Abstract
miR169 is a conserved microRNA (miRNA) family involved in plant development and stress-induced responses. However, how miR169 functions in rice immunity remains unclear. Here, we show that miR169 acts as a negative regulator in rice immunity against the blast fungus Magnaporthe oryzae by repressing the expression of nuclear factor Y-A (NF-YA) genes. The accumulation of miR169 was significantly increased in a susceptible accession but slightly fluctuated in a resistant accession upon M. oryzae infection. Consistently, the transgenic lines overexpressing miR169a became hyper-susceptible to different M. oryzae strains associated with reduced expression of defense-related genes and lack of hydrogen peroxide accumulation at the infection site. Consequently, the expression of its target genes, the NF-YA family members, was down-regulated by the overexpression of miR169a at either transcriptional or translational level. On the contrary, overexpression of a target mimicry that acts as a sponge to trap miR169a led to enhanced resistance to M. oryzae. In addition, three of miR169’s target genes were also differentially up-regulated in the resistant accession upon M. oryzae infection. Taken together, our data indicate that miR169 negatively regulates rice immunity against M. oryzae by differentially repressing its target genes and provide the potential to engineer rice blast resistance via a miRNA.
Keywords: microRNA, miR169, nuclear factor Y-A, target mimicry, rice blast
Introduction
Small RNAs are a type of short non-coding RNAs involved in regulation of gene expression either by chromatin methyl modification or by mRNA cleavage or/and translation inhibition (Baulcombe, 2004). Based on the difference of origin and function, small RNAs are classified into microRNAs (miRNAs) and small interfering RNAs (siRNAs). Both miRNAs and siRNAs are involved in the regulation of diverse biological processes, including growth, development and responses to biotic and abiotic stresses (Katiyar-Agarwal and Jin, 2010; Chen, 2012; Khraiwesh et al., 2012). Currently, more than 28000 miRNAs are listed in miRNA database1. Increasing evidence indicates that miRNAs are involved in fine-tuning plant immunity against pathogen invasion (Padmanabhan et al., 2009; Katiyar-Agarwal and Jin, 2010). The first identified resistance-related miRNA is miR393 in Arabidopsis, which can be induced by the pathogen-associated molecular pattern (PAMP) peptide flg22 and positively contributes to resistance against avirulent Pseudomonas syringae DC3000 by silencing auxin receptors to suppress auxin signaling (Navarro et al., 2006). miR160a was functionally characterized as positive regulator, whereas, miR398b and miR773 as negative regulators of plant PAMP-triggered immunity (PTI) in Arabidopsis (Li Y. et al., 2010). In addition, several miRNA families act as resistance regulators to direct the silence of nucleotide-binding leucine-rich-repeat (NB-LRR) type disease resistance (R) genes in plants (Baldrich and San Segundo, 2016). For example, nta-miR6019 and nta-miR6020 in tobacco guide the cleavage of transcripts of the Toll and Interleukin-1 receptor-NB-LRR immune receptor N that confers resistance to tobacco mosaic virus (TMV) (Li et al., 2012). In potato, three highly abundant miRNA families, miR1507, miR2109 and miR2118, target the conserved sequences in the transcripts of NB-LRRs, and trigger the production of trans-acting siRNAs, most of which are matched to over 60% of the NB-LRR-encoding genes in potato (Zhai et al., 2011). In tomato, miR482 targets the coiled-coil (CC)-NB-LRR-encoding genes and causes production of secondary siRNAs, which target other defense-related genes (Shivaprasad et al., 2012). In addition, miR482f and miR5300 also mediate silencing of the expression of resistance-related NB domain genes, but both the miRNAs are repressed in resistant cultivars during infection of the fungus Fusarium oxysporum f. sp. lycopersici, which causes vascular wilt disease in tomato (Ouyang et al., 2014). In rice, many miRNAs are also proposed to be involved in regulation of immunity against Magnaporthe oryzae, although the mechanism is largely unknown (Li et al., 2014, 2016).
Rice is the staple food for more than half of the world’s population (Liu et al., 2014). Rice blast caused by the fungal pathogen M. oryzae is one of the most devastating diseases threatening rice production worldwide. Understanding the mechanism of resistance to this disease can underpin disease control and many breakthroughs have been achieved in the past decades. On one hand, it is well-known that rice mounts two-layered innate immunity to defend against the invasion of M. oryzae. The first layer of innate immunity is activated upon recognition of PAMPs, such as chitin, by the cell-surface located pattern recognition receptors (PRRs), such as CEBiP, OsCERK1, LYP4 and LYP6, which is known as PTI (Kaku et al., 2006; Shimizu et al., 2010; Liu et al., 2012). This layer of immunity is suppressed by effector proteins of M. oryzae, some of which are recognized by R proteins, leading to the second layer of immunity, called effector-triggered immunity (ETI) (Liu et al., 2014). To date, more than 25 R genes have been functionally characterized and some of them are widely exploited in rice blast-resistant breeding and production2 (Liu et al., 2014; Ma et al., 2015). On the other hand, increasing reports indicate that miRNA signaling pathway is involved in rice immunity against M. oryzae and miRNAs can act as either positive or negative regulators to fine-tune PTI and ETI. For example, silencing OsDCL1, the key gene for miRNA biogenesis, results in enhancement of resistance to virulent rice blast strains due to the constitutive expression of 13 pathogenesis-related (PR) genes and two PTI-related genes OsKS4 and OsNAC4 (Zhang et al., 2015). Recently, nine new miRNAs were identified by deep sequencing of small RNA libraries derived from leaves treated with M. oryzae elicitors (Baldrich et al., 2015). In addition, the M. oryzae elicitor-responsive miRNA, Osa-miR7695, was identified as a positive regulator for rice resistance against M. oryzae by down-regulating the expression of OsNramp6 (Natural resistance-associated macrophage protein 6) (Campo et al., 2013).
In a previous study, we identified more than 30 miRNAs that are differentially responsive to M. oryzae infection by comparing deep sequencing data of small RNA libraries from resistant and susceptible accessions (Li et al., 2014). Among these miRNAs, miR169 family members were differentially accumulated in the resistant accession IRBLkm-Ts and the susceptible accession Lijiang xin Tuan Heigu (LTH) upon M. oryzae infection (Li et al., 2014). While the accumulation of miR169a and miR169b/c were increased in both accessions upon M. oryzae infection, miR169f/g and miR169h/i/j/k/l/m were increased in LTH but decreased in IRBLkm-Ts. Therefore, it is intriguing to investigate the integrative role of miR169 because all the miR169 isoforms share high sequence identity and target to the same batch of genes encoding nuclear transcription factor Y (NF-Y) (Wu et al., 2009; Li Y. F. et al., 2010; Zhou et al., 2010).
Nuclear transcription factor Y is a family of transcription factors specifically binding the CCAAT-box in the promoters of eukaryotic genes via heterotrimer comprised of three subunits: NF-YA, NF-YB and NF-YC (Mantovani, 1999). In plants, each subunit of NF-Ys is respectively encoded by a group of genes. For example, there are 11 NF-YA, 11 NF-YB and seven NF-YC in rice and 10 NF-YA, 10 NF-YB, and 10 NF-YC in Arabidopsis (Petroni et al., 2012), resulting in the formation of a flexible and complicated transcription factor system that may regulate plant responses to different environmental conditions. In rice, eight of the 11 NF-YA genes are identified to be the authentic targets of miR169, including from NF-YA1 to NF-YA6, NF-YA10 and NF-YA11 (Wu et al., 2009; Li Y. F. et al., 2010; Zhou et al., 2010; Petroni et al., 2012). In fact, miR169 is a big microRNA family containing 17 known members representing nine different mature isoforms in rice (Zhao et al., 2007). Among all the isoforms, only miR169g is drought-induced, whereas, both miR169g and miR169n/o are inducible by high salinity, leading to down-regulation of the transcripts of NF-YA2 (Os03g29760) (Zhao et al., 2007, 2009). In Arabidopsis, miR169/NF-YA module is linked with drought stress (Li et al., 2008), nitrogen (N) stress (Zhao et al., 2011; Liang et al., 2012), and associated with carbohydrate metabolism and cell expansion (Leyva-Gonzalez et al., 2012). In alfalfa, miR169/NF-YA module is involved in regulating symbiotic nitrogen fixation (SNF) process (Kaur et al., 2014) and freezing tolerance (Shu et al., 2016). In maize, the expression of Zma-miR169 and its target genes ZmNF-YAs is conversely responsive to drought, salt, and hormone stresses (Luan et al., 2014, 2015). In wheat, the expression of miR169 is reduced, whereas the expression of NF-YAs, are induced by N and P starvation (Qu et al., 2015). In rice, OsNF-YA7 is induced by drought stress and the transgenic plants overexpressing OsNF-YA7 exhibit ABA-independent tolerance to drought stress (Lee et al., 2015). These literatures indicate that miR169/NF-YA modules regulate tolerance to abiotic stresses in both monocots and dicots.
Recently, emerging evidence indicates that miR169/NF-YA modules also play roles in regulation of plant responses to biotic stresses. For example, in Arabidopsis, overexpression of miR169 abrogates the resistance phenotypes of clv1 and clv2, the mutants of the LRR-receptor-like kinases CLAVATA1 and CLAVATA2, to the bacterial wilt pathogen Ralstonia solanacearum via the suppression on NF-YA expressions (Hanemian et al., 2016). In rice, different miR169 isoforms are differentially accumulated in the resistant and susceptible accessions upon M. oryzae infection (Li et al., 2014, 2016), and are differentially responsive to elicitor treatment (Campo et al., 2013; Baldrich et al., 2015), indicating the involvement of miR169/NF-YA modules in rice responses to M. oryzae. However, it is unclear what roles the miR169/NF-YA regulation modules play in rice immunity against M. oryzae.
To figure out the integrative role of miR169 in rice immunity against the blast fungus, we first examined the abundance of different miR169 isoforms in the susceptible accession LTH and the resistant accession IRBL9-W upon M. oryzae infection. Then we constructed transgenic rice plants overexpressing miR169a and its target mimicry, respectively, and examined blast disease phenotypes of the transgenic lines. By examining the expression of miR169 target genes in transgenic lines and in susceptible/resistant accessions, we identified candidate NF-YA genes that might act as positive regulators for rice immunity. Taken together, our data demonstrate that miR169 acts as a negative regulator for rice immunity against M. oryzae.
Materials and Methods
Plant Materials and Growth Conditions
Rice (Oryza sativa) plants used in this study include the susceptible accession LTH, the resistant accession IRBL9-W, the japonica accession Taipei 309 (TP309) and its transgenic lines overexpressing miR169a, and the indica accession Kasalath and its transgenic lines overexpressing mimicry of miR169a (MIM169a). All rice plants were grown in a growth room maintained at 26°C and 70% relative humidity with a 14/10-h day/night regime. Nicotiana benthamiana plants were planted at 22°C with a 16/8 h light/dark photoperiod in a growth room and used for agro-infiltration experiments.
Plasmid Construction and Genetic Transformation
To generate miR169a overexpressing transgenic plants, its genomic sequence containing 421 bp upstream and 500 bp downstream sequences was amplified from Nipponbare (NPB) DNA with primers OsmiR169a-F and OsmiR169a-R (Supplementary Table S2), and the amplified fragment was cloned into KpnI-SalI sites of the binary vector 35S-pCAMBIA1300, resulting in the overexpression construct p35S:miR169a. To make artificial target mimicry construct, we exploited the Arabidopsis gene IPS1 (INDUCED BY PHOSPHATE STARVATION1) that contains sequences complement to miR399 with a mismatched loop at the expected miRNA cleavage site (Franco-Zorrilla et al., 2007). Artificial target mimicry sequences of miR169a were inserted into the IPS1 to replace the miR399 target site with primers miR169-IPS1-F, miR169-IPS1-R, miR169mimic-F, and miR169mimic-R (Supplementary Table S2) as described previously (Franco-Zorrilla et al., 2007), and cloned into KpnI-SpeI sites of binary vector 35S-pCAMBIA1300, resulting in the overexpression construct p35S:MIM169a. Construct p35S:miR169a was transformed into TP309 and construct p35S:MIM169a was transformed into Kasalath via Agrobacterium tumefaciens (strain GV3101)-mediated transformation and the transgenic plants were screened as previously described (Li et al., 2014). To express the YFP-tagged 3′-UTRs of target genes, the 3′-UTRs of Os03g29760 (NF-YA2), Os03g44540 (NF-YA3), and Os03g48970 (NF-YA4) containing the target site of miR169 were amplified from Nipponbare (NPB) cDNAs using gene-specific primers (Supplementary Table S2). The isolated fragments were then fused to the C-terminus of YFP and inserted into KpnI-SpeI sites of binary vector 35S-pCAMBIA1300. Agrobacterium strain GV3101 was used for agroinfection assay in N. benthamiana (Huang et al., 2014).
Pathogen Infection and Microscopy Analysis
Three M. oryzae strains, Y34, Guy11 and eGFP-tagged Zhong8-10-14 (GZ8), were used in this study. M. oryzae strains were cultured in complete medium at 28°C with 12-h/12-h light/dark cycles for sporulation. After 2 weeks, spores were collected and the inoculum concentration was adjusted to 5 × 105 spores mL–1 for spray inoculation on three-leaf-stage plants (Qu et al., 2006). T3 seedlings of miR169a transgenic plants were used for spray inoculation. Disease phenotypes on the leaf two were recorded at 5 days post inoculation (dpi). The blast disease lesions were examined and classified into six types (0–5 type; 0–2 type is classified as resistant phenotype, and 3–5 type is classified as susceptible phenotype) based on the sporulation rate on the lesions in infected rice leaves following a previous report (Bonman et al., 1986). In brief, 0 = no disease lesions observed; 1 = small pinpoint-like disease lesions between two small vascular bundles; 2 = lesions with diameter 0.5–1 mm and develop over the two small vascular bundles but do not reach the big vascular bundles; 3 = disease lesions with diameter about 1–3 mm and develop between the two big vascular bundles; 4 = disease lesions with diameter about 3–4 mm and develop over the two big vascular bundles; 5 = disease lesions with diameter over 4 mm and develop over the main vein. For evaluation of the transgenic lines expressing the target mimicry of miR169a, 8-week old T0 plants were inoculated following the drop inoculation method (Park et al., 2012), and the negative transgenic plants were used as control. In brief, leaves from 8-week-old plants were slightly wounded with a mouse ear punch, and 5 μL of spore suspension (5 × 105 spores mL–1) was added to the wound. Lesions were measured at 5 dpi. Relative fungal mass was calculated using the DNA level of M. oryzae Pot2 against the rice genomic ubiquitin DNA level by qPCR (Park et al., 2012). To observe the infection process of M. oryzae, the eGFP-tagged M. oryzae strain GZ8 was inoculated on 5-cm-long leaf sheaths as described (Kankanala et al., 2007). The inoculated epidermal layer was excised and analyzed by Laser Scanning Confocal Microscopy (Nikon A1) at 12 and 36 hpi, respectively.
For examining H2O2 accumulation in infected rice leaves, we followed the procedure published previously by Xiao et al. (2003). DAB and trypan blue were used to stain H2O2 and fungal structures, respectively. Images were acquired with a microscope (Zeiss imager A2).
Quantitative Reverse Transcription PCR Assay
Three-leaf-stage plants were inoculated with M. oryzae by spraying spore suspensions at a concentration of 5 × 105 spores mL–1, and samples were collected at 0, 6, 12, 24, and 48 hpi. TP309 is used as the control of miR169a transgenic lines. For examining the target gene expression in transgenic lines, negative transgenic lines were used as the control of MIM169a transgenic lines. Total RNA was extracted from collected samples using TRIzol reagent (Invitrogen) and was reverse transcribed to cDNA using the SuperScript first-strand synthesis system primed by oligo dT (Invitrogen). To test the expression of miRNAs, NCode miRNA first-strand cDNA module (Invitrogen) was selected to elongate and reverse transcribe miRNA following the manufacturer’s instructions. Quantitative RT-PCR was performed using isoform-specific primers (Supplementary Table S2) and SYBR Green mix (TaKaRa). snRNA U6 served as internal reference for the detection of miRNAs in quantitative RT-PCR. The rice ubiquitin (UBQ) gene was selected as an internal reference for data normalization. The data were determined by a one-way ANOVA followed by post hoc Tukey HSD analysis with significant differences (P < 0.01).
Agrobacterium-Mediated Transient Expression Assay in N. benthamiana
Agrobacterium strain GV3101 harboring the respective expression constructs in the binary vector pCAMBIA1300 was incubated at 28°C overnight in LB media containing rifampin (50 μg/mL), kanamycin (50 μg/mL), and carbenicillin (50 μg/mL) at a 250 r/min shaking table. The bacteria were collected at 3000 rpm for 5 min and resuspended in an MMA buffer (10 mM MES, 10 mM MgCl2, 100 μM AS), respectively. The Agrobacteria harboring the expression constructs were infiltrated into leaves of N. benthamiana for transient expression assay. Leaves were examined between 36 and 80 hpi for image acquisition using a Nikon A1 Confocal Laser Scanning Microscope (Nikon Instruments, Inc., Chengdu, China) as previously described (Huang et al., 2014). Western blotting analyses were performed following a previous protocol (Chen et al., 2008). In brief, 15 micrograms of total protein were electrophoresed on 10% SDS-PAGE gel, and the protein blot was reacted with anti-GFP sera (BBI Life Science) to detect and determine YFP accumulation.
Results
Different miR169 Isoforms Are Differentially Accumulated in the Susceptible and Resistant Accessions
To address the question of how different miR169 isoforms are differentially responsive to M. oryzae infection, we examined the accumulation of the five most abundant miR169 isoforms in the susceptible accession LTH and the monogenic resistant accession IRBL9-W that contains the resistance (R) gene Pi-9 (Tsumematsu et al., 2000) and confers high resistance to M. oryzae (Khanna et al., 2015). LTH is widely used as a susceptible reference in blast inoculation experiments for it is susceptible to all M. oryzae strains ever tested (Lin et al., 2001). We first confirmed the disease phenotypes by inoculating the virulent strain Guy11 on three-leaf-stage seedlings. While large disease lesions were observed in LTH, small lesions were occasionally found in IRBL9-W (Figure 1A), indicating susceptibility and resistance, respectively. We next performed a time-course assay to examine the accumulation of miR169 isoforms in LTH and IRBL9-W by miRNA quantitative RT-PCR. According to the small RNA deep sequencing data in a previous report (Li et al., 2014), the accumulation of miR169a, miR169b/c, miR169f.1/g, miR169h/i/j/k/l/m, and miR169n/o is mostly abundant and thus was examined in this study, whereas, the accumulation of miR169d, miR169e, miR169p, and miR169q is rarely or not detected (below 50 reads or no reads per one million total reads) and thus not included in this study. Our data confirmed that the abundance of different miR169 isoforms was quite different, with the least abundant being for miR169f/g and miR169n/o, the most abundance being for miR169b/c, and the middle being for miR169a and miR169h/i/j/k/l/m (Figures 1B,C). Moreover, the accumulation pattern of miR169b/c was quite similar to the sum accumulation pattern of miR169 (Figure 1D). It is intriguing that the accumulation of all the tested miR169 isoforms was increased in LTH upon M. oryzae infection, whereas, the accumulation of miR169b/c was quite stable in IRBL9-W, although the accumulation of the other miR169 isoforms was also increased at certain time points (Figures 1B,C). However, the high abundance of miR169b/c might mask the fluctuation of the other isoforms so that its pattern is similar to the sum of miR169 (Figure 1D). Particularly, the total accumulation of all miR169 isoforms was highly up-regulated upon M. oryzae inoculation in LTH, but significantly down-regulated at 12 hpi, up-regulated at 24 hpi and back to the background level at 48 hpi in IRBL9-W (Figure 1D), which was consistent with the total reads in LTH and IRBLKm-Ts (in Supplementary Table S1 of Li et al., 2014). These data imply that miR169 may integratively act as a negative regulator in rice immunity against M. oryzae.
FIGURE 1.
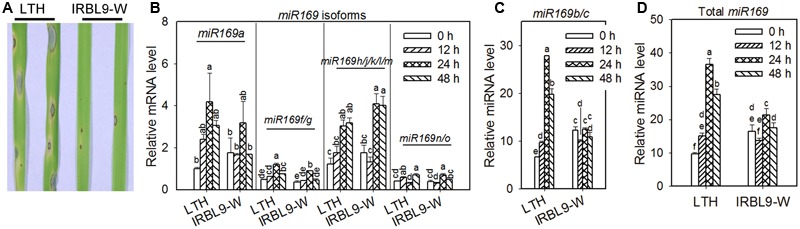
Differential accumulation of miR169 in susceptible and resistant accessions upon Magnaporthe oryzae infection. (A) Representative leaf sections from the susceptible accession LTH and the resistant accession IRBL9-W show the blast disease phenotype. Three-leaf-stage seedlings were inoculated with Guy11 spore suspensions, and disease phenotypes were recorded at 5 dpi. Similar results were obtained in at least two independent experiments. (B,C) Differential accumulation of different miR169 isoforms in the indicated accessions upon M. oryzae infection. RNA was extracted at the indicated time points for qRT-PCR analysis. The transcriptional level of miR169 family members was normalized to miR169a in untreated LTH (0 h). SnRNA U6 served as an internal reference. (D) The sum accumulation of total miR169 in the indicated accessions upon M. oryzae infection. Values in (B–D) are means of three replications. Error bars indicate SD. The letters above the bars indicate significant differences (P < 0.01) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Similar results were obtained in at least two independent experiments.
Overexpressing miR169a Leads to Enhanced Susceptibility to M. oryzae
All miR169 isoforms target the same eight NF-YA genes (Wu et al., 2009; Li Y. F. et al., 2010; Zhou et al., 2010) because of their high sequence identity (Supplementary Figure S1). Thus, it is feasible to investigate the integrative role of all miR169 isoforms in rice immunity against M. oryzae by over expressing one of the miR169 isoforms, although the authentic miR169-NF-YA regulation module could be much more complicated due to different miR169 isoform has different expression pattern and is differentially responsive to different stimuli (Zhao et al., 2007, 2009). Following this scenario, we constructed rice transgenic lines overexpressing miR169a within the TP309 background. Two independent transgenic lines with high miR169a accumulation were identified and used for phenotypic analyses (Figure 2A). Three-leaf-stage seedlings were separately spray-inoculated with three M. oryzae strains with different virulence. While Y34 is an incompatible strain to TP309, Guy11 and the enhanced GFP-tagged Zhong8-10-14 (GZ8) are virulent strains. Disease phenotypes were recorded at 5 dpi. All the three strains formed more and larger disease lesions in the transgenic lines than those in the control plants (Figures 2B–D), indicating enhanced susceptibility by overexpression of miR169a. Then, the composition of lesion types was quantitatively analyzed. While the Y34-infected lesions in the control plants were mostly scored as types 1–2 (resistant phenotype), the lesions in the transgenic lines overexpressing miR169a were scored as types 3–5 (susceptible phenotype, Figure 2B), indicating that overexpression of miR169a converted the partial resistance into fully susceptibility to Y34. Moreover, the susceptible lesion types (3–5) of GZ8 and Guy11 in the control plants were all exacerbated in the transgenic lines overexpressing miR169a. The percentage of susceptible lesion types (types 3–4) of GZ8 was increased from 50% in the control plants to 80% in the transgenic lines (Figure 2C). Guy11-infected susceptible lesion types accounting for more than 80% in the control plants were exacerbated into 100% in the transgenic lines overexpressing miR169a, in which type 5 was increased from 10% in the control plants to more than 50% in the transgenic lines overexpressing miR169a (Figure 2D). These data demonstrate that overexpressing miR169a makes the susceptibility of TP309 to these strains into hyper-susceptibility.
FIGURE 2.
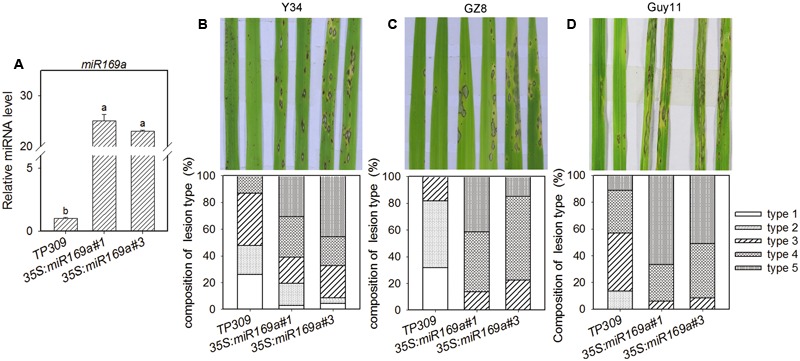
Overexpressing miR169a results in enhanced susceptibility to M. oryzae. (A) Comparison of miR169a accumulation in the transgenic lines harboring 35S:miR169a. SnRNA U6 served as the internal reference. The accumulation level of miR169a was normalized to that of the control plants. Values are means of three replications. Error bars indicate SD. The letters above the bars indicate significant differences (P < 0.01) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Similar results were obtained in at least two independent experiments. Comparison of disease phenotypes and composition of disease lesion types caused by the M. oryzae strain Y34 (B), GZ8 (C) and Guy11 (D), respectively. Lesions in 30 diseased leaves were analyzed at 5 dpi. Similar results were obtained in at least two independent experiments.
Overexpressing miR169a Results in Reduced Defense Responses
To explain why overexpression of miR169a leads to enhanced susceptibility, we examined typical defense responses, including expression levels of defense-related genes, production of hydrogen peroxide (H2O2) and microscopic pathogenesis of M. oryzae. First, three-leaf-stage seedlings were inoculated with M. oryzae strains Guy11. Then, the expression levels of three defense-related genes were examined by quantitative RT-PCR, including the two defense marker genes, Oryza sativa pathogenesis-related protein10b (OsPR10b) (Yamaguchi et al., 2013) and Os04g10010 (Schwessinger et al., 2015), and OsNAC4, an earlier induced basal defense-related gene (Park et al., 2012). The three genes were significantly induced in the wild type at 6 hpi of M. oryzae (Figures 3A–C), in which OsNAC4 was peaked at 12 hpi, whereas OsPR10b and Os04g10010 were further increased along with the time points. On the contrary, the expression levels of these genes were much lower and the induced time was delayed in the transgenic lines, although their expressions were also induced (Figures 3A–C). Next, we examined the production of H2O2 upon inoculation of M. oryzae. Consistent with the reduced expression of defense-related genes, H2O2 was hardly detected in the infected sheath cells of transgenic lines at 24 hpi, while easily observed in the control plants at the same time point (Figure 3D). These data indicate that overexpression of miR169a compromises M. oryzae-related defense responses in rice plants.
FIGURE 3.
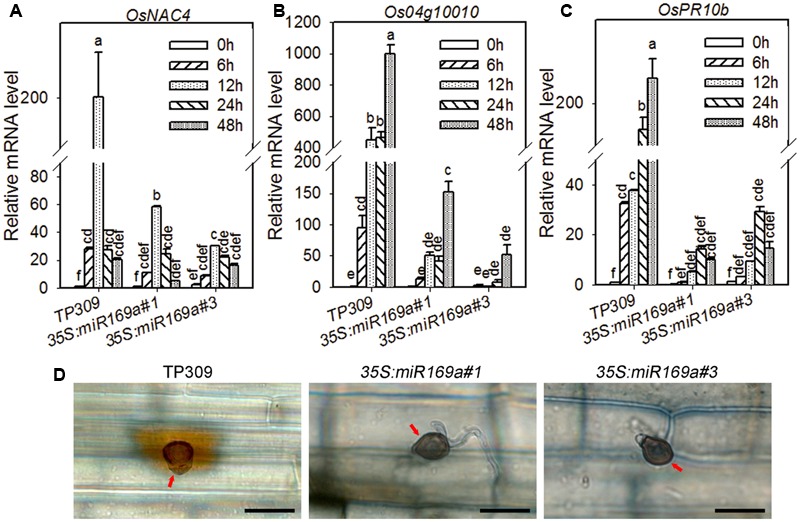
Overexpressing miR169a results in compromised defense responses. (A–C) Expression pattern of the indicated defense-related genes in the transgenic lines harboring 35S:miR169a and the control plants, respectively, upon M. oryzae infection. RNA was extracted at the indicated time points for qRT-PCR analysis. The indicated mRNA level was normalized to that in the untreated control plants (0 h). Values are means of three replications. Error bars indicate SD. The letters above the bars indicate significant differences (P < 0.01) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Similar results were obtained in at least two independent experiments. (D) Representative leaf sections from the indicated lines show H2O2 accumulation at 2 dpi, respectively. Note that there was no or trace amounts of H2O2 accumulation in the transgenic lines overexpressing miR169a, but high level of H2O2 was observed around the appressorium (arrow) in the leaf cell from the control plants. The fungal structure and accumulated H2O2 were stained by Trypan blue and DAB, respectively. Bars = 10 μm. Similar results were obtained in at least two independent experiments.
In addition, we compared the infection process of M. oryzae strain GZ8 on leaf sheath of the wild type and the transgenic lines by using Laser Scanning Confocal Microscopy. At 12 hpi, more than half of the inoculated spores germinated and formed appressoria on the sheath from the transgenic lines overexpressing miR169a, which was in contrast to no or seldom appressoria formation on the sheath from the control plants (Figures 4A,B). At 36 hpi, more than 70% spores formed the invasive hyphae in the transgenic lines, which was in contrast to about 30% invasive hyphae formed in the control plants (Figures 4A,B). These results indicate that overexpressing miR169 can compromise rice defense responses so as to facilitate M. oryzae invasion at the early infection stage.
FIGURE 4.
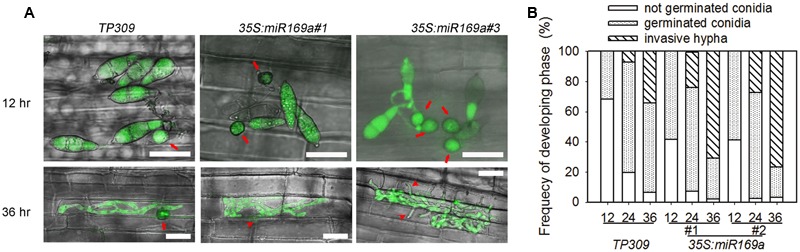
Overexpressing miR169a leads to increased M. oryzae growth. (A) Representative Laser Scanning Confocal Microscopy (LSCM) images show the growth of the M. oryzae strain GZ8 at 12 hours post inoculation (hpi) and 36 hpi on sheath cells of the indicated transgenic lines harboring 35S:miR169a and the control plants, respectively. Note that the appressoria (arrows) were formed at 12 hpi on miR169a-overexpression plants but delayed on control plants. Invasive hyphae were formed at 36 hpi on both control and miR169a-overexpressing plants, but only extended into the neighbor cells (arrowheads) of miR169a-overexpressing plants. Bars = 20 μm. (B) The quantitative analysis of M. oryzae growth. More than 200 conidia in each lines were analyzed. Similar results were obtained in at least two independent experiments.
miR169a Down-Regulates the Expression of Target Genes at Both Transcriptional and Translational Levels
All of the known target genes of miR169a encode NF-YA (Supplementary Table S1) (Wu et al., 2009; Li Y. F. et al., 2010; Zhou et al., 2010). As the role of miRNAs is to suppress target gene expression, we speculated that the expression of the target genes should be reduced in the lines overexpressing miR169a. To this end, we examined the mRNA levels of target genes in the lines overexpressing miR169a by quantitative RT-PCR. As anticipated, the transcription levels of six target genes, including Os02g53620 (NF-YA11), Os03g07880 (NF-YA1), Os03g44540 (NF-YA3), Os03g48970 (NF-YA4), Os07g41720 (NF-YA6) and Os12g42400 (NF-YA10), were repressed significantly in the transgenic lines; particularly, the expression of Os03g44540 (NF-YA3) and Os12g42400 (NF-YA10) was down-regulated to less than 20% of that in the control plants (Figure 5A). However, the transcription level of Os03g29760 (NF-YA2) was significantly increased in one line and did not change in the other line (Figure 5A), and the simplest explanation is that miR169a may repress its translation or that Os03g29760 (NF-YA2) is feedback regulated by its products. Then, we established an YFP-based reporter system to examine whether miR169a inhibits translation of Os03g29760. To this end, we made a construct expressing the Yellow Fluorescent protein (YFP) with the 3′-UTR of Os03g29760 as the terminator (YFP-3′-UTROs03g29760) because the target sites of miR169a on all of the eight NF-YA genes are located in the 3′-UTR (Supplementary Figure S2). Next, YFP-3′-UTROs03g29760 was separately expressed or co-expressed with miR169a in N. benthamiana and the protein levels were compared by both Western blotting analysis and examining the intensity of YFP. The results showed that when expressed alone, YFP-3′-UTROs03g29760 was highly accumulated (Figure 5B) and expressed in both the cytoplasm and the nucleus (Figure 5C). However, its expression level was obviously reduced when miR169a was co-expressed, particularly at higher OD value of the infiltrated Agrobacteria. By contrast, the co-expression of YFP that did not contain the 3′-UTR of Os03g29760 and miR169a did not affect the protein accumulation of YFP (Figures 5B,C). To further confirm the translation inhibition by miR169 on 3′-UTR of Os03g29760, we constructed the target mimicry of miR169a, MIM169a, which could act as a sponge to trap miR169a because of the insertion of three nucleotides between positions 10 and 11, and as a result, the cleavage on the formed double strand RNA was inhibited because of the loop formation at the mismatched nucleotides (Franco-Zorrilla et al., 2007). When YFP-3′-UTROs03g29760, miR169a and MIM169a were co-expressed, the protein accumulation of YFP-3′-UTROs03g29760 was obviously increased with the addition of MIM169a, indicating that the suppression on YFP-3′-UTROs03g29760 by miR169a was recovered by MIM169a (Figures 5B,D). These results suggest that miR169a may authentically repress the expression of Os03g29760 at translational level.
FIGURE 5.
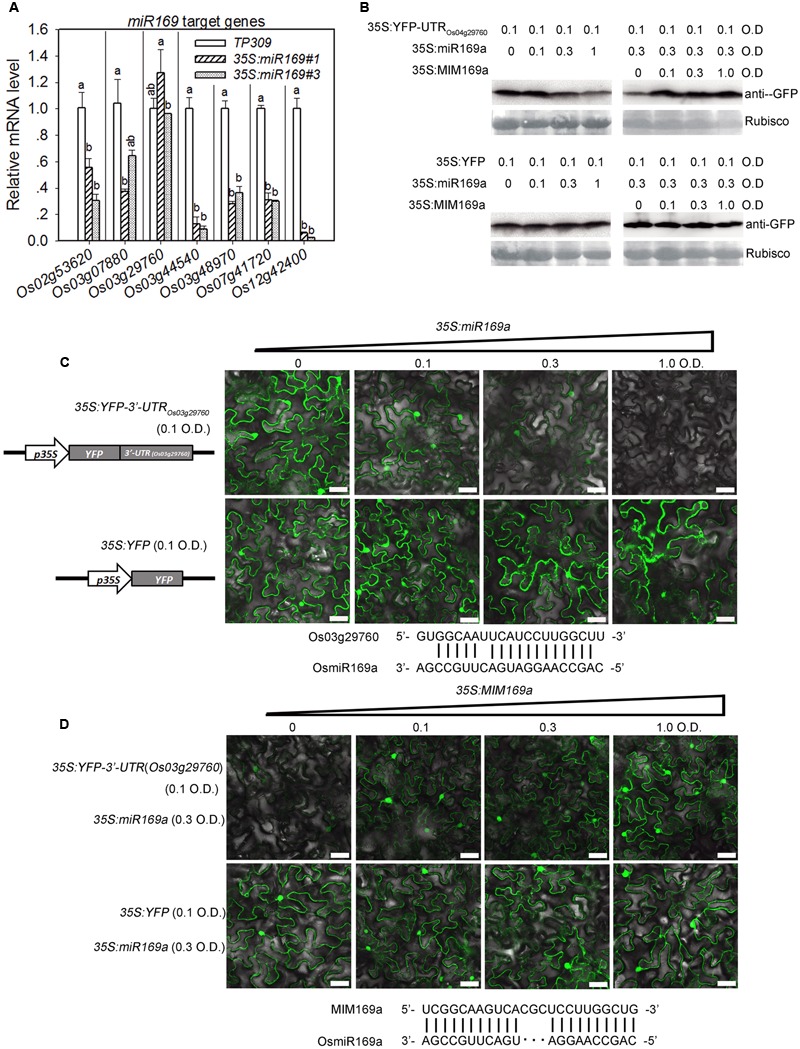
MiR169a represses the expression of its target genes at transcriptional or translational level. (A) Comparison of the relative mRNA levels of the indicated miR169’s target genes in the indicated lines. RNA was extracted in the indicated lines for qRT-PCR analysis. The mRNA level was normalized to that in the control plants. Values are means of three replications. Error bars indicate SD. The letters above the bars indicate significant differences (P < 0.01) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Western blotting analysis (B) and confocal images (C,D) show that miR169a suppressed the protein accumulation of Os03g29760 but did not affect the protein level of YFP control. The indicated YFP-3′-UTROs03g29760 and YFP-based reporter constructs were transiently expressed alone or co-expressed with miR169a (C) or/and the miR169 target mimicry MIM169a (D) in Nicotiana benthamiana leaves using Agrobacterium-mediated infiltration at the indicated optical density (O.D.) concentration. Protein extracts from the same amount of infiltrated leaves were subjected to Western blot analysis using anti-GFP sera. The Coomassie Blue stained Rubisco indicated the equal sample loading (B). The alignments of miR169a with Os03g29760 target sequence (C) and MIM169a with miR169a (D) were listed below the images, respectively. Size bars, 40 μm. Similar results were obtained in at least two independent experiments.
To figure out whether miR169 suppresses the expression of the other target genes at protein levels, we also tested the expression of two target genes by using the reporter system. As anticipated, the YFP intensity of YFP-3′-UTROs03g44540 and YFP-3′-UTROs03g48970 were both reduced by co-expression of miR169a, whereas when MIM169a was co-expressed, the suppression on YFP-3′-UTROs03g44540 and YFP-3′-UTROs03g48970 by miR169a was recovered (Supplementary Figure S3). These results are consistent with the transcriptional level of the two target genes, and demonstrate that miR169a can indeed repress the expression of the two target genes at protein levels.
Unfortunately, we did not detect the expression of Os07g06470 (NF-YA5) by qRT-PCR and failed to amplify this gene to make a transient expression construct. It is unclear whether and how the expression of Os07g06470 (NF-YA5) is regulated by miR169a.
Expressing Target Mimicry of miR169a Leads to Enhanced Resistance to M. oryzae
To further test the role of miR169a in rice immunity against the blast fungus, we constructed the target mimicry of miR169a, MIM169a, and obtained transgenic lines expressing MIM169a. Because overexpressing miR169a significantly repressed the expression of its target genes and led to enhanced susceptibility to M. oryzae, expressing MIM169a is predicted to up-regulate these target genes and result in enhanced resistance. Therefore, we first examined the expression of the target genes in the transgenic lines. As anticipated, the expression of all the detected target genes were significantly up-regulated in the two transgenic lines harboring the MIM169a construct (Figure 6A). Particularly, the expression of Os12g42400 (NF-YA10) was up-regulated about sixfold in the transgenic line #12 and 2-fold in the transgenic line #13, whereas, the mRNA levels of all the other six target genes were generally up-regulated from 2- to 4-fold in the two independent MIM169a transgenic lines compared with that in the control plants. Next, we tested the resistance of the transgenic lines to M. oryzae strains GZ8. Indeed, the lesions on the leaves from transgenic lines were obviously smaller than that on the leaves from the control plants, and the fungal biomass on the transgenic lines were also decreased significantly than that on the control plants (Figure 6B), indicating that the up-regulation of miR169’s target genes alleviated rice susceptibility to M. oryzae. Taken together, these results confirmed that miR169 natively regulates rice immunity, whereas, its target genes, or some of its target genes, positively regulate rice immunity against M. oryzae.
FIGURE 6.
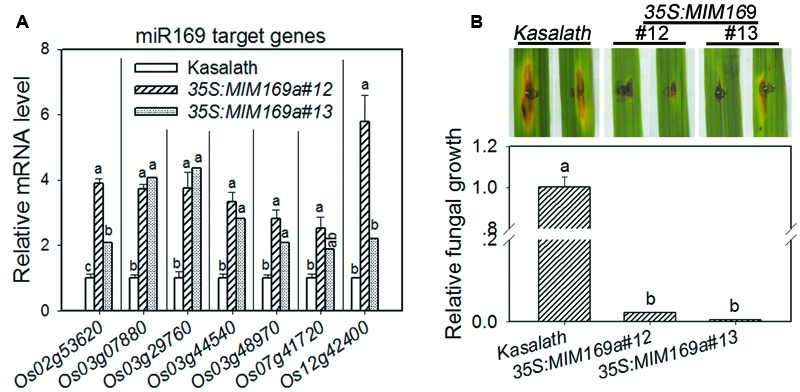
Expression of the miR169a target mimicry results in enhanced resistance to M. oryzae. (A) Comparison of the transcriptional level of miR169’s target genes in control plants and the transgenic lines expressing the miR169a target mimicry MIM169a. RNA was extracted in indicated lines for qRT-PCR analysis. mRNA level was normalized to that in control plants. Values are means of three replications. Error bars indicate SD. The letters above the bars indicate significant differences (P < 0.01) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. (B) MIM169a transgenic lines display enhanced resistance to the M. oryzae strain GZ8. The relative fungal mass on the inoculated leaves of the indicated transgenic lines was measured by using the ratio of DNA level of M. oryzae Pot2 against the rice genomic ubiquitin DNA level. Values are means of three replications. Error bars indicate SD. The letters above the bars indicate significant differences (P < 0.01) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Similar results were obtained in at least two independent experiments.
Different Target Genes of miR169 are Differentially Expressed in Susceptible and Resistant Rice Accessions
Now that all the detected seven target genes can be down-regulated by miR169 at either transcriptional or translational level, and up-regulated by the miR169’s target mimicry, it is questionable whether all of them similarly or equally regulate rice immunity against M. oryzae. To address this question, we performed a time course examination of their expression in LTH and IRBL9-W upon M. oryzae inoculation. According to the dynamic expression pattern, we divided the target genes into three groups. The first group contained one gene, Os02g53620 (NF-YA11), its transcription level was decreased significantly upon M. oryzae infection in both LTH and IRBL9-W (Figure 7A). The second group included three genes, Os03g44540 (NF-YA3), Os03g48970 (NF-YA4), and Os12g42400 (NF-YA10), their expression levels were decreased significantly and rapidly in LTH, but decreased slightly and slowly in IRBL9-W with Os03g44540 (NF-YA3) and Os03g48970 (NF-YA4) being transiently increased at 12 hpi and then decreased at the later time points of M. oryzae inoculation (Figure 7B). The third group included Os03g07880 (NF-YA1), Os03g29760 (NF-YA2) and Os07g41720 (NF-YA6), their transcription levels were kept stable or decreased in LTH, but increased significantly at one or all examined time points in IRBL9-W (Figure 7C). Therefore, different target genes of miR169 are differently responsive to M. oryzae infection in the resistant and susceptible accessions and thus may act differently in rice immunity against M. oryzae. The three genes, Os03g07880 (NF-YA1), Os03g29760 (NF-YA2), and Os07g41720 (NF-YA6) most likely contribute to the resistant phenotype of IRBL9-W.
FIGURE 7.
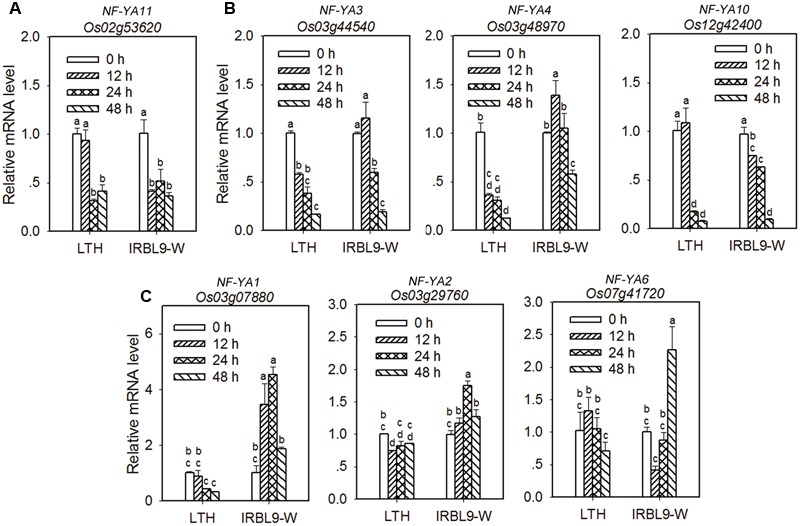
Differential expression of miR169’s target genes in susceptible and resistant accessions upon M. oryzae infection. (A–C) Expression of the indicated target genes in LTH and IRBL9-W upon M. oryzae infection. RNA was extracted at the indicated time points for quantitative RT-PCR analysis. mRNA level was normalized to that in untreated samples (0 h). Values are means of three replications. Error bars indicate SD. The letters above the bars indicate significant differences (P < 0.01) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Similar results were obtained in at least two independent experiments.
Discussion
The miR169 family is conserved and contains 17 members in rice (Zhao et al., 2007). Here, we provide data to show that miR169 acts as a negative regulator for rice immunity against the blast fungus M. oryzae. First, the sum accumulation of all miR169 isoforms was increased in the susceptible accession LTH, but fluctuated slightly in the resistant accession IRBL9-W (Figure 1), which was consistent with the previous high-throughput sequencing data in LTH and another resistant accession IRBLTs-Km (in Supplementary Table S1 of Li et al., 2014). Second, the transgenic lines overexpressing miR169a exhibited enhanced susceptibility to M. oryzae as indicated by the more severe disease phenotypes, down-regulated expression of defense-related genes, reduced defense response upon inoculation of M. oryzae, and accelerated M. oryzae invasion during the early infection period (Figures 2–4). Third, the transgenic plants expressing target mimicry of miR169a displayed enhanced resistance against M. oryzae (Figures 6A,B). In addition, the expression of all the tested seven NF-YA target genes were depressed at transcriptional or translational level by overexpressing miR169a, but up-regulated by the expression of a target mimicry of miR169a (Figures 5 and 6). Therefore, miR169 seems to negatively regulate rice immunity via multiple NF-YAs.
