing antibodies and is probably an indispensable component of a dengue vaccine candidate. Importantly, the findings with DENV-4 E VLPs essentially mirror the findings of the other three DENV E VLPs. This can translate into more homogeneity in formulating tetravalent mixture of the four DENV-E VLPs. Next, we envisage evaluation of tetravalent mixture of the four DENV E VLPs in mice, and optimize the dose of each of the four components to generate a balanced immune response against the four DENV serotypes. With all the four monovalent P. pastoris–expressed E proteins eliciting EDIII-directed homotypic neutralizing immune response, we expect the tetravalent mixture to be a potential candidate in our quest to develop a safe, affordable, and efficacious dengue vaccine candidate. Additional studies are warranted to evaluate if the proposed tetravalent E-based VLP vaccine formulation would elicit EDIII-directed serotype-specific neutralizing antibodies in primates and humans.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Aravinda de Silva for access to DENV mAbs and EDIII-MBPs. BEI resources are also acknowledged for access to several DENV mAbs.
Footnotes
Financial support: The funding for this work was received from Department of Biotechnology, Government of India (Grant no. BT/PR11807/MED/29/871/2014) to Navin Khanna.
Authors' addresses: Niyati Khetarpal, Rahul Shukla, Ravi Kant Rajpoot, Ankur Poddar, Meena Pal, Upasana Arora, and Navin Khanna, Recombinant Gene Products Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, India, E-mails: 27.niyati@gmail.com, rahulshukla.2802@gmail.com, rajpootravikant@gmail.com, ankurgemini29@gmail.com, maina2u@gmail.com, upasanaaro@gmail.com, and navinkhanna5@gmail.com. Sathyamangalam Swaminathan, Department of Biological Sciences, Birla Institute of Technology and Sciences, Hyderabad, India, E-mail: ssn225@gmail.com.
References
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanaway JD, Shepard DS, Undurraga EA, Halasa YA, Coffeng LE, Brady OJ, Hay SI, Bedi N, Bensenor IM, Castaneda-Orjuela CA, Chuang TW, Gibney KB, Memish ZA, Rafay A, Ukwaja KN, Yonemoto N, Murray CJ. The global burden of dengue: an analysis from the Global Burden of Disease Study 2013. Lancet Infect Dis. 2016;16:712–723. doi: 10.1016/S1473-3099(16)00026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler DJ, Kuno G, Markoff L. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th edition. Philadelphia, PA: Wolters Kluwer and Lippincott Williams and Wilkins; 2007. pp. 1153–1252. [Google Scholar]
- 4.Swaminathan S, Khanna N. Dengue: recent advances in biology and current status of translational research. Curr Mol Med. 2009;9:152–173. doi: 10.2174/156652409787581592. [DOI] [PubMed] [Google Scholar]
- 5.WHO Fact Sheet on Dengue and Severe Dengue. 2016. http://www.who.int/mediacentre/factsheets/fs117/en/ Available at. Accessed June 20, 2016.
- 6.Halstead SB. Dengue antibody-dependent enhancement: knowns and unknowns. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.AID-0022-2014. doi:10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- 7.Swaminathan S, Batra G, Khanna N. Dengue vaccines: state of the art. Expert Opin Ther Pat. 2010;20:819–835. doi: 10.1517/13543771003767476. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz J, Roehrig J, Barrett A, Hombach J. Next generation dengue vaccines: a review of candidates in preclinical development. Vaccine. 2011;29:7276–7284. doi: 10.1016/j.vaccine.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Coller BA, Clements DE. Dengue vaccines: progress and challenges. Curr Opin Immunol. 2011;23:391–398. doi: 10.1016/j.coi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Thomas SJ, Endy TP. Vaccines for the prevention of dengue: development update. Hum Vaccin. 2011;7:674–684. doi: 10.4161/hv.7.6.14985. [DOI] [PubMed] [Google Scholar]
- 11.Guy B, Briand O, Lang J, Saville M, Jackson N. Development of the Sanofi Pasteur tetravalent dengue vaccine: one more step forward. Vaccine. 2015;33:7100–7111. doi: 10.1016/j.vaccine.2015.09.108. [DOI] [PubMed] [Google Scholar]
