Abstract
The aim of this study was to reveal the biological function of Cj1199 which was overexpressed in the laboratory induced erythromycin resistant strains. The Cj1199 deletion mutant (ΦCj1199) was constructed via insertional inactivation from its parent strain Campylobacter jejuni NCTC11168. The ΦCj1199 and NCTC11168 were then subjected to microarray and real-time PCR to find gene pathway of Cj1199. The antimicrobial susceptibility, antimicrobial resistance development, growth characteristics and leucine metabolism were examined to confirm the biological function of Cj1199. Our result showed that a total of 20 genes were down-regulated in ΦCj1199. These genes were mainly involved in leucine biosynthesis, amino acid transport and periplasmic/membrane structure. Compared to NCTC11168, ΦCj1199 was difficult to acquire higher-level erythromycin resistance during the in vitro step-wise selection. The competition growth and leucine-dependent growth assays demonstrated that ΦCj1199 imposed a growth disadvantage under pressure of erythromycin and in the leucine-free medium. In conclusion, Cj1199 gene may directly regulate the leucine biosynthesis and transport and indirectly affect the development of erythromycin resistance in C. jejuni.
Keywords: Cj1199, C. jejuni, biological function, leucine biosynthesis, erythromycin resistance
Introduction
Campylobacter jejuni (C. jejuni), is an important foodborne pathogen whose infection often results in acute, self-limiting enteritis, but can lead to more serious complications such as Guillain-Barre Syndrome (Butzler, 2004; Hughes and Cornblath, 2005). Macrolides (mainly erythromycin), which can bind to the 50S subunit of bacterial ribosome and interfere with protein synthesis by inhibiting the elongation of peptide chains (Weisblum, 1995), are the valuable antimicrobials of choice to treat severe Campylobacter infections (Gibreel et al., 2005; EFSA, 2009). Point mutations in domain V of 23S rRNA (Taylor et al., 1997), in the ribosome protein L4/L22 and overexpression of efflux pumps are known as the major causes of resistance in Campylobacter (Lin et al., 2002; Gibreel et al., 2005). Despite the improved understanding of its general physiology and biochemistry, the molecular mechanisms involved in Campylobacter fitness and resistance development are largely unknown.
New approaches, like microarray and RNA-seq, have been used to determine the transcriptome changes associated with some gene mutations and the adaptive mechanisms of C. jejuni to erythromycin treatment (Chaudhuri et al., 2011; Xia et al., 2013). Our previous study, using a microarray analysis to find the transcriptional change involved in the development of macrolide resistance in Campylobacter, showed that a large number of genes were responsible to the development of macrolide resistance in Campylobacter, ranging from genes involved in energy metabolism to macromolecular metabolism to cell processes (Hao et al., 2013). In these differentially expressed genes, a gene of unknown function, Cj1199, was overexpressed in the erythromycin resistant C. jejuni mutants selected from their parent strains NCTC11168 and 81176. The expression level of Cj1199 was dose dependent to the level of resistance in the series erythromycin resistant C. jejuni mutants selected from NCTC11168. Bioinformatic analyses suggested that Cj1199 may encode putative iron/ascorbate-dependent oxidoreductase which may be involved in leucine biosynthesis and consequently affect the growth of the strains. Many previous studies revealed that leucine is a key amino acid of erythromycin resistant short peptides (E-Peptide) in E. coli (Dam et al., 1996; Tenson et al., 1997; Tripathi et al., 1998; Tenson and Mankin, 2001; Vimberg et al., 2004). Therefore, Cj1199 may also be associated with E-Peptide mediated erythromycin resistance. The function of Cj1199 on the development of macrolide resistance and fitness has yet to be determined. The objectives of this study were to decipher functionality and active network of this gene in C. jejuni.
Materials and Methods
Bacterial Strains and Growth Conditions
The C. jejuni NCTC11168 was kindly provided by Chinese Center for Disease Control. C. jejuni strains were routinely cultured on Mueller-Hinton (MH) agar (Oxoid, Basingstoke, UK) or blood agar plates at 42°C under microaerophilic conditions. E. coli DH5α was grown aerobically in Luria-Bertani medium at 37°C. The Ham’s F-12 nutrient powder mixture (SH30010, Thermo Fisher, Waltham, MA, USA) and Ham’s F-12m nutrient powder mixture (RR13033.01, Thermo Fisher, Waltham, MA, USA) lacking leucine were used for leucine biosynthesis test.
Construction of Cj1199 Deletion Mutant
A 2.5-Kb fragment containing Cj1199 and its flanking region was amplified with primers of cj1199F and cj1199R (Table 1) by PCR and cloned into pGEM-T easy vector (Promega, Madison, WI, USA) to yield pL1199. A chloramphenicol resistance cassette (Cmr marker) was amplified from pUOA18 using primers of ecatF and ecatR. Primers rF and rR were used to inversely amplify pL1199. The PCR product of the inverse amplification of pL1199 was digested with SmaI and BamHI and ligated with the Cmr marker to yield plasmid pL1199T. The plasmid pL1199T was transformed into E. coli DH5α and then introduced to C. jejuni NCTC11168 by natural transformation (Jeon et al., 2010). The Cj1199 deletion mutant (ΦCj1199) was selected on MH agar containing chloromycetin (20 μg/ml) and confirmed by PCR and DNA sequencing.
Table 1.
Primers used for construction of Cj1199 mutant and for qRT-PCR
| Primer name | Primer sequence | Product size (bp) |
|---|---|---|
| Primers used for construction and confirmation of Cj1199 mutant | ||
| Cj1199F | TAGCGTATTTGATTTGCGTTTT | 2536 |
| Cj1199R | TATTTGCACCTATTCTTGGGTAA | |
| rF | ACCCCGGGAAATGGCTTAGTATACCCCCACT SmaI site | 5140 |
| rR | ACGGATCCGCTAGTATAACCTCTAAATTGAGG BamHI site | |
| ecatF | ACGGATCCAAAGAGTGACCGCCGAGA BamHI | 810 |
| ecatR | ACCCCGGGCAGTGCGACAAACTGGGA SmaI site | |
| 16SF | CTGGCTCAGAGTGAACGC | 533 |
| 16SR | CCCTTTACGCCCAGTGATT | |
| hipOF | AGAAGCCATCATCGCACCT | 148 |
| hipOR | TGCTGAAGAGGGTTTGGGT | |
| catF | AAAGAGTGACCGCCGAGATA | 0/798a |
| catR | CAGTGCGACAAACTGGGATT | |
| Cj1199F | TGAACTTACCTATACTTGATT | 954/1353b |
| Cj1199R | AAATACTTGCCACATCAG | |
| Primers used for real-time qRT-PCR | ||
| cj0560F | GCATTTAGGTTTAGGCAAGAAG | 159 |
| cj0560R | GTCATCGTTTTAATCTCATCGCT | |
| cj0920F | CTATTGGTGTATTGGGTGTTGG | 184 |
| cj0920R | GCTTCAAACTGACCTCTTGGC | |
| cj1241F | AGTTACTGGAGTGCCGCTTG | 147 |
| cj1241R | AGATTGGTTGGGCTAGGATGT | |
| cj1388F | CAACTTCCTATCAACCCTGCTTC | 221 |
| cj1388R | GCACTTCTTGCTGGATAAGGAG | |
| cj1646F | CAGGCTGCTTACCAACTCG | 275 |
| cj1646R | GCATCGCTAATGGCAACG | |
| cj1717cF | GCACAGCAAAAGGCACAGG | 232 |
| cj1717cR | TCATCACTTCTTAAACTCTTCCAAT | |
| q16SF | GCTCGTGTCGTGAGATGTTG | 199 |
| q16SR | GCGGTATTGCGTCTCATTGTAT | |
aThere should be no amplification product for NCTC11168, while 798 bp product for ΦCj1199; bProduct length of NCTC11168 should be 954 bp, product length of ΦCj1199 should be 1353 bp.
RNA Extraction
The cells of ΦCj1199 or C. jejuni NCTC11168 were grown in MH agar plate for 24 h at 42°C under microaerophilic conditions. For RNA extraction, RNAprotect Bacteria Reagent (Qiagen, Valencia, CA, USA) was added to the cultures immediately after the incubation to stabilize mRNA. The total RNA from each sample was extracted using the RNeasy Protect Mini Kit (Qiagen, Valencia, CA, USA) following the manufacture’s protocol. RNA samples were extracted from four independent experiments.
DNA Microarray and Data Analysis
The differential gene expression between ΦCj1199 and C. jejuni NCTC11168 was identified using DNA microarray which was supplied by CapitalBio Corporation (Santa Clara, CA, USA). Briefly, iScript cDNA synthesis kit (BioRad, Hercules, CA, USA) was used for synthesis of cDNA from an RNA template via reverse transcription. The cDNA was then labeled with Cy5 or Cy3 dye. The labeled cDNA probes were co-hybridized onto one microarray slide (Roche NimbleGen 4 × 72K, Indianapolis, IN, USA). Hybridized slides were scanned using NimbleGen MS200, and the fluorescence intensities were collected with NimbleGen Scan Software. The linear normalization method was used for data analysis based on expression of the housekeeping genes. Normalized data was log transformed and loaded into MANOVA under R environment. Microarray spots with false discovery rate (FDR)-corrected q-values < 0.01 and fold change ≥2 in the T-test were regarded as differentially expressed genes. The differentially expressed genes were classified based on the genomic annotation in NCBI and then subjected to KEGG database for pathway analysis.
Quantitative Reverse Transcriptase PCR (qRT-PCR)
The surplus cDNA of ΦCj1199 and C. jejuni NCTC11168 was subjected to real-time quantitative reverse transcriptase PCR (qRT-PCR) analysis to confirm the expression of some respective genes identified by DNA microarray. The primers (Table 1) of respective genes were designed using Primer 5 software. The qRT-PCR was performed using SYBR Green Ex TaqTM kit (Takara, Madison, WI, USA) in IQ5 Multicolor Real-time PCR Detection System (Bio-Rad) following method described in previous study (Hao et al., 2010). The qRT-PCR was initiated with a 30 s denaturation at 95°C, followed by 38 cycles of amplification with 5 s of denaturation at 95°C, 30 s of annealing according to the melting temperatures of amplifications. The melting curve was performed from 65 to 95°C (1 s hold per 0.2°C increase) to check the specificity of the amplified product.
Determination of Antimicrobial Resistance (AMR)
Minimum inhibitory concentrations (MICs) of azithromycin (AZM), erythromycin (ERY), tylosin (TYL), ciprofloxacin (CIP), nalidixic acid (NAL), olaquindox (OLA), florfenicol (FFC), doxycycline (DOX), gentamicin (GEN), and levofloxacin (LEV) against ΦCj1199 and C. jejuni NCTC11168 were determined with broth microdilution method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2006). The concentrations of the antimicrobial agents tested ranged from 0.01 to 16 μg/mL. The microdilution plates (96 well plates) were incubated at 42°C for 24 h and the MIC was determined as the lowest concentration where visible bacterial growth was inhibited at the end of the incubation period. C. jejuni ATCC 33560 was used as a quality control strain.
For the minimal bactericidal concentration (MBC) of drugs against C. jejuni strains, 100 μL from each well was successively diluted 1:10 in MH broth. Then 10 μL was spread on MH agar plates and incubated at 42°C for 24 h for colony forming unit (CFU) counting. The MBC was defined as the lowest drug concentration that resulted in a 99.9% reduction in the bacterial density. The final result was expressed as mean of five independent experiments.
The mutant prevention concentration (MPC) of drugs was determined by agar method (Blondeau et al., 2001). The inoculum of C. jejuni was concentrated to 1010 CFU/mL. Bacterial suspensions were inoculated on the agar plates containing serial dilutions of drugs and cultured for 96 h. The MPC was the lowest drug concentration on agar plates without bacterial growth under micro-aerobic conditions.
Erythromycin Tolerance Tested by Flow Cytometry (FCM)
The concentration of bacteria was adjusted to a 0.5 McFarland density. Serial twofold dilutions of erythromycin were prepared with MH broth. The final concentrations of erythromycin ranged from 0.0625 to 8 μg/mL. A 100 μL aliquot of bacteria was added to 900 μL MH broth containing different concentrations of erythromycin and incubated at 42°C in a micro-aerobic condition for 3 h. Each dilution was then centrifuged and the supernatant was discarded. The pellet was washed twice with sterile PBS and propidium iodide (PI), a membrane-impermeable DNA-intercalating dye, was used to stain the erythromycin-treated bacterial cells. One milliliter of buffer and 5 μL of PI were added to each sample, and then incubated for 30 min at 4°C. FCM analysis was conducted on CyAn ADPTM FC500 flow cytometer with SummitTM software (Beckman Coulter, Miami, FL, USA). The intensity of fluorescence of 10,000 cells labeled with PI was analyzed to obtain the mean channel fluorescence (MCF).
In vitro Selection of Erythromycin Resistance
ΦCj1199 and NCTC11168 were subjected to step-wise selection in MH agar plate containing erythromycin. In the first step of selection, cells were plated on MH agar containing 0.25 μg/mL erythromycin. After 3–5 days of incubation under microaerophilic condition at 42°C, colonies were selected and repeatedly incubated with same concentration of erythromycin up to five times. The mutants obtained from first-step selection were then subjected to next step selection by exposing the cells to a twofold increased concentration of erythromycin. The procedure of selection was repeated up to 10 times to obtain highly erythromycin-resistant mutants. All in vitro-selected mutants were subjected to MIC test using the broth microdilution method as described above.
Single Growth Curve and Pair-Wise Competition Experiments
Growth curves of ΦCj1199 and C. jejuni NCTC11168 were measured individually and repeated three times. Briefly, a fresh C. jejuni culture was diluted 100-fold into 10 mL MH broth and incubated for up to 36 h. Viable C. jejuni counts were determined at 4, 12, 24, and 36 h.
Pair-wise competition experiments were used to estimate the growth rate of ΦCj1199 compared with C. jejuni NCTC11168 in the erythromycin containing (1/2 MIC) or drug-free MH broth. Briefly, the same densities of the parent and mutant cultures were prepared before experiments. Equal volumes (0.1 mL) of adjusted cultures were mixed into 10 mL of fresh medium with or without erythromycin. After 24 h incubation, 0.1 mL bacterial culture was then transferred to another 10 mL fresh medium. The passage was repeated up to 10 times and the initial and final concentrations of parent and mutant were determined by standard plate counting on MH plate with (for the mutant population) or without 10 μg/mL chloromycetin (for the total population), respectively. The assays were repeated three times with three technical replicates.
Leucine-Dependent Growth Test
Cultures of ΦCj1199 and C. jejuni NCTC11168 were adjusted to the same densities at the beginning of the experiments. Equal volumes (0.1 ml) of adjusted cultures were added into 10 mL Ham’s F-12 and F-12m individually. After 48 h incubation under microaerophilic conditions at 42°C, 0.1 mL of bacterial culture was then transferred to another 10 mL of fresh medium. The passage was repeated up to five times. Viable C. jejuni counts were determined by dilution and spatula method on MH plates (ICC, 1992). The experiment was performed in triplicate.
Results
Differentially Expressed Genes in ΦCj1199
The 20 genes down-regulated in ΦCj1199 are shown in Table 2. Five genes (leuA, leuB, leuC, leuD, and trpE) were involved in amino acid biosynthesis. The leuABCD genes participated in leucine biosynthesis pathway. Five genes (Cj0919c, Cj0920c, peb1A, pebC, and ceuC) encoded transporters or binding proteins. The permease protein (Cj0919c-Cj0920c), substrate-binding protein (Peb1A), and ATP-binding protein (PebC) worked cooperatively in an ABC-type amino acid transport system. The ceuC encoded enterochelin uptake permease which was also transporters or binding protein. The Cj0246c was involved in signal transduction. Six genes (Cj0560, Cj1356c, Cj0423, Cj0424, Cj0425, and Cj0200c) encoded putative integral membrane or periplasmic proteins. Another three (Cj1025c, Cj1722c, and Cj1388) were hypothetical genes.
Table 2.
Differentially expressed genes in the mutant of ΦCj1199 comparing to its parent strain NCTC 11168.
| Function class | Gene name | Gene function | Fold change |
|---|---|---|---|
| Signal tranduction | Cj0246c | putative MCP-domain signal transduction protein | 0.3624 |
| Transporters/binding proteins | Cj0919c | putative ABC-type amino-acid transporter permease | 0.4997 |
| Cj0922c/pebC | amino-acid ABC transporter ATP-binding protein | 0.461 | |
| Cj0921c/peb1A | bifunctional adhesin/ABC transporter aspartate/glutamate-binding protein | 0.4597 | |
| Cj0920c | putative ABC-type amino-acid transporter permease | 0.3920 | |
| Cj1353/ceuC | enterochelin uptake permease | 0.4557 | |
| Cj1241 | putative MFS transport protein/Arabinose efflux permease | 2.3237 | |
| Amino acid biosynthesis | Cj1716c/leuD | isopropylmalate isomerase small subunin | 0.3996 |
| Cj1717c/leuC | isopropylmalate isomerase large subunin | 0.378 | |
| Cj1718c/leuB | 3-isopropylmalate dehydrogenase | 0.348 | |
| Cj1719c/leuA | 2-isopropylmalate synthase | 0.3167 | |
| Cj0345/trpE | putative anthranilate synthase componentI | 0.4076 | |
| Cell envelop | Cj0560 | putative MATE family transport protein | 0.3564 |
| Cj1356c | putative integral membrane protein | 0.4983 | |
| Cj0423 | putative integral membrane protein | 0.4534 | |
| Cj0425 | putative periplasmic protein | 0.4918 | |
| Cj0424 | putative acidic periplasmic protein | 0.4483 | |
| Cj0200c | putative periplasmic protein | 0.4689 | |
| Hypothetical proteins and unknown function proteins | Cj1025c | hypothetical protein | 0.4974 |
| Cj1722c | hypothetical protein | 0.4727 | |
| Cj1388 | putative endoribonuclease L-PSP | 0.3849 |
Only Cj1241 gene encoding a putative major facilitator superfamily (MFS) transport protein was up-regulated in ΦCj1199. Using qRT-PCR to verify the microarray results, Cj1241 gene was confirmed to be up-regulated, while other four respective genes (Cj0920c, Cj1388, Cj0560, and Cj1717c) were confirmed to be down-regulated (Figure 1).
FIGURE 1.
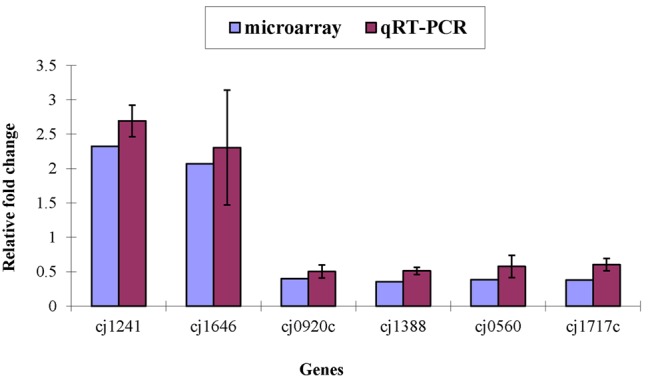
Differential expression of target genes in ΦCj1199 and NCTC11168 determined by real-time qRT-PCR and microarray. The genes were randomly selected. RNA used for real-time qRT-PCR and microarray were prepared in the same time.
Antimicrobial Susceptibility of ΦCj1199
The antimicrobial susceptibility of the parent and mutant strains are presented in Table 3. There was no significant difference between the MIC of 11 antimicrobials in the ΦCj1199 and its parent strain NCTC11168. Both of the strains exhibited very low MIC (MIC ≤ 0.5 μg/mL) to the 14, 15, and 16-membered ring macrolides (erythromycin, azithromycin, and tylosin). The MBC and MPC of five antimicrobials in ΦCj1199 were similar (erythromycin, azithromycin, tylosin, olaquindox, and cydox) with that in NCTC11168 (Table 3).
Table 3.
Minimum inhibitory concentration (MIC)/minimal bactericidal concentration (MBC)/mutant prevention concentration (MPC) of wild-type and mutant strains to different types of drugs.
| Drug | Strains |
|||||
|---|---|---|---|---|---|---|
| Mutant |
NCTC 11168 |
|||||
| MIC (μg/mL) | MBC (μg/mL) | MPC (μg/mL) | MIC (μg/mL) | MBC (μg/mL) | MPC (μg/mL) | |
| ERY | 0.125 | 0.25 | 0.25 | 0.125 | 0.125 | 0.25 |
| TYL | 0.5 | 1 | 1 | 0.5 | 1 | 1 |
| AZI | 0.015 | / | 0.031 | 0.015 | / | 0.031 |
| OLA | 0.25 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 |
| CYA | 0.25 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 |
| FFC | 0.5 | / | / | 0.5 | / | / |
| GEN | 0.125 | / | / | 0.125 | / | / |
| NAL | 8 | / | / | 8 | / | / |
| CIP | 0.062 | / | / | 0.062 | / | / |
| LEV | 0.062 | / | / | 0.062 | / | / |
| DOX | 0.125 | / | / | 0.125 | / | / |
ERY, erythromycin; TYL, tylosin; AZM, azithromycin; FFC, florfenicol; GEN, gentamicin; NAL, nalidixic acid; CIP, ciprofloxacin; LEV, levofloxacin; DOX, doxycycline; OLA, olaquindox; CYA, cydox. The “/” means not detected.
Erythromycin Tolerance of ΦCj1199
The erythromycin tolerance of ΦCj1199 to increased concentration of erythromycin was detected by flow cytometer analysis. As shown in Figure 2, the mean value of the fluorescence intensity was increased following the increase of erythromycin concentration. However, when the concentration of erythromycin reached to their MIC (0.125 μg/mL), the mean PI intensity of C. jejuni NCTC11168 was significantly decreased, while the mean PI intensity of ΦCj1199 kept at its highest level. Under the treatment of different concentrations of erythromycin, the mean PI intensity of ΦCj1199 was higher than that of parent strain, which indicated that ΦCj1199 had higher mortality and lower tolerance to erythromycin.
FIGURE 2.
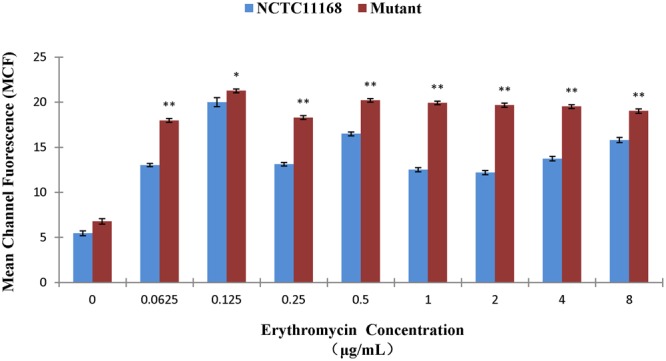
Mean values of PI stain of ΦCj1199 and NCTC11168 cultured with different concentrations of erythromycin. The asterisk (∗) and (∗∗) represent statistical significant difference with P ≤ 0.05 and P ≤ 0.01 comparing with C. jejuni NCTC11168, respectively.
Development of Erythromycin Resistance In vitro
There was no significant difference between ΦCj1199 and NCTC11168 in the first and second steps of selections. When transferred to third step selection by 1 μg/mL erythromycin, ΦCj1199 descendants exhibited less growth than NCTC11168 descendants. When transferred to fourth step of selection, ΦCj1199 descendants could not grow on MH agar containing 2 μg/mL erythromycin, while C. jejuni NCTC11168 descendants grew well and successfully promoted to fifth step of selection by 4 μg/mL erythromycin. The erythromycin MIC of mutants selected from NCTC11168 was equal to or higher than 128 μg/mL, which was significantly higher than erythromycin MIC (4 μg/mL) of mutants selected from ΦCj1199.
Growth Disadvantage of ΦCj1199
The ΦCj1199 and NCTC11168 were separately cultured in macrolide-free MH broth. No significant difference in growth rates was observed between the two strains in single cultures (Figure 3A).
FIGURE 3.
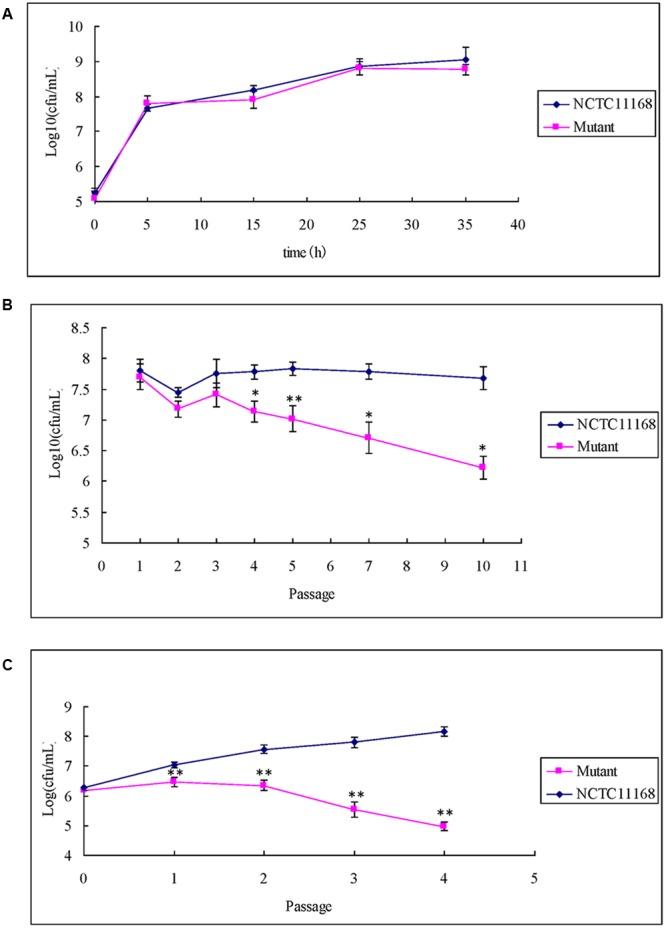
Single growth curve and competitive growth of ΦCj1199 and NCTC11168 in the culture with or without erythromycin. (A) Bacterial culture grown in MH for 36 h. Viable colonies were determined at 5, 15, 25, and 35 h. (B,C) Pair-wise competition of C. jejuni NCTC11168 and mutant was determined during 5–10 passages in the mixed culture. The Initial and final concentrations were determined by standard plate count on MH or MH with chloromycetin. The asterisk (∗) and (∗∗) represent statistical significant difference with P ≤ 0.05 and P ≤ 0.01 comparing with C. jejuni NCTC11168, respectively.
When ΦCj1199 and NCTC11168 were mixed in pairs into drug-free MH broth, population of ΦCj1199 was decreased after the fifth passage and reduced by twofolds of magnitude after the 10th passage (Figure 3B).
In the mixture culture containing 1/2 MIC erythromycin, population of ΦCj1199 was reduced during passages. The ΦCj1199 was outcompeted by NCTC11168 in the drug-containing culture after the fourth passage (Figure 3C).
Leucine-Dependent Growth
There is no significant difference of the growth between ΦCj1199 and NCTC11168 in the medium F-12. Both of them grew well after five passages in the medium F-12 (Figure 4). However, the number of ΦCj1199 was remarkably decreased during the five times of passage in the medium F-12m. After the fifth passages in the medium F-12m, the number of ΦCj1199 mutant had been reduced by two orders of magnitude comparing with its parent strain NCTC11168 which kept at the level of 105 CFU/mL. Conclusively, in absence of leucine, the growth of ΦCj1199 was much slower than NCTC11168.
FIGURE 4.
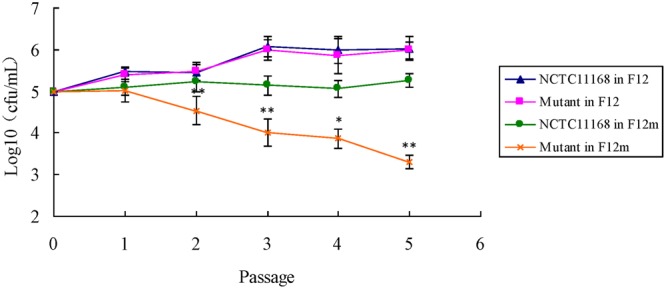
The leucine-dependent growth test. The growth curve of NCTC11168 and ΦCj1199 in Ham’s F12 and Ham’s F12m were determined. The asterisk (∗) and (∗∗) represent statistically significant difference with P ≤ 0.05 and P ≤ 0.01 comparing with C. jejuni NCTC11168, respectively.
Discussion
To our knowledge, this is the first study of the function of Cj1199 on the development of macrolide resistance and biological characteristics of C. jejuni. Previously, Cj1199, a gene of unknown function, was up-regulated in the macrolide resistant C. jejuni mutants selected by step-wise increased erythromycin in vitro from their parent strains NCTC11168 and 81176 (Hao et al., 2013). Bioinformatic analyses suggested that Cj1199 may encode putative iron/ascorbate-dependent oxidoreductase. This gene was located in the downstream of luxS and over-expressed in the luxS null mutation (He et al., 2008). Therefore, the function of Cj1199 attracted our attention. The gene knockout method, microarray assay, antimicrobial susceptibility test, FCM, and growth determination were used to find out the function and molecular mechanism of Cj1199.
From the microarray data, four genes (Cj0921c-Cj0919c-Cj0920c-Cj0922c) involved in amino acid transporters were obviously down-regulated in ΦCj1199. The Cj0921c gene was known to encode a bifunctional adhesin/ABC transporter aspartate/glutamate-binding protein (Peb1A; Pei and Blaser, 1993; Leon-Kempis Mdel et al., 2006). The Peb1A had higher affinity to both aspartate and glutamate, thus it could bind aspartate and glutamate as specific substrates to initiate the nutrient transport systems in gram-negative species (Pei and Blaser, 1993; Leon-Kempis Mdel et al., 2006). The glutamine could modulate bacterial metabolism of the arginine family as well as serine and aspartate families of amino acid and could reduce catabolism of most nutritionally essential amino acids including lysine, leucine, valine and serine, thus it may initiate signaling pathways related to amino acid metabolism in the intestinal bacteria (Dai et al., 2012, 2013). As shown in previous studies, peb1A was essential for the growth, invasion and intestinal colonization of Campylobacter species (Kervella et al., 1993; Pei et al., 1998; Leon-Kempis Mdel et al., 2006; Muller et al., 2007). Under a microaerobic environment, peb1A deficient mutants cannot grow on dicarboxylic amino acids which were the major nitrogen source (Leon-Kempis Mdel et al., 2006). In C. jejuni, the mutation in peb1A locus could reduce the interactions with epithelial cells and intestinal colonization of mice (Pei et al., 1998). There was no report about the characteristic of the other three genes (Cj0919c-Cj0920c-Cj0922c). The string 9.0 and KEGG analysis revealed that these four genes have close interaction with each other and they may work synergistically in the amino acid transport system. The gene cluster (Cj0921c-Cj0919c-Cj0920c-Cj0922c) may be essential for the uptake of nitrogen source which was indispensable for the growth and fitness of Campylobacter species. However, it is still unknown that how Cj1199 effects the expression of Cj0921c-Cj0919c-Cj0920c-Cj0922c gene cluster.
The leuABCD gene cluster was involved in leucine biosynthesis (Labigne et al., 1992). The leuA encoded alpha-isopropylmalate synthase (alpha-IPMS) which was key enzyme to catalyze the first committed step in the leucine biosynthetic pathway (Yindeeyoungyeon et al., 2009). The leuB gene encoded 3-isopropylmalate dehydrogenase, while leuC and leuD genes encoded the large and small subunits of isopropylmalate isomerase, respectively (Tamakoshi et al., 1998). The leuB gene was reported to be essential for the survival in starvation, colony formation and growth in the Enterobacteriaceae (Matsumoto et al., 2011). In the sequences of leuC and leuD, cysteine residues for iron-sulfur binding and other amino acid residues involved in isomerase activity were highly conserved (Tamakoshi et al., 1998). The transcriptional derepresssion of a leuC-defective allele leaded to accumulation of Leu (+) mutations (Martin et al., 2011). Recent studies revealed that the leuABCD gene cluster also participated in the formation of norvaline and norleucine from 2-ketobutyrate and potentially contributed to non-polymeric carbon-chain elongation, which was essential in the synthesis of non-native metabolites in microorganisms (Sycheva et al., 2007; Shen and Liao, 2011). The coordinated expression of the functionally related leuABCD gene clusters may be mediated by LeuO and governed by transcription-driven DNA supercoiling (Chen et al., 2003; Wu and Fang, 2003). Our study indicated that the inactivation of Cj1199 impacted the coordinated expression of leuABCD genes. However, the molecular mechanism involved in the regulation is still unknown. The leucine biosynthesis was versatile for many biogenic processes in prokaryotic and eukaryotic organisms. In the yeast and human cells, leucine biosynthesis regulated cytoplasmic iron-sulfur enzyme biogenesis in an atm1p (ABC transporter)-independent manner (Bedekovics et al., 2011). In many enteric bacteria and fungi, leucine, or a precursor of leucine can be used to regulate peripheral nitrogen metabolism with other cellular processes and can be selected directly or indirectly as a signal molecule in a wider metabolic context by very different mechanisms (Kohlhaw, 2003). Thus, leucine biosynthesis acts as a back door for effecting metabolism in cells (Kohlhaw, 2003).
Little is known about the gene cluster encoding putative membrane proteins (Cj0423-Cj0424-Cj0425). However, the Cj0425 gene was considered to be an important contributor to the oxygen requirement and tolerance of C. jejuni (Kaakoush et al., 2007). The function of the other three genes involved in cell envelop (Cj0560, Cj1356c, and Cj0200c) is still unknown, but they may affect the fitness of ΦCj1199.
The down-regulation of the genes involved in amino acid transport and biosynthesis, and the genes encoding cell envelop may result in the disadvantage of nutrient utilization and growth in ΦCj1199. This hypothesis was confirmed by a series of competitive growth tests in the present study. Although there was no significant change in the short-term and single growth tests, the remarkable disadvantage of ΦCj1199 was observed in the long-term and competitive growth tests. Therefore, the Cj1199 gene was essential for the fitness (growth) of C. jejuni by controlling the expression of the several genes involved in amino acid transport and leucine biosynthesis pathway.
According to the previous literature, a mechanism of macrolide resistance based on the expression of a specific short peptide in the cell was discovered several years ago (Tenson and Mankin, 1995; Tenson et al., 1996). The Shine-Delgarno region of the mini-gene encoding the short peptide was located in the 23S rRNA secondary structure (Tenson and Mankin, 2001). The peptide rendering C. jejuni resistance to erythromycin was one of the C-peptides. It bound erythromycin and removed the drug from its binding site of the ribosome and then sequestered it in an inactive complex (Tripathi et al., 1998). C-peptides showed a very strong bias in amino acid composition with a prevalence of the hydrophobic amino acid residues Leu, Ile, Val, Ala, Phe, and Trp. The most conserved position was the third, which was most commonly occupied by leucine. Therefore, it was presumable that the obstruction of leucine biosynthesis may lead to the failure of short peptide synthesis in the environment containing erythromycin. However, the role of Cj1199 on the formation of C-peptides was not verified. No difference of the MIC between ΦCj1199 and NCTC11168 was observed in our study. Considering the limited sensitivity of MIC determination by broth microdilution method, the erythromycin susceptibility was detected by FCM, the result indicated that the tolerance of ΦCj1199 to erythromycin was decreased under the treatment by a series concentration of erythromycin. Additionally, ΦCj1199 showed remarkable disadvantage on the development of erythromycin resistance in vitro step-wise selection. Therefore, Cj1199 may not directly impact on the erythromycin resistance, but it can contribute to the initial survival rate of the bacteria during erythromycin treatment and may increase the chances of acquiring resistance.
Conclusion
ΦCj1199 exhibited a weak competition of growth in a variety of competitive environments, especially in the medium with erythromycin. ΦCj1199 possessed lower risk for development of erythromycin resistance. Based on functions of differentially expressed genes, Cj1199 has an impact on the biosynthesis of leucine and utilization of amino acid in gut, and thus may play an important role in the synthesis of short peptide and in the provision of energy and carbon. The genotype and phenotype change of ΦCj1199 indicates that Cj1199 may contribute to the initial survival and colonization of the C. jejuni under the antibiotic treatment. Cj1199 may impact the resistance acquirement or resistance development in a long term but not in a short term of growth. The role of Cj1199 on the development of erythromycin resistance is indirectly dependent on the regulation of the expression of genes involved in amino acid biosynthesis and utilization. This study widened our understanding on the molecular mechanism of resistance and provides scientific reference for drug research and application.
Ethical Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Author Contributions
Conceived and designed the experiments: FL, HH, MD, XW, and ZY. Performed the experiments: FL, HH, YW, and YS. Analyzed the data: FL, HH, MD, JH, SF, LH, and ZY. Contributed reagents/materials/analysis tools: ZY, MD, HH, and ZL. Wrote the paper: HH, FL, JH, SF, and ZY.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks the curious review by Dr. Huizhong Chen and Kidon Sung from FDA National Center for Toxicological Research.
Footnotes
Funding. This work was supported by National Basic Research Program of China (2013CB127200), National Key research and development program of China (2016YFD0501302), Morning program of Wuhan in China (2015070404010191), Fundamental Research Funds for the Central Universities (2662015PY035), National Natural Science Foundation of China (31101856), National Key Technology R&D Program (2012BAK01B00), and National Program for Risk Assessment of Quality and Safety of Livestock and Poultry Products (GJFP2016008). The funders had no role in study design, data analysis, decision to publish, or preparation of the manuscript. The opinions expressed in this manuscript are solely the responsibility of the authors and do not necessarily represent the official views and policy of the US Food and Drug Administration. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration.
References
- Bedekovics T., Li H., Gajdos G. B., Isaya G. (2011). Leucine biosynthesis regulates cytoplasmic iron-sulfur enzyme biogenesis in an Atm1p-independent manner. J. Biol. Chem. 286 40878–40888. 10.1074/jbc.M111.270082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau J. M., Zhao X., Hansen G., Drlica K. (2001). Mutant prevention concentrations of fluoroquinolones for clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45 433–438. 10.1128/AAC.45.2.433-438.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzler J. P. (2004). Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect 10 868–876. 10.1111/j.1469-0691.2004.00983.x [DOI] [PubMed] [Google Scholar]
- Chaudhuri R. R., Yu L., Kanji A., Perkins T. T., Gardner P. P., Choudhary J., et al. (2011). Quantitative RNA-seq analysis of the Campylobacter jejuni transcriptome. Microbiology 157 2922–2932. 10.1099/mic.0.050278-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. C., Ghole M., Majumder A., Wang Z., Chandana S., Wu H. Y. (2003). LeuO-mediated transcriptional derepression. J. Biol. Chem. 278 38094–38103. 10.1074/jbc.M300461200 [DOI] [PubMed] [Google Scholar]
- CLSI (2006). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Dai Z. L., Li X. L., Xi P. B., Zhang J., Wu G., Zhu W. Y. (2012). Regulatory role for L-arginine in the utilization of amino acids by pig small-intestinal bacteria. Amino Acids 43 233–244. 10.1007/s00726-011-1067-z [DOI] [PubMed] [Google Scholar]
- Dai Z. L., Li X. L., Xi P. B., Zhang J., Wu G., Zhu W. Y. (2013). L: -Glutamine regulates amino acid utilization by intestinal bacteria. Amino Acids 45 501–512. 10.1007/s00726-012-1264-4 [DOI] [PubMed] [Google Scholar]
- Dam M., Douthwaite S., Tenson T., Mankin A. S. (1996). Mutations in domain II of 23 S rRNA facilitate translation of a 23 S rRNA-encoded pentapeptide conferring erythromycin resistance. J. Mol. Biol. 259 1–6. 10.1006/jmbi.1996.0296 [DOI] [PubMed] [Google Scholar]
- EFSA (2009). Joint Opinion on antimicrobial resistance (AMR) focused on zoonotic infections. Scientific opinion of the European centre for disease prevention and control; scientific opinion of the panel on biological hazards; opinion of the committee for medicinal products for veterinary use; scientific opinion of the scientific committee on emerging and newly identified health risks. EFSA J. 7 1–78. [Google Scholar]
- Gibreel A., Kos V. N., Keelan M., Trieber C. A., Levesque S., Michaud S., et al. (2005). Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob. Agents Chemother. 49 2753–2759. 10.1128/AAC.49.7.2753-2759.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Dai M., Wang Y., Chen D., Yuan Z. (2010). Quantification of mutated alleles of 23S rRNA in macrolide-resistant Campylobacter by TaqMan real-time polymerase chain reaction. Foodborne pathog. Dis. 7 43–49. 10.1089/fpd.2009.0339 [DOI] [PubMed] [Google Scholar]
- Hao H., Yuan Z., Shen Z., Han J., Sahin O., Liu P., et al. (2013). Mutational and transcriptomic changes involved in the development of macrolide resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 57 1369–1378. 10.1128/AAC.01927-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Frye J. G., Strobaugh T. P., Chen C. Y. (2008). Analysis of AI-2/LuxS-dependent transcription in Campylobacter jejuni strain 81-176. Foodborne Pathog. Dis. 5 399–415. 10.1089/fpd.2008.0106 [DOI] [PubMed] [Google Scholar]
- Hughes R. A., Cornblath D. R. (2005). Guillain-barre syndrome. Lancet 366 1653–1666. 10.1016/S0140-6736(05)67665-9 [DOI] [PubMed] [Google Scholar]
- ICC (1992). Enumeration of Bacteria (Spatula Method). Available at: https://www.icc.or.at/standard_methods/147 [Google Scholar]
- Jeon B., Wang Y., Hao H., Barton Y. W., Zhang Q. (2010). Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J. Antimicrob. Chemother. 66 79–85. 10.1093/jac/dkq418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush N. O., Miller W. G., De Reuse H., Mendz G. L. (2007). Oxygen requirement and tolerance of Campylobacter jejuni. Res. Microbiol. 158 644–650. 10.1016/j.resmic.2007.07.009 [DOI] [PubMed] [Google Scholar]
- Kervella M., Pages J. M., Pei Z., Grollier G., Blaser M. J., Fauchere J. L. (1993). Isolation and characterization of two Campylobacter glycine-extracted proteins that bind to HeLa cell membranes. Infect. Immun. 61 3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaw G. B. (2003). Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67 1–15. 10.1128/MMBR.67.1.1-15.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labigne A., Courcoux P., Tompkins L. (1992). Cloning of Campylobacter jejuni genes required for leucine biosynthesis, and construction of leu-negative mutant of C. jejuni by shuttle transposon mutagenesis. Res. Microbiol. 143 15–26. 10.1016/0923-2508(92)90030-R [DOI] [PubMed] [Google Scholar]
- Leon-Kempis Mdel R., Guccione E., Mulholland F., Williamson M. P., Kelly D. J. (2006). The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60 1262–1275. 10.1111/j.1365-2958.2006.05168.x [DOI] [PubMed] [Google Scholar]
- Lin J., Michel L. O., Zhang Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46 2124–2131. 10.1128/AAC.46.7.2124-2131.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H. A., Pedraza-Reyes M., Yasbin R. E., Robleto E. A. (2011). Transcriptional de-repression and Mfd are mutagenic in stressed Bacillus subtilis cells. J. Mol. Microbiol. Biotechnol. 21 45–58. 10.1159/000332751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Ito Y., Tsuru S., Ying B. W., Yomo T. (2011). Bacterial cells carrying synthetic dual-function operon survived starvation. J. Biomed. Biotechnol. 2011:489265 10.1155/2011/489265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A., Leon-Kempis Mdel R., Dodson E., Wilson K. S., Wilkinson A. J., Kelly D. J. (2007). A bacterial virulence factor with a dual role as an adhesin and a solute-binding protein: the crystal structure at 1.5 A resolution of the PEB1a protein from the food-borne human pathogen Campylobacter jejuni. J. Mol. Biol. 372 160–171. 10.1016/j.jmb.2007.06.041 [DOI] [PubMed] [Google Scholar]
- Pei Z., Blaser M. J. (1993). PEB1 the major cell-binding factor of Campylobacter jejuni, is a homolog of the binding component in gram-negative nutrient transport systems. J. Biol. Chem. 268 18717–18725. [PubMed] [Google Scholar]
- Pei Z., Burucoa C., Grignon B., Baqar S., Huang X. Z., Kopecko D. J., et al. (1998). Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66 938–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. R., Liao J. C. (2011). A synthetic iterative pathway for ketoacid elongation. Methods Enzymol. 497 469–481. 10.1016/B978-0-12-385075-1.00020-2 [DOI] [PubMed] [Google Scholar]
- Sycheva E. V., Iampol’skaia T. A., Preobrazhenskaia E. S., Novikova A. E., Matrosov N. G., Stoinova N. V. (2007). [Overproduction of noncanonical amino acids by Escherichia coli cells]. Mikrobiologiia 76 805–812. [PubMed] [Google Scholar]
- Tamakoshi M., Yamagishi A., Oshima T. (1998). The organization of the leuC, leuD and leuB genes of the extreme thermophile Thermus thermophilus. Gene 222 125–132. 10.1016/S0378-1119(98)00482-X [DOI] [PubMed] [Google Scholar]
- Taylor D. E., Ge Z., Purych D., Lo T., Hiratsuka K. (1997). Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob. Agents Chemother. 41 2621–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T., DeBlasio A., Mankin A. (1996). A functional peptide encoded in the Escherichia coli 23S rRNA. Proc. Natl. Acad. Sci. U.S.A. 93 5641–5646. 10.1073/pnas.93.11.5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T., Mankin A. (1995). Comparison of functional peptide encoded in the Escherichia coli 23S rRNA with other peptides involved in cis-regulation of translation. Biochem. Cell Biol. 73 1061–1070. 10.1139/o95-114 [DOI] [PubMed] [Google Scholar]
- Tenson T., Mankin A. S. (2001). Short peptides conferring resistance to macrolide antibiotics. Peptides 22 1661–1668. 10.1016/S0196-9781(01)00501-0 [DOI] [PubMed] [Google Scholar]
- Tenson T., Xiong L., Kloss P., Mankin A. S. (1997). Erythromycin resistance peptides selected from random peptide libraries. J. Biol. Chem. 272 17425–17430. 10.1074/jbc.272.28.17425 [DOI] [PubMed] [Google Scholar]
- Tripathi S., Kloss P. S., Mankin A. S. (1998). Ketolide resistance conferred by short peptides. J. Biol. Chem. 273 20073–20077. 10.1074/jbc.273.32.20073 [DOI] [PubMed] [Google Scholar]
- Vimberg V., Xiong L., Bailey M., Tenson T., Mankin A. (2004). Peptide-mediated macrolide resistance reveals possible specific interactions in the nascent peptide exit tunnel. Mol. Microbiol. 54 376–385. 10.1111/j.1365-2958.2004.04290.x [DOI] [PubMed] [Google Scholar]
- Weisblum B. (1995). Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39 577–585. 10.1128/AAC.39.3.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Fang M. (2003). DNA supercoiling and transcription control: a model from the study of suppression of the leu-500 mutation in Salmonella typhimurium topA- strains. Prog. Nucleic Acid Res. Mol. Biol. 73 43–68. 10.1016/S0079-6603(03)01002-X [DOI] [PubMed] [Google Scholar]
- Xia Q., Muraoka W. T., Shen Z., Sahin O., Wang H., Wu Z., et al. (2013). Adaptive mechanisms of Campylobacter jejuni to erythromycin treatment. BMC Microbiol. 13:133 10.1186/1471-2180-13-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yindeeyoungyeon W., Likitvivatanavong S., Palittapongarnpim P. (2009). Characterization of alpha-isopropylmalate synthases containing different copy numbers of tandem repeats in Mycobacterium tuberculosis. BMC Microbiol. 9: 122 10.1186/1471-2180-9-122 [DOI] [PMC free article] [PubMed] [Google Scholar]


