Abstract
CYCLOIDEA-like genes are involved in the symmetry gene network, limiting cell proliferation in the dorsal regions of bilateral flowers in core eudicots. CYC-like and closely related TCP genes (acronym for TEOSINTE BRANCHED1, CYCLOIDEA, and PROLIFERATION CELL FACTOR) have been poorly studied in Asparagales, the largest order of monocots that includes both bilateral flowers in Orchidaceae (ca. 25.000 spp) and radially symmetrical flowers in Hypoxidaceae (ca. 200 spp). With the aim of assessing TCP gene evolution in the Asparagales, we isolated TCP-like genes from publicly available databases and our own transcriptomes of Cattleya trianae (Orchidaceae) and Hypoxis decumbens (Hypoxidaceae). Our matrix contains 452 sequences representing the three major clades of TCP genes. Besides the previously identified CYC specific core eudicot duplications, our ML phylogenetic analyses recovered an early CIN-like duplication predating all angiosperms, two CIN-like Asparagales-specific duplications and a duplication prior to the diversification of Orchidoideae and Epidendroideae. In addition, we provide evidence of at least three duplications of PCF-like genes in Asparagales. While CIN-like and PCF-like genes have multiplied in Asparagales, likely enhancing the genetic network for cell proliferation, CYC-like genes remain as single, shorter copies with low expression. Homogeneous expression of CYC-like genes in the labellum as well as the lateral petals suggests little contribution to the bilateral perianth in C. trianae. CIN-like and PCF-like gene expression suggests conserved roles in cell proliferation in leaves, sepals and petals, carpels, ovules and fruits in Asparagales by comparison with previously reported functions in core eudicots and monocots. This is the first large scale analysis of TCP-like genes in Asparagales that will serve as a platform for in-depth functional studies in emerging model monocots.
Keywords: Cattleya trianae, CINCINNATA, CYCLOIDEA, Hypoxidaceae, Hypoxis decumbens, floral symmetry, Orchidaceae, PROLIFERATION CELL FACTOR
Introduction
As currently circumscribed the order Asparagales is a species-rich group comprising ca. 50% of all monocots, corresponding to 10–15% of flowering plants (Chase et al., 2009, 2016; Chen et al., 2013; Givnish et al., 2016). The most recent phylogenetic analyses in the monocots place Orchidaceae as sister to all other Asparagales (Chen et al., 2013). The family is divided into five subfamilies: Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae, and Epidendroideae (Chase et al., 2015; Endress, 2016). The floral groundplan in Asparagales varies primarily in the floral symmetry and the number of stamens (Simpson, 2006). The floral morphology of Asparagales outside orchids consists of radially symmetrical, trimerous flowers with tepaloid perianth and free floral organs, although a few exceptions have been documented in Aspidistra (Asparagaceae), Gethyllis (Amaryllidaceae), Neoastelia (Asteliaceae) and Pauridia (Hypoxidaceae) (Rudall, 2002; Rudall and Bateman, 2002, 2004; Kocyan, 2007). Conversely, orchid flowers are variously bilateral and undergo extreme elaboration of some organs, including differentiation of perianth parts, stamen abortion, and fusion of floral parts from the same whorl or from different whorls (Rudall, 2002). In the bilateral resupinated orchid flowers the two dorsal petals are very similar to each other, whereas the ventral one (the lip or labellum) often undergoes extreme elaboration in shape, color, size and epidermal specializations (Rudall and Bateman, 2004; Pabón-Mora and González, 2008; Mondragón-Palomino and Theißen, 2009; Rudall et al., 2013; Endress, 2016). In the inner floral whorls bilateral symmetry is evident by the formation of a gynostemium that results from the congenital fusion between the single fertile stamen (sometimes two fertile stamens) and stigmas (Rudall and Bateman, 2002; Pabón-Mora and González, 2008; Endress, 2016). Such floral elaboration has been linked to extremely specialized pollination mechanisms and the exceedingly high diversification rates in Orchidaceae (Gong and Huang, 2009; Mondragón-Palomino and Theißen, 2009; Mondragón-Palomino, 2013).
The genetic network underlying bilateral floral symmetry has been assessed using Antirrhinum majus floral symmetry mutants (Luo et al., 1996). This network includes the differential dorsiventral expression of four transcription factors in the two-lipped flowers of this species. Three transcription factors, CYCLOIDEA (CYC), DICHOTOMA (DICH), and RADIALIS (RAD) regulate cell division on the dorsal portion of the flower primordium and during dorsal petal and stamen primordia initiation. Additionally, RAD outcompetes DIVARICATA (DIV) for binding proteins in the dorsal side of the flower, restricting DIV function to the ventral and lateral petals (Almeida et al., 1997; Galego and Almeida, 2002; Raimundo et al., 2013). Thus, cyc/dich mutants show radially symmetrical ventralized flowers (Luo et al., 1996, 1999). Both, CYC and DICH genes belong to the TCP gene family (acronym for TEOSINTE BRANCHED 1 -TB1- from Z. mays, CYCLOIDEA -CYC- from A. majus y PROLIFERATION CELL FACTOR 1 and 2 -PCF1 and PCF2- from Oryza sativa) (Doebley et al., 1997; Kosugi and Ohashi, 1997; Luo et al., 1999). RAD and DIV belong to the MYB (Myeloblastosis) gene family (Luo et al., 1999; Galego and Almeida, 2002; Corley et al., 2005; Costa et al., 2005).