Nevertheless, each miR169 isoform may have its preferential target genes so that their expressions will be preferentially repressed when overexpressing a miR169 isoform. In Arabidopsis, overexpressing miR169d/e/f/g repressed the transcription of seven AtNF-YA genes, of which the three target genes, AtNF-YA2, AtNF-YA8, and AtNF-YA10, were down-regulated more significantly than the other four genes, including AtNF-YA1, AtNF-YA3, AtNF-YA5, and AtNF-YA9 (Xu et al., 2014). On the contrary, expressing the target mimicry MIM169defg up-regulates the accumulation of NF-YA2 and NF-YA10 transcripts to levels significantly higher than that of other NF-YA target genes, suggesting that the miR169d/e/f/g isoform preferentially represses NF-YA2 and NF-YA10 (Sorin et al., 2014). In addition, co-expression of miR169 isoforms and NF-YA5 in N. benthamiana revealed that miR169a was more efficient than miR169c in repressing NF-YA5 at transcriptional level (Li et al., 2008). In the present study, overexpressing miR169a leads to repression of all the target genes tested. Particularly, the two target genes, Os03g44540 (NF-YA3) and Os12g42400 (NF-YA10), were repressed more than the other five NF-YA target genes (Figure 5A), indicating that miR169a may preferentially target Os03g44540 (NF-YA3) and Os12g42400 (NF-YA10).
In addition, different miR169 isoforms may differentially regulate their target genes through transcript cleavage or translation inhibition. The drought/salinity inducible miRNA isoforms, miR169g and miR169n/o, seems to preferentially mediate cleavage of Os03g29760 (NF-YA2) and Os07g41720 (NF-YA6) as indicated by the down-regulation of their transcripts when miR169g and miR169n/o were induced to higher levels (Zhao et al., 2009). In the present study, the transcription level of Os03g29760 (NF-YA2) was not repressed in the transgenic lines overexpressing miR169a (Figure 5A). Potential reasons are that miR169a does not target Os03g29760 (NF-YA2) or does not repress this target gene by transcript cleavage, or that the expression of Os03g29760 (NF-YA2) is subjected to self-regulation to complement the down-regulation of the other NF-YA target genes. Therefore, we designed a transient expression reporter system to clarify whether miR169a could repress its target genes at protein level. Indeed, miR169a could repress the protein intensity of YFP-UTROs03g29760 when transiently co-expressed in N. benthamiana, implying that miR169a can regulate the expression of Os03g29760 (NF-YA2) via translation inhibition. On the other hand, the expression of Os03g29760 (NF-YA2) could also be suppressed by other miR169 isoforms via transcript cleavage, because its transcription was slightly decreased in the susceptible accession LTH upon M. oryzae infection, which was reversely correlated with the induced accumulation of miR169 isoforms (Figure 1C).
Finally, although miR169 targets the same batch of NF-YA genes, different target genes may function differently. Rice contains 11 NF-YA genes that are classified into several clades in a phylogenetic tree (Petroni et al., 2012), eight of the NF-YA genes are identified to be the target of miR169 (Wu et al., 2009; Li Y. F. et al., 2010; Zhou et al., 2010). Os03g29760 (NF-YA2) and Os07g41720 (NF-YA6) are rapidly down-regulated upon high salinity treatment, while the other NF-YA genes are not obviously changed (Zhao et al., 2009), indicating different NF-YA genes are differentially responsive to salt stress. In addition, different NF-YA genes are differentially induced by drought, salt and temperature stresses, but only OsNF-YA7 (Os08g09690), a non-target of miR169, is highly induced by drought and salt treatment (Lee et al., 2015). In the present study, seven target NF-YA genes of miR169a were also differentially responsive to M. oryzae infection in LTH and IRBL9-W (Figure 7A). These target genes belong to different clades in a phylogenetic tree. While Os02g53620 (NF-YA11) is not reported before, Os03g07880 (NF-YA1), Os03g29760 (NF-YA2), and Os07g41720 (NF-YA6) are classified into class 1 clade, Os03g48970 (NF-YA4) belongs to class 2, and Os03g44540 (NF-YA3) and Os12g42400 (NF-YA10) belong to class 3 clade (Petroni et al., 2012; Lee et al., 2015). It is interesting that the genes in the same clade displayed similar transcription patterns upon M. oryzae infection in LTH and IRBL9-W, suggesting that the NF-YAs in the different clade may play different roles in rice immunity against M. oryzae. For example, Os02g53620 (NF-YA11) might act as a negative regulator in rice immunity against M. oryzae because its expression was rapidly and significantly decreased in the resistant accession IRBL9-W upon M. oryzae infection (Figure 7A), whereas, the three target genes, Os03g07880 (NF-YA1), Os03g29760 (NF-YA2) and Os07g41720 (NF-YA6), might function as positive regulators because their expression levels were significantly up-regulated in the resistant accession IRBL9-W (Figure 6C). However, whether the three target genes, Os03g48970 (NF-YA4), Os03g44540 (NF-YA3), and Os12g42400 (NF-YA10) function in rice immunity as positive or negative regulators requires further investigation because their expressions are differentially fluctuated in both the resistant and susceptible accessions upon M. oryzae infection. However, functional characterization of each of these genes via knockout approach could be difficult because of possible functional redundance among them. On the other hand, because all the seven target genes are significantly up-regulated in MIM169a transgenic lines which display enhanced resistance to M. oryzae, constructing transgenic lines overexpressing single target genes will be informative to understand the role of each target gene in regulation of rice immunity against M. oryzae. Taken together, our data demonstrated that miR169 acts as a negative regulator in rice immunity against M. oryzae. Overexpressing miR169a led to suppression of its target genes and enhanced susceptibility to M. oryzae. To the contrast, expressing the target mimicry of miR169a resulted in up-regulation of its target genes and enhanced resistance to M. oryzae. Therefore, it is possible to engineer rice blast resistance via managing the accumulation of miR169.
Author Contributions
YL, S-LZ, J-LL, HW, X-LC, Y-JX, Z-XZ, Z-YX, and NY conducted the experiments. X-HH and FH conducted the field experiment. YL and JF supervised the study. YL wrote the manuscript. W-MW coordinated the overall study and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Cai-Lin Lei (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the monogenic resistant lines IRBL9-W.
Funding. This work was supported by the National Natural Science Foundation of China (grant 31430072 to W-MW, 31471761 and 31101406 to YL).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00002/full#supplementary-material
References
- Baldrich P., Campo S., Wu M. T., Liu T. T., Hsing Y. I., San Segundo B. (2015). MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 12 847–863. 10.1080/15476286.2015.1050577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrich P., San Segundo B. (2016). MicroRNAs in rice innate immunity. Rice (N. Y.) 9 6 10.1186/s12284-016-0078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silence in plants. Nature 431:8 10.1038/nature02874 [DOI] [PubMed] [Google Scholar]
- Bonman J. M., Dios T. I. V. D., Khin M. M. (1986). Physiologic specialization of Pyricularia oryzae in the Philippines. Plant Dis. 70 767–769. 10.1094/PD-70-767 [DOI] [Google Scholar]
- Campo S., Peris-Peris C., Sire C., Moreno A. B., Donaire L., Zytnicki M., et al. (2013). Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 199 212–217. 10.1111/nph.12292 [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146 368–376. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2012). Small RNAs in development – insights from plants. Curr. Opin. Genet. Dev. 22 361–367. 10.1016/j.gde.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., Rubio-Somoza I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39 1033–1037. 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- Hanemian M., Barlet X., Sorin C., Yadeta K. A., Keller H., Favery B., et al. (2016). Arabidopsis CLAVATA1 and CLAVATA2 receptors contribute to Ralstonia solanacearum pathogenicity through a miR169-dependent pathway. New Phytol. 211 502–515. 10.1111/nph.13913 [DOI] [PubMed] [Google Scholar]
- Huang Y. Y., Shi Y., Lei Y., Li Y., Fan J., Xu Y. J., et al. (2014). Functional identification of multiple nucleocytoplasmic trafficking signals in the broad-spectrum resistance protein RPW8.2. Planta 239 455–468. 10.1007/s00425-013-1994-x [DOI] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103 11086–11091. 10.1073/pnas.0508882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 706–724. 10.1105/tpc.106.046300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Jin H. (2010). Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48 225–246. 10.1146/annurev-phyto-073009-114457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Sharma B., Gupta A. K., Kaur S., Kaur J. (2014). Nodule metabolism in cold stress tolerant and susceptible chickpea cultivars. Symbiosis 64 33–42. 10.1007/s13199-014-0300-4 [DOI] [Google Scholar]
- Khanna A., Sharma V., Ellur R. K., Shikari A. B., Gopala Krishnan S., Singh U. D., et al. (2015). Development and evaluation of near-isogenic lines for major blast resistance gene(s) in Basmati rice. Theor. Appl. Genet. 128 1243–1259. 10.1007/s00122-015-2502-4 [DOI] [PubMed] [Google Scholar]
- Khraiwesh B., Zhu J. K., Zhu J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819 137–148. 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. K., Kim H. I., Jang G., Chung P. J., Jeong J. S., Kim Y. S., et al. (2015). The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. 241 199–210. 10.1016/j.plantsci.2015.10.006 [DOI] [PubMed] [Google Scholar]
- Leyva-Gonzalez M. A., Ibarra-Laclette E., Cruz-Ramirez A., Herrera-Estrella L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7:e48138 10.1371/journal.pone.0048138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Pignatta D., Bendix C., Brunkard J. O., Cohn M. M., Tung J., et al. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 109 1790–1795. 10.1073/pnas.1118282109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Oono Y., Zhu J., He X., Wu J., Iida K., et al. (2008). The Arabidopsis NFYA5 transcription factors regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20 2238–2251. 10.1105/tpc.108.059444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu Y. G., Shi Y., Wu L., Xu Y. J., Huang F., et al. (2014). Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 164 1077–1092. 10.1104/pp.113.230052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Zhang J., Wu L., Qi Y., Zhou J. M. (2010). Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 152 2222–2231. 10.1104/pp.109.151803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Zheng Y., Addo-Quaye C., Zhang L., Saini A., Jagadeeswaran G., et al. (2010). Transcriptome-wide identification of microRNA targets in rice. Plant J. 62 742–759. 10.1111/j.1365-313X.2010.04187.x [DOI] [PubMed] [Google Scholar]
- Li Z. Y., Xia J., Chen Z., Yu Y., Li Q. F., Zhang Y. C., et al. (2016). Large-scale rewiring of innate immunity circuitry and microRNA regulation during initial rice blast infection. Sci. Rep. 6:25493 10.1038/srep25493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., He H., Yu D. (2012). Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE 7:e48951 10.1371/journal.pone.0048951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z. Z., Jiang W. W., Wang J. L., Lei C. L. (2001). Research and utilization of universally susceptible property of japonica rice variety Lijiangxintuanheigu. Sci. Agric. Sin. 34 116–117. [Google Scholar]
- Liu B., Li J. F., Ao Y., Qu J., Li Z., Su J., et al. (2012). Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24 3406–3419. 10.1105/tpc.112.102475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu J., Triplett L., Leach J. E., Wang G. L. (2014). Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52 213–241. 10.1146/annurev-phyto-102313-045926 [DOI] [PubMed] [Google Scholar]
- Luan M., Xu M., Lu Y., Zhang L., Fan Y., Wang L. (2015). Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 555 178–185. 10.1016/j.gene.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Luan M., Xu M., Lu Y., Zhang Q., Zhang L., Zhang C. (2014). Family-wide survey of miR169s and NF-YAs and their expression profiles response to abiotic stress in maize roots. PLoS ONE 9:e91369 10.1371/journal.pone.0091369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Lei C., Xu X., Hao K., Wang J., Cheng Z., et al. (2015). Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in Rice. Mol. Plant Microbe Interact. 28 558–568. 10.1094/MPMI-11-14-0367-R [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239 15–27. 10.1016/S0378-1119(99)00368-6 [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436–439. 10.1126/science.1126088 [DOI] [PubMed] [Google Scholar]
- Ouyang S., Park G., Atamian H. S., Han C. S., Stajich J. E., Kaloshian I., et al. (2014). MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 10:e1004464 10.1371/journal.ppat.1004464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan C., Zhang X., Jin H. (2009). Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 12 465–472. 10.1016/j.pbi.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Park C. H., Chen S., Shirsekar G., Zhou B., Khang C. H., Songkumarn P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24 4748–4762. 10.1105/tpc.112.105429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R. W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., et al. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24 4777–4792. 10.1105/tpc.112.105734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu B., He X., Wang J., Zhao Y., Teng W., Shao A., et al. (2015). A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 167 411–423. 10.1104/pp.114.246959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Liu G., Zhou B., Bellizzi M., Zeng L., Dai L., et al. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172 1901–1914. 10.1534/genetics.105.044891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Bahar O., Thomas N., Holton N., Nekrasov V., Ruan D., et al. (2015). Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog. 11:e1004809 10.1371/journal.ppat.1004809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64 204–214. 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P. V., Chen H. M., Patel K., Bond D. M., Santos B. A., Baulcombe D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24 859–874. 10.1105/tpc.111.095380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y. J., Liu Y., Li W., Song L. L., Zhang J., Guo C. H. (2016). Genome-wide investigation of micrornas and their targets in response to freezing stress in Medicago sativa L., based on high-throughput sequencing. G3 (Bethesda) 6 755–765. 10.1534/g3.115.025981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C., Declerck M., Christ A., Blein T., Ma L., Lelandais-Briere C., et al. (2014). A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 202 1197–1211. 10.1111/nph.12735 [DOI] [PubMed] [Google Scholar]
- Tsumematsu H., Yanoria M. J. T., Ebron L. A., Hayashi N., Ando I., Kato H., et al. (2000). Development of monogenic lines of rice for blast resistance. Breed. Sci. 50 229–234. 10.1270/jsbbs.50.229 [DOI] [Google Scholar]
- Wu L., Zhang Q., Zhou H., Ni F., Wu X., Qi Y. (2009). Rice MicroRNA effector complexes and targets. Plant Cell 21 3421–3435. 10.1105/tpc.109.070938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Brown S., Patrick E., Brearley C., Turner J. G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15 33–45. 10.1105/tpc.006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Zhang L., Li W., Hu X., Wang M., Fan Y., et al. (2014). Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J. Exp. Bot. 65 89–101. 10.1093/jxb/ert353 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Yamada K., Ishikawa K., Yoshimura S., Hayashi N., Uchihashi K., et al. (2013). A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13 347–357. 10.1016/j.chom.2013.02.007 [DOI] [PubMed] [Google Scholar]
- Zhai J., Jeong D. H., De Paoli E., Park S., Rosen B. D., Li Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25 2540–2553. 10.1101/gad.177527.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Liu M., Tang M., Dong B., Wu D., Zhang Z., et al. (2015). Repression of microRNA biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice. Plant Sci. 237 24–32. 10.1016/j.plantsci.2015.05.002 [DOI] [PubMed] [Google Scholar]
- Zhao B., Ge L., Liang R., Li W., Ruan K., Lin H., et al. (2009). Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 10:29 10.1186/1471-2199-10-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Liang R., Ge L., Li W., Xiao H., Lin H., et al. (2007). Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 354 585–590. 10.1016/j.bbrc.2007.01.022 [DOI] [PubMed] [Google Scholar]
- Zhao M., Ding H., Zhu J. K., Zhang F., Li W. X. (2011). Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190 906–915. 10.1111/j.1469-8137.2011.03647.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Gu L. F., Li P. C., Song X. W., Wei L. Y., Chen Z. Y., et al. (2010). Degradome sequencing reveals endogenous small RNAtargets in rice (Oryza sativa L. ssp. indica). Front. Biol. 5 67–90. 10.1007/s11515-010-0007-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