- 12.Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, Tran HN, Bouckenooghe A, Chansinghakul D, Cortes M, Fanouillere K, Forrat R, Frago C, Gailhardou S, Jackson N, Noriega F, Plennevaux E, Wartel TA, Zambrano B, Saville M, CYD-TDV Dengue Vaccine Working Group Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med. 2015;373:1195–1206. doi: 10.1056/NEJMoa1506223. [DOI] [PubMed] [Google Scholar]
- 13.Halstead SB, Russell PK. Protective and immunological behavior of chimeric yellow fever dengue vaccine. Vaccine. 2016;34:1643–1647. doi: 10.1016/j.vaccine.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Edelman R. Unique challenges faced by the clinical evaluation of dengue vaccines. Expert Rev Vaccines. 2011;10:133–136. doi: 10.1586/erv.10.159. [DOI] [PubMed] [Google Scholar]
- 15.Thomas SJ. The necessity and quandaries of dengue vaccine development. J Infect Dis. 2011;203:299–303. doi: 10.1093/infdis/jiq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swaminathan S, Khanna N, Herring B, Mahalingam S. Dengue vaccine efficacy trial: does interference cause failure? Lancet Infect Dis. 2013;13:191–192. doi: 10.1016/S1473-3099(13)70028-8. [DOI] [PubMed] [Google Scholar]
- 17.Chackerian B. Virus-like particles: flexible platforms for vaccine development. Expert Rev Vaccines. 2007;6:381–390. doi: 10.1586/14760584.6.3.381. [DOI] [PubMed] [Google Scholar]
- 18.Wang PG, Kudelko M, Lo J, Siu LY, Kwok KT, Sachse M, Nicholls JM, Bruzzone R, Altmeyer RM, Nal B. Efficient assembly and secretion of recombinant subviral particles of the four dengue serotypes using native prM and E proteins. PLoS One. 2009;4:e8325. doi: 10.1371/journal.pone.0008325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Jiang H, Zhou J, Yang X, Tang Y, Fang D, Jiang L. Recombinant dengue virus-like particles from Pichia pastoris: efficient production and immunological properties. Virus Genes. 2010;40:53–59. doi: 10.1007/s11262-009-0418-2. [DOI] [PubMed] [Google Scholar]
- 20.Kuwahara M, Konishi E. Evaluation of extracellular subviral particles of dengue virus type 2 and Japanese encephalitis virus produced by Spodoptera frugiperda cells for use as vaccine and diagnostic antigens. Clin Vaccine Immunol. 2010;17:1560–1566. doi: 10.1128/CVI.00087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang YX, Jiang LF, Zhou JM, Yin Y, Yang XM, Liu WQ, Fang DY. Induction of virus-neutralizing antibodies and T cell responses by dengue virus type 1 virus-like particles prepared from Pichia pastoris. Chin Med J (Engl) 2012;125:1986–1992. [PubMed] [Google Scholar]
- 22.Bachmann MF, Jennings GT. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 23.Yildiz I, Shukla S, Steinmetz NF. Applications of viral nanoparticles in medicine. Curr Opin Biotechnol. 2011;22:901–908. doi: 10.1016/j.copbio.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrone LA, Ahmad A, Veguilla V, Lu X, Smith G, Katz JM, Pushko P, Tumpey TM. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. J Virol. 2009;83:5726–5734. doi: 10.1128/JVI.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, Nabel GJ. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med. 2010;16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindenbach BD, Thiel HJ, Rice CM. Knipe DM, Howley PM. Fields Virology. 5th edition. Philadelphia, PA: Wolters Kluwer and Lippincott Williams and Wilkins; 2007. Flaviviridae: The viruses and their replication; pp. 1101–1152. [Google Scholar]
- 27.Gromowski GD, Barrett AD. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology. 2007;366:349–360. doi: 10.1016/j.virol.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 28.Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O'Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, Chawansuntati K, Malasit P, Mongkolsapaya J, Screaton G. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodenhuis-Zybert IA, van der Schaar HM, da Silva Voorham JM, van der Ende-Metselaar H, Lei HY, Wilschut J, Smit JM. Immature dengue virus: a veiled pathogen? PLoS Pathog. 2010;6:e1000718. doi: 10.1371/journal.ppat.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mani S, Tripathi L, Raut R, Tyagi P, Arora U, Barman T, Sood R, Galav A, Wahala W, de Silva A, Swaminathan S, Khanna N. Pichia pastoris-expressed dengue 2 envelope forms virus-like particles without pre-membrane protein and induces high titer neutralizing antibodies. PLoS One. 2013;8:e64595. doi: 10.1371/journal.pone.0064595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poddar A, Ramasamy V, Shukla R, Rajpoot RK, Arora U, Jain SK, Swaminathan S, Khanna N. Virus-like particles derived from Pichia pastoris-expressed dengue virus type 1 glycoprotein elicit homotypic virus-neutralizing envelope domain III-directed antibodies. BMC Biotechnol. 2016;16:50. doi: 10.1186/s12896-016-0280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripathi L, Mani S, Raut R, Poddar A, Tyagi P, Arora U, de Silva A, Swaminathan S, Khanna N. Pichia pastoris-expressed dengue 3 envelope-based virus-like particles elicit predominantly domain III-focused high titer neutralizing antibodies. Front Microbiol. 2015;6:1005. doi: 10.3389/fmicb.2015.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- 35.Kraus AA, Messer W, Haymore LB, de Silva AM. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol. 2007;45:3777–3780. doi: 10.1128/JCM.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henchal EA, Gentry MK, McCown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- 37.Brien JD, Austin SK, Sukupolvi-Petty S, O'Brien KM, Johnson S, Fremont DH, Diamond MS. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J Virol. 2010;84:10630–10643. doi: 10.1128/JVI.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol. 2010;84:9227–9239. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, Diamond MS. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J Virol. 2013;87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, de Silva AM. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog. 2010;6:e1000821. doi: 10.1371/journal.ppat.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Alwis R, Beltramello M, Messer WB, Sukupolvi-Petty S, Wahala WM, Kraus A, Olivarez NP, Pham Q, Brien JD, Tsai WY, Wang WK, Halstead S, Kliks S, Diamond MS, Baric R, Lanzavecchia A, Sallusto F, de Silva AM. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis. 2011;5:e1188. doi: 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, Crowe JE., Jr Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol. 2012;86:2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith SA, de Alwis AR, Kose N, Harris E, Ibarra KD, Kahle KM, Pfaff JM, Xiang X, Doranz BJ, de Silva AM, Austin SK, Sukupolvi-Petty S, Diamond MS, Crowe JE., Jr The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. MBio. 2013;4:e00873–e13. doi: 10.1128/mBio.00873-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arora U, Tyagi P, Swaminathan S, Khanna N. Virus-like particles displaying envelope domain III of dengue virus type 2 induce virus-specific antibody response in mice. Vaccine. 2013;31:873–878. doi: 10.1016/j.vaccine.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 45.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp M and Dohme Corp. A phase I randomized, double-blind, placebo-controlled, dose-escalation study to evaluate the safety, tolerability, and immunogenicity of a tetravalent recombinant subunit dengue vaccine (V180) in Healthy adults. https://clinicaltrials.gov/ct2/show/NCT01477580 ClinicalTrials.gov Identifier NCT01477580. Available at. Accessed 20, 2016.
- 47.Clements DE, Coller BA, Lieberman MM, Ogata S, Wang G, Harada KE, Putnak JR, Ivy JM, McDonell M, Bignami GS, Peters ID, Leung J, Weeks-Levy C, Nakano ET, Humphreys T. Development of a recombinant tetravalent dengue virus vaccine: immunogenicity and efficacy studies in mice and monkeys. Vaccine. 2010;28:2705–2715. doi: 10.1016/j.vaccine.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Govindarajan D, Meschino S, Guan L, Clements DE, ter Meulen JH, Casimiro DR, Coller BA, Bett AJ. Preclinical development of a dengue tetravalent recombinant subunit vaccine: immunogenicity and protective efficacy in nonhuman primates. Vaccine. 2015;33:4105–4116. doi: 10.1016/j.vaccine.2015.06.067. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe S, Chan KW, Wang J, Rivino L, Lok SM, Vasudevan SG. Dengue virus infection with highly neutralizing levels of cross-reactive antibodies causes acute lethal small intestinal pathology without a high level of viremia in mice. J Virol. 2015;89:5847–5861. doi: 10.1128/JVI.00216-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.