The TCP genes encode putative basic-Helix-Loop-Helix (bHLH) transcription factors (Cubas et al., 1999). The bHLH domain recognizes a consensus sequence GGNCCCAC/GTGGNCCC required for DNA binding and activation or repression of transcription (Kosugi and Ohashi, 2002; Martín-Trillo and Cubas, 2010). Gene evolution analyses have identified two large groups of TCP genes, namely Class I (which include PCF homologs) and Class II (containing the CIN/CYC/TB1-like genes) (Cubas et al., 1999; Damerval and Manuel, 2003; Reeves and Olmstead, 2003; Broholm, 2009; Mondragón-Palomino and Trontin, 2011). Additional large scale duplications (i.e., those occurring prior to the diversification of major inclusive hierarchical groupings) have been found within the CYC genes. Two rounds of duplication occurred specifically in core eudicots, resulting in CYC1, CYC2, and CYC3 clades, and one duplication specific to monocots resulting in the RETARDED PALEA 1 (REP1) and TEOSINTE BRANCHED 1 (TB1) clades (Vieira et al., 1999; Damerval and Manuel, 2003; Howarth and Donoghue, 2006; Navaud et al., 2007; Yao et al., 2007; Mondragón-Palomino and Trontin, 2011). In addition, species specific duplications (i.e., those occurring in a single species) have also been reported, often linked to polyploidy (Ma et al., 2016). Non-core eudicot homologs are known as the CYC-like genes (Damerval et al., 2007; Preston and Hileman, 2012; Horn et al., 2015). Functional characterization has concentrated in CYC2 orthologs in eudicots, including Asterales, Brassicales, Dipsacales, Fabales, Lamiales and Malpighiales, among others (Busch and Zachgo, 2007; Gao et al., 2008; Preston et al., 2009; Wang et al., 2010; Zhang et al., 2010, 2013; Howarth et al., 2011; Tähtiharju et al., 2012; Yang et al., 2012). These studies have found CYC2 expression restricted to the same dorsal floral domain and a conserved role as cell proliferation repressors resulting in bilateral symmetry (reviewed in Hileman, 2014). Fewer studies have been made in basal eudicots, but dissymmetric Fumarioids (Papaveraceae) do have asymmetric expression of CYC-like genes, suggesting that CYC-like recruitment to form bilateral flowers has occurred independently several times in eudicots (Damerval et al., 2007, 2013).
Less is known about the role of pre-duplication CYC-like genes in monocots (Bartlett and Specht, 2011; Mondragón-Palomino and Trontin, 2011; Preston and Hileman, 2012). Expression analyses of CYC-like genes in Costus (Costaceae; Zingiberales) and Commelina (Commelinaceae; Commelinales) suggest that they play a role in bilateral symmetry (Bartlett and Specht, 2011; Preston and Hileman, 2012). Functional studies in O. sativa (Poaceae, Poales) confirm that these genes contribute to the asymmetric growth of the dorsal versus the ventral portions of the flower, as shown by the rep1 mutants which exhibit a smaller palea due to cell division arrest (Yuan et al., 2009). The only two studies available in Orchidaceae are particularly intriguing as they show very different expression patterns of CYC/TB1-like orthologs. Whereas, the only copy of CYC/TB1-like in Orchis italica (OitaTB1) is expressed exclusively in leaves (De Paolo et al., 2015), two of three CYC/TB1-like copies in Phalaenopsis equestris, PeCYC1 and PeCYC2, seem to be expressed in higher levels (2–10 times more) in the dorsal sepals and the labellum compared to the ventral sepal and the lateral petals (Lin et al., 2016). Furthermore, some authors have hypothesized that the expression gradient of TCP genes is largely controlled by upstream expression of the AP3/DEF petal-stamen identity genes, resulting in higher concentrations of CYC/TB1-like genes in the dorsal floral regions; however, more experimental data is needed to support this (Mondragón-Palomino and Theißen, 2009).
It is unclear whether closely related TCP-like CINCINNATA (CIN) and PROLIFERATION CELL FACTOR (PCF) genes play any role in floral symmetry. CIN was originally characterized in A. majus and more recently in Arabidopsis thaliana (Crawford et al., 2004; Nag et al., 2009; Sarvepalli and Nath, 2011; Danisman et al., 2013). In both species CIN controls cellular proliferation in petals and cellular arrest in leaves (Crawford et al., 2004; Nag et al., 2009). On the other hand, O. sativa PCF1 and PCF2 are involved in axillary meristem repression, likely via the activation of PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA), which encodes a protein involved in DNA replication and repair, maintenance of chromatin structure, chromosome segregation, and cell-cycle progression (Kosugi and Ohashi, 1997). Other studies in A. thaliana suggest that PCF-like genes are also involved in gametophyte development, transduction of hormonal signals, mitochondrial biogenesis, leaf and flower morphogenesis, seed germination, branching, and even circadian clock regulation (Koyama et al., 2007; Pruneda-Paz et al., 2009; Giraud et al., 2010; Kieffer et al., 2011; Resentini et al., 2015). Recent studies in the model orchid P. equestris have found that PeCIN8 (CIN-like) and PePCF10 (PCF-like) control cell proliferation and cell shape in petals, ovules and leaves (Lin et al., 2016).
In order to study the contribution of CYC/TB1-like and the closely related CIN-like and PCF-like genes to floral patterning in Asparagales, we first determined copy number and characteristic protein motifs and then assessed gene lineage evolution including a vast sampling of TCP-like genes across angiosperms and particularly of Asparagales monocots. Next, we evaluated the expression patterns of all TCP-like genes in dissected floral organs, young leaves, and fruits of Hypoxis decumbens (Hypoxidaceae) which has typical asparagalean radial trimerous flowers with free parts, and Cattleya trianae (Orchidaceae), that has bilateral flowers, and a single fertile stamen fused with the tree stigmas (i.e., gynostemium). Finally, we propose hypotheses on functional evolution based on previous literature reports and comparisons with our results that suggest different trends among gene clades when comparing Asparagales to model core eudicots.
Materials and methods
Gene isolation and phylogenetic analyses
In order to isolate putative TCP-like homologs in Asparagales, searches were performed using previously reported TCP genes from eudicots, monocots and in particular Orchidaceae as queries (Mondragón-Palomino and Trontin, 2011; Preston and Hileman, 2012; De Paolo et al., 2015; Horn et al., 2015). Searches included homologs from all the three main clades of TCP genes: CYC-like, CIN-like and PCF-like. Searches were done using BLAST tools (Altschul et al., 1990) in the orchid specific available databases including Orchidbase 2.0 (http://orchidbase.itps.ncku.edu.tw/) (Tsai et al., 2013), Orchidstra (http://orchidstra2.abrc.sinica.edu.tw/orchidstra2/index.php) (Su et al., 2013), as well as the more inclusive OneKP database (http://www.bioinfodata.org/Blast4OneKP/). All core eudicot sequences, were isolated from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and genbank (https://www.ncbi.nlm.nih.gov/genbank/).
In addition to the available databases we generated two transcriptomes from C. trianae (Orchidaceae) and H. decumbens (Hypoxidaceae). The transcriptome for each species was generated from mixed material from 3 biological replicates and included vegetative and reproductive meristems, floral buds, young leaves and fruits in as many developmental stages as possible. Total RNA was purified and used for the preparation of one mRNA (polyA) HiSeq library for each species. RNA-seq experiments were conducted using truseq mRNA library construction kit (Illumina, San Diego, California, USA) and sequenced in a HiSeq 2000 instrument reading 100 base paired end reads.
The transcriptome was assembled de novo with Trinity v2 following default settings. Read cleaning was performed with prinseq-lite v0.20.4 with a quality threshold of Q35 and a minimum read length of 50 bases. Contig metrics are as follows: (1) H. decumbens total assembled bases: 73,787,751; total number of contigs (>101 bases): 157,153; average contig length: 469 bp; largest contig: 15,554 bp; contig N50: 1075 bp; contig GC%: 46,42. (2) C. trianae total assembled bases: 63,287,862 bp; total number of contigs (>101 bases): 109,708; average contig length: 576 bp; largest contig: 9321 bp; contig N50: 1401 bp; contig GC%: 42,73. Homologous gene search was performed using BLASTN with the query sequences downloaded from GenBank and other databases (see above). In order to estimate the relative abundance of the assembled contigs, cleaned reads were mapped against the de novo assembled dataset and counted with two different strategies. The first one involved the mapping algorithm of the software Newbler v2.9 where only the unique matching read pairs were accepted as positive counts. The second one involved the mapping algorithm BOWTIE2 and raw reads counts as well as RPKM were calculated to each assembled contig (Table 1).
Table 1.
Selected features of TCP and AP3-like gene contigs taken from sequences and the transcriptomes of Cattleya trianae and Hypoxis decumbens.
| Gene CDS | Contig | CDS size (in bp) | Protein size (in AA) | URP* Newbler | RRC** RPKM | RPKM | RPKM rank |
|---|---|---|---|---|---|---|---|
| CtrAP3a | c19830_g1_i1 | 684 | 227 | 4047 | 3877 | 91,02338579 | 1689 |
| CtrAP3b | c16430_g1_i2 | 696 | 231 | 15 | 719 | 16,64198541 | 7551 |
| CtrAP3c | c16430_g1_i1 | 669 | 222 | 184 | 1002 | 23,72478436 | 5775 |
| CtrAP3d | c14728_g2_i1 | 628 | 208 | 1218 | 1228 | 38,72393988 | 3786 |
| CtrAP3e | c13983_g1_i1 | 601 | 199 | 321 | 269 | 9,445413314 | 11262 |
| CtrCIN1 | c10815_g1_i1 | 945 | 314 | 565 | 540 | 7,473218106 | 13078 |
| CtrCIN2 | c20571_g1_i1 | 975 | 324 | 5413 | 5455 | 58,71380149 | 2597 |
| CtrCIN3 | c20515_g2_i1 | 1158 | 385 | 2065 | 2091 | 28,27740553 | 4987 |
| CtrCIN4 | c20515_g1_i1 | 1116 | 371 | 1436 | 880 | 13,36057701 | 8882 |
| CtrCIN5 | c19791_g1_i1 | 1116 | 371 | 530 | 491 | 6,141835864 | 14794 |
| CtrCIN6 | c18629_g1_i1 | 957 | 319 | 453 | 411 | 8,008505294 | 12506 |
| CtrPCF1 | c14669_g1_i1 | 930 | 309 | 260 | 258 | 6,560658931 | 14228 |
| CtrPCF2 | c10284_g1_i1 | 759 | 252 | 306 | 308 | 8,318287473 | 12198 |
| CtrPCF3 | c22906_g1_i1 | 882 | 293 | 1109 | 1118 | 23,04243274 | 5910 |
| CtrPCF4 | c10751_g2_i1 | 561 | 187 | 315 | 230 | 11,37525956 | 9984 |
| CtrPCF5 | c21320_g2_i1 | 900 | 299 | 493 | 401 | 6,065023082 | 14927 |
| CtrPCF6 | c23464_g2_i1 | 828 | 275 | 5821 | 928 | 15,39001893 | 7984 |
| CtrPCF7 | c13613_g1_i1 | 684 | 227 | 1023 | 1050 | 25,7820899 | 5397 |
| CtrPCF8 | c8381_g1_i1 | 642 | 213 | 136 | 136 | 3,198372774 | 21993 |
| CtrPCF9 | c16850_g1_i1 | 1032 | 343 | 1021 | 529 | 6,68019211 | 14059 |
| CtrPCF10 | c24243_g4_i1 | 1107 | 368 | 269 | 140 | 2,935430718 | 23098 |
| CtrPCF11 | c20858_g1_i1 | 963 | 320 | 181 | 171 | 3,452818247 | 21033 |
| CtrTB1 | c62044_g1_i1/c74262_g1_i1 | 330 | 110 | 6/15 | 7/14 | 0,507584894922/0,584445576 | 58492/52988 |
| HydDEF1a | c29923_g1_i2 | 181 | 59 | 175 | 8 | 1,074179469 | 58639 |
| HydDEF1b | c29923_g1_i4 | 684 | 227 | 80 | 84 | 3,992750518 | 25641 |
| HydDEF2 | c23342_g1_i1 | 672 | 223 | 533 | 513 | 19,98971549 | 6376 |
| HydCIN1 | c26829_g2_i1 | 717 | 239 | 605 | 420 | 12,7762255 | 9853 |
| HydCIN2 | c25446_g2_i3 | 1272 | 424 | 496 | 589 | 17,24524272 | 7370 |
| HydCIN3 | c25446_g2_i4 | 651 | 217 | 2655 | 338 | 17,37878284 | 7322 |
| HydCIN4 | c25446_g2_i2 | 858 | 286 | 1355 | 1314 | 44,60569165 | 2753 |
| HydPCF1 | c25819_g1_i2 | 1038 | 345 | 12 | 266 | 7,587936907 | 15736 |
| HydPCF2 | c25819_g1_i1 | 939 | 312 | 11 | 236 | 8,050262478 | 14962 |
| HydPCF3 | c29001_g5_i3 | 300 | 100 | 75 | 64 | 5,51807531 | 20195 |
| HydPCF4 | c29001_g5_i1 | 387 | 129 | 23 | 20 | 1,853254689 | 41814 |
| HydPCF5 | c29001_g5_i4 | 408 | 136 | 32 | 43 | 3,099053674 | 30449 |
| HydPCF6 | c29001_g5_i2 | 411 | 137 | 42 | 57 | 4,208770608 | 24720 |
| HydPCF7 | c27872_g4_i1 | 771 | 256 | 732 | 896 | 26,84914255 | 4743 |
| HydPCF8 | c27872_g3_i2 | 762 | 253 | 334 | 528 | 16,30864126 | 7759 |
| HydPCF9 | c27872_g3_i1 | 756 | 251 | 108 | 181 | 6,950127044 | 16867 |
| HydPCF10 | c27872_g2_i1 | 675 | 225 | 138 | 157 | 9,763071436 | 12576 |
| HydTB1 | c52607_g1_i1 | 258 | 85 | 3 | 3 | 0,364509027 | 119496 |
Unique read pairs - number of specific/unique read pairs supporting each contig.
Raw read counts.
All sequences isolated were compiled with Bioedit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Sequences shorter than 200 bp lacking similarity with a region of the putative bHLH motif were discarded. Nucleotide sequences were subsequently aligned using the online version of MAFFT (http://mafft.cbrc.jp/alignment/software/) (Katoh et al., 2002) with a gap open penalty of 3.0, offset value of 1.0 and all other default settings. The alignment was then refined by hand using Bioedit considering as a reference the 60–70 aa reported as conserved in the TCP protein domain (Cubas et al., 1999). To better understand the evolution of the TCP gene lineage, and to integrate previous eudicot and monocot specific phylogenetic analyses (Damerval and Manuel, 2003; Hileman and Baum, 2003; Reeves and Olmstead, 2003; Howarth and Donoghue, 2006; Damerval et al., 2007; Bartlett and Specht, 2011; Mondragón-Palomino and Trontin, 2011; Preston and Hileman, 2012; Horn et al., 2015), we performed Maximum likelihood (ML) phylogenetic analyses using the nucleotide sequences with RaxML-HPC2 BlackBox through the CIPRES Science Gateway (https://www.phylo.org/) (Miller et al., 2010). Bootstrapping (BS) was performed according to the default criteria in RAxML (200–600 replicates). The PCF-like gene from Amborella trichopoda (AtrTCP4) as well as all other PCF-like sequences were used as the outgroup. To find the molecular evolution model that best fit our data, we used the jModelTest package implemented in MEGA6 (Posada and Crandall, 1998). Trees were observed and edited using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) (Rambaut, 2014). Newly isolated sequences from our own generated transcriptomes from C. trianae (Orchidaceae) and H. decumbens (Hypoxidaceae) can be found under Genbank numbers KY296315–KY296347. All sequences included in the phylogenetic analyses can be found in the Supplementary Table 1.
Identification of new protein motifs
In order to detect previously reported, as well as to identify new, conserved motifs, 77 TCP-like genes were selected representing major model eudicot and monocot groups of this study (i.e., A. majus, O. sativa, Aloe vera, P. equestris, C. trianae and H. decumbens). Sequences were permanently translated and uploaded as amino acids to the online MEME server (http://meme-suite.org/tools/meme) and run with all the default options (Bailey et al., 2006). Specific analyses for each of the TCP clades (i.e., CYC-like, CIN-like, PCF-like) were also performed.
Expression analyses by RT-PCR
To examine and compare the expression patterns of TCP-like genes we used floral buds, dissected floral organs, leaves, and fruits of C. trianae and H. decumbens. Preanthetic floral buds of H. decumbens were dissected into sepals, petals, stamens and carpels. In addition whole floral buds, inmature fruits (F1, right after tepals shed off), mature fruits (F2, before lignification), and young leaves were also collected. Preanthetic floral buds of C. trianae were dissected into sepals, lateral petals, labellum (or lip), gynostemium, and ovary. Young leaves were also collected. Total RNA was isolated from each organ collected using the SV Total RNA Isolation System kit (Promega, Madison, WI, USA), and resuspended in 20 μl of DEPC water. RNA was treated with DNAseI (Roche, Basel, Switzerland) and quantified with a NanoDrop 2000 (Thermo Scientific, Waltham, MA) (Wilfinger et al., 1997). Three Micrograms of RNA were used as a template for cDNA synthesis (SuperScriptIII RT, Invitrogen) using OligodT primers. The cDNA was diluted 1:4 for amplification reactions by RT-PCR. Primers were designed in specific regions like portions flanking the conserved domains for each copy found in C. trianae and H. decumbens (Supplementary Table 2). Each amplification reaction incorporated 9 μl of EconoTaq (Lucigen, Middleton, WI), 6 μl of nuclease free water, 1 μl of BSA (5 μg/ml), 1 μl of Q solution (5 μg/μl) 1 μl fwd primer (10 mM), 1 μl rev primer (10 mM), and 1 μl of template cDNA for a total of 20 μl. Thermal cycling profiles followed an initial denaturation step (94°C for 30 s), an annealing step (50–59°C for 30 s) and an extension step with polymerase (72°C for 10 min), all by 30–38 amplification cycles. ACTIN2 was used as a load control. PCR products were run on a 1.0% agarose gel stained with ethidium bromide and digitally photographed using a Whatman Biometra® BioDoc Analyzer.
Results
Exhaustive search from available databases retrieved 452 TCP Class I and Class II sequences from flowering plants. From these 138 belong to the Orchidaceae, including 18 homologs from C. trianae; and 110 sequences belong to non-Orchidaceae Asparagales, including 15 homologs from H. decumbens (Supplementary Table 1).
ML analyses were performed using the complete nucleotide sequences of all TCP-like genes isolated. The A. trichopoda AtrTCP4 together with all other isolated PCF-like genes were used as the outgroup. The analysis recovered two clades previously reported in TCP genes, namely the CYC/TB1-like clade (with a Bootstrap Support, BS = 98) and the CIN-like clade (BS = 67) (Figure 1). We will discuss our results for each clade separately.
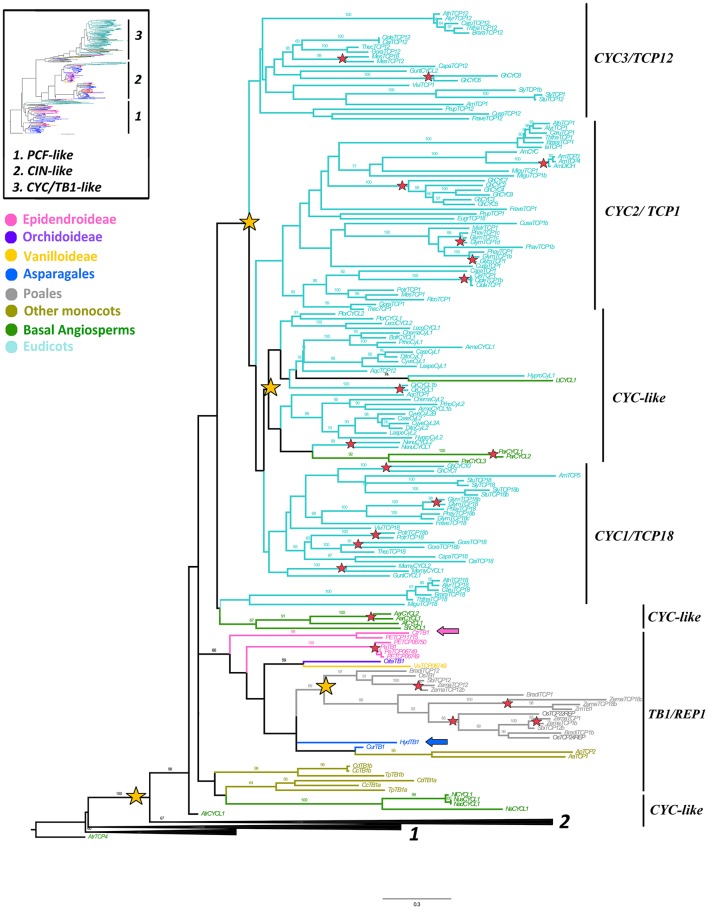
Figure 1.
ML analysis of CYC/TB1-like genes. Overview (upper left) summary Maximum Likelihood (ML) analysis of all TCP-like genes, where (1) corresponds to PCF-like homologs, (2) to the CIN-like clade and (3) to the CYC/TB1-like clade. Detailed (right) gene tree expanded in the CYC/TB1-like clade (3); yellow stars indicate large-scale duplication events in core, basal eudicots and monocots; resulting in the core eudicot clades CYC3/TCP12, CYC2/TCP1 and CYC1/TCP18, the basal eudicot CYC-like clades and the TB1 and REP1 clades in Poales; red stars indicate species-specific duplication events; blue and pink arrows indicate H. decumbens and C. trianae homologs, respectively; branch and taxa colors correspond to those in the overview tree to the upper left. BS values ≥ 50 are shown.
CYC/TB1-like gene evolution
We were able to isolate 168 sequences belonging to the CYC/TB1-like clade (Supplementary Table 1). Our sampling includes 15 sequences from four species of Poales, 10 sequences from seven species of Asparagales (incl. Orchidaceae), eight from five species of Commelinales (Commelina, Alstroemeria, Tradescantia), 14 from nine species of basal angiosperms and 121 from 46 species of eudicots. Only one homolog from Curculigo spp. (CurTB1) and one homolog from H. decumbens (HydTB1) were recovered from our blast searches. Moreover, an exhaustive search in Orchidaceae specific databases resulted in eight additional copies: three from P. equestris (PETCP06750, PETCP06749, PETCP11715), two from P. aphrodite (PaTB1, PaTCP06749) and one from C. trianae (CtrTB1) (Epidendroideae); one homolog from O. italica (OitaTB1) (Orchidoideae); and one homolog from Vanilla shenzhenica (VaTCP06749) (Vanilloideae). CYC/TB1-like homologs seem to have undergone size reduction in Asparagales, and only the searches made with monocot TB1 genes yielded positive hits. Interestingly, in the two transcriptomes newly obtained in the present research, the CYC/TB1-like contigs were supported by fewer reads (when compared to MADS-box APETALA3 floral organ identity genes and other TCP-like genes; Table 1) suggesting low expression of these transcripts.
The resulting ML topology recovered the three previously established core eudicot subclades (Howarth and Donoghue, 2006) with very low support (BS < 50), namely CYC1/TCP18, CYC2/TCP1, and CYC3/TCP12 (Figure 1). Our analysis also recovers the previously identified duplication of CYC-like genes in basal eudicots (Citerne et al., 2013), and another in Poales, the latter resulting in the REP1/TB1 clades (Mondragón-Palomino and Trontin, 2011). Most basal angiosperms and many monocots outside Poales have single copy CYC-like genes that predate the independent duplications in eudicots and Poales (Figure 1). Intraspecific duplications in monocots have occurred in Zea mays, as well as in the orchids P. equestris and P. aphrodite (Figure 1; BS = 100). Outside of the monocots, local specific duplications have also occurred in the basal angiosperms Aristolochia ringens and Persea americana, in the basal eudicots Circaeaster and Nelumbo, and in core eudicots such as Gerbera, Antirrhinum, Citrus, Glycine, Populus, and Gossypium (Figure 1).
Members of the CYC/TB1-like clade show very little variation in the ca. 60 amino acid TCP domain (sensu Cubas et al., 1999) consisting of a putative basic-Helix-Loop-Helix (bHLH) domain (Figure 2). We were able to identify the highly conserved residues previously reported for the putative bipartite Nuclear Localization Signal (NLS) at the N-terminus flanking the bHLH, which provide hydrophobicity in the α-helices and in the loop region between the two. The loop itself is highly conserved in all CYC/TB1-like sequences except for AmDICH and AmCYC that have an A > P change at position 42. The second helix contains the LxxLL motif in all CYC2 proteins; this motif is modified into a VxWLx motif in other CYC-like proteins.
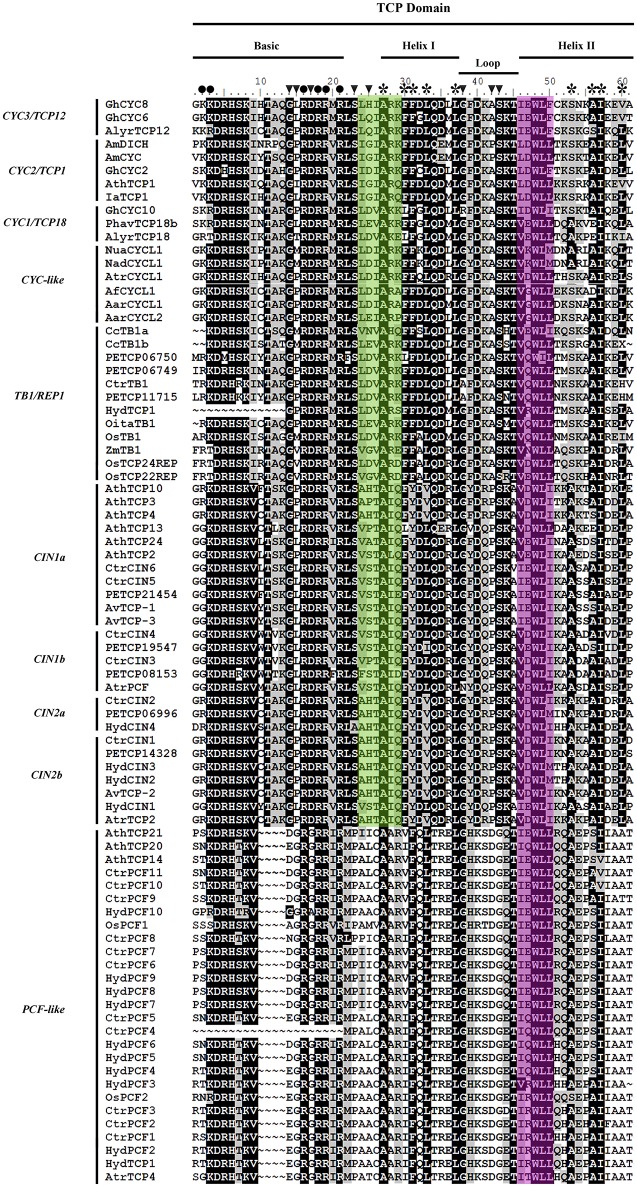
Figure 2.
TCP protein domain alignment with Asparagales representative sequences. Oryza sativa (Poales) was used for reference. Names to the left indicate the clade to which sequences belong according to Figures 1, 4, 5 and Supplementary Table 1. The upper bars point to the putative structure bHLH (basic-Helix-Loop-Helix) at the TCP domain. Circles indicate residues forming part of the putative bipartite NLS; asterisks indicate conserved hydrophobic residues in the helices; black arrowheads point to residues (glycine or proline) that disrupt α-helix formation (modified following Cubas et al., 1999). The green box indicates changes in residues between CYC-like and CIN-like proteins. The pink box indicates the LxxLL motif with significant variations outside the CYC2 clade.
Our MEME analysis identified motifs 1 and 2 corresponding to the TCP domain (Supplementary Figure 1). At the start of Helix I, between positions 24–29 we found specific amino acids exclusive to CYC protein homologs. Outside the TCP domain, motifs 7 and 10 (reported also by Bartlett and Specht, 2011) and motifs 36–40, 42, and 43 (reported also by De Paolo et al., 2015) were recovered in our analysis as conserved in all CYC proteins. In addition, the protein interaction R domain (motif 11) putatively involved in hydrophilic α-helix formation in TCP Class II genes (shared between CYC and CIN proteins) was also identified (Supplementary Figure 1; Cubas et al., 1999). However, this motif is absent from CtrTB1 and REP-1 (Supplementary Figure 1; Yuan et al., 2009). Previously unidentified motifs include motif 4, exclusive to Epidendroideae and motif 5, exclusive to Phalaenopsis species. Whereas, most orchid CYC-like proteins (including O. italica OitaTB1 and C. trianae CtrTB1) do not share any common motifs with the canonical A. majus paralogs outside the TCP domain, the Phalaenopsis CYC-like homolog (PETCP11715) shares motifs 6, 9, and 12 with AmDICH or AmCYC.
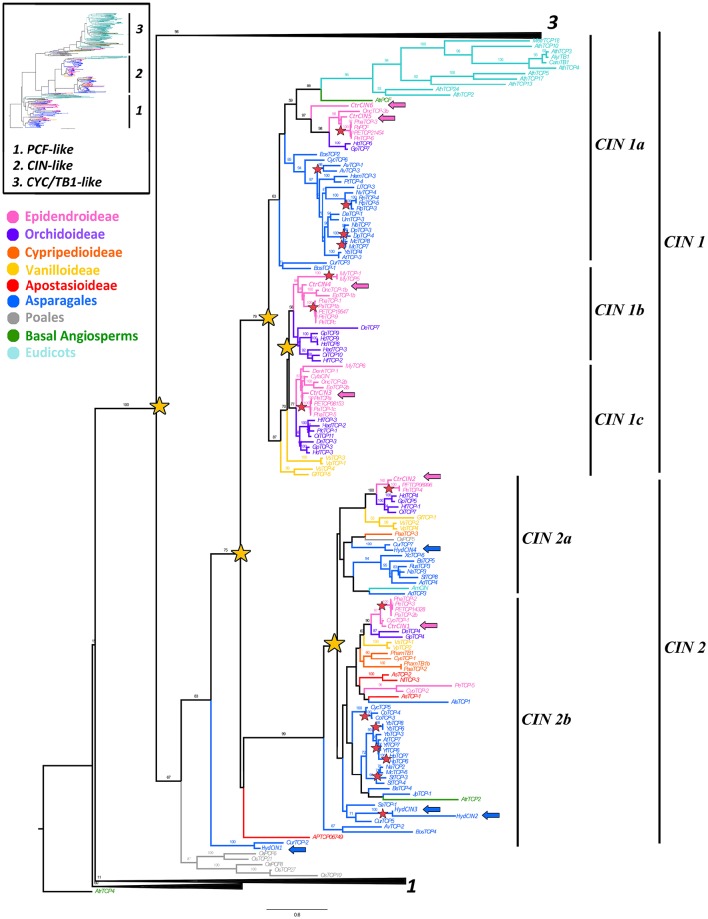
CIN-like gene evolution
A total of 155 CIN-like homologs were recovered and unlike the CYC-like sampling, most CIN-like sequences belong to the Asparagales (Supplementary Table 1). Our sampling contains 57 sequences from 35 non-Orchidaceae Asparagales species, including four paralogs from H. decumbens labeled HydCIN1-HydCIN4. A total of 78 CIN-like homologs were isolated, including four homologs from two Apostasioideae species, nine homologs from three Vanilloideae species, five homologs from three Cypripedioideae species, 22 homologs from seven Orchidoideae species and 38 homologs from 11 Epidendroideae species. Furthermore, six paralogs were identified in C. trianae labeled CtrCIN1-CtrCIN6. Searches outside Asparagales were restricted to six homologs from O. sativa, two from A. trichopoda and 12 homologs from five eudicots species, including the canonical A. majus CINCINNATA (AmCIN).
The CIN-like ML analysis shows a duplication (BS = 75) that predates the diversification of angiosperms resulting in the CIN1 (BS = 79) and CIN2 clades (BS = 99) (Figure 3). This is confirmed by the position of the two A. trichopoda CIN paralogs, AtrPCF and AtrTCP2, each in its own clade (Figure 3). Additional support for this early duplication is found in the topology yielded by a second complementary analysis that includes 11 Solanaceae CIN homologs, where CIN1 and CIN2 clades have monocot and core eudicot representatives and, at least CIN2 is well supported (BS = 93) (Supplementary Figure 2). The CIN1 clade has undergone at least two additional duplications resulting in the CIN1a-c clades. It is likely that the duplication resulting in CIN1a and CIN1b/c occurred exclusively in monocots, although the exact timing is unclear. The other duplication resulting in CIN1b and CIN1c appears to be Orchidaceae-specific, prior to the diversification of Orchidoideae and Epidendroideae (Figure 3). On the other hand, the CIN2 clade underwent an independent duplication predating the diversification of Asparagales, resulting in CIN2a and CIN2b subclades. Intraspecific duplications were identified in Hesperaloe, Disporopsis, Maianthemum, Rhodophiaia, Sansevieria and Yucca, (Figure 3). Poales CIN homologs form a clade, with a low BS, in the first analysis, with the exception of OsPCF5 clustered with AmCIN (Figure 3). However, our second analysis shows two Poales clades nested in each angiosperm paralogous CIN1 and CIN2 clades (with low BS), suggesting that the two Poales clades likely resulted from the angiosperm CIN1/2 duplication (Supplementary Figure 2).
Figure 3.
ML analysis of CIN-like genes. Overview (upper left), summary tree as in Figure 1. To the right, ML phylogenetic analysis of TCP genes expanded to show the CIN-like clade (2). Yellow stars indicate large scale duplication events: one prior to the diversification of angiosperms yielding clades CIN1 and CIN2, two before the radiation of Asparagales and one prior to the origin of Epidendroideae and Orchidoideae; red stars indicate species-specific duplication events; blue and pink arrows indicate H. decumbens and C. trianae homologs, respectively. Branch and taxa colors correspond to those in the conventions to the left. BS values ≥ 50 are shown.
CIN-like sequences show high degree of conservation at the N-flank of the TCP domain (Figure 2). The only changes with respect to the key aminoacids in the bHLH domain in CYC proteins are at the second helix where the LxxLL motif shifts to V/IxxLL (Figure 2). Toward the 3′ end of the TCP domain proteins are highly variable, except for motifs 13, 14, 16–18 and 21, reported also by De Paolo et al. (2015) (Supplementary Figure 1). The R domain (motif 11) in CIN proteins is only present in the CIN1a clade and O. sativa homologs OsTCP21, OSTCP8 and OsTCP10. Motif 14, which corresponds to the miR319 binding site is present in most CIN-like sequences. The miR319 binding motif is lacking in HydCIN2, HydCIN3, OsTCP21, OsTCP27, and OsTCP10. All CIN1 subclade sequences share motifs 13, 17, 19, 20 21, and 25; the CIN1a subclade shares motifs 11, 28, 29, 34 and 35, while the CIN1b subclade shares motifs 15, 23, 26, and 30. Synapomorphies for the CIN2 subclade include motifs 16, 18, 22, 24 and only motif 27 is exclusive to Orchidaceae. Finally, CIN2b homologs share motifs 32 and 33. The most divergent C. trianae sequence is CtrCIN6, which only has the motifs 1, 2, 3, 11, 13, 14, 17, and 21.
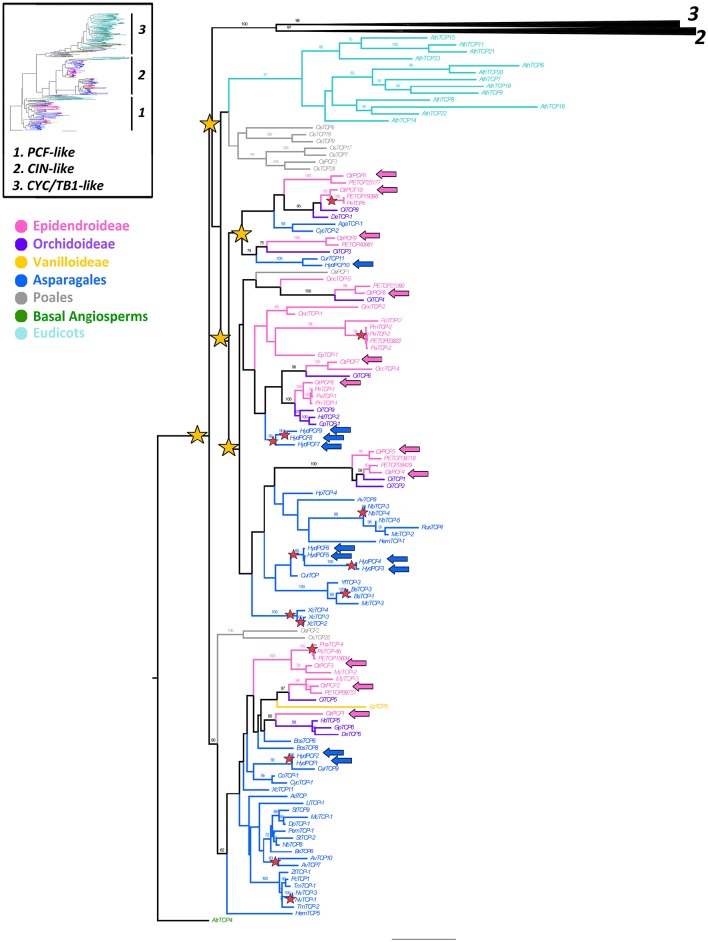
PCF-like gene evolution
Our analysis recovered 129 PCF-like homologs (Supplementary Table 1). Similarly to CIN-like genes most sampling is concentrated in Asparagales, thus 53 homologs belong to 35 species of non-Orchidaceae Asparagales and 52 sequences correspond to Orchidaceae. H. decumbens has 10 PCF-like copies (HydPCF1–HydPCF10). Sampling in Orchidaceae includes one homolog from one Vanilloideae species, 14 homologs from seven Orchidoideae species and 37 homologs from 11 Epidendroideae species. We recovered 11 PCF-like homologs from C. trianae (CtrPCF1–CtrPCF11). Sampling in monocots outside Asparagales include 10 homologs from O. sativa (Poales) and sequences that are not monocots are restricted to one homolog from A. trichopoda and 13 homologs from A. thaliana.
Our analysis detected at least five duplication events of PCF-like prior to the diversification of Asparagales, however support is low for all clades (BS = < 50) (Figure 4). In addition, our complementary analysis including 18 Solanaceae PCF-like genes also shows support for at least two rounds of core eudicot specific PCF-like duplications (Supplementary Figure 2).
Figure 4.
ML analysis of PCF-like genes. Overview (upper left) summary tree as in Figure 1. To the right, ML phylogenetic analysis of TCP genes expanded to show the PCF-like clade (1); yellow stars indicate large scale duplication events at least three before the diversification of Asparagales; red stars indicate species-specific duplication events; blue and pink arrows indicate H. decumbens and C. trianae homologs, respectively. Branch and taxa colors correspond to those in the conventions to the left. BS values ≥ 50 are shown.
PCF-like sequences exhibit the shortest basic motif in TCP proteins, with a deletion in the bipartite NLS between positions 10 and 13 (Figure 2; Cubas et al., 1999). The TCP domain shows little conservation in comparison to the TCP II class (CYC and CIN) proteins. For instance, there is a four amino acid deletion in the middle of the basic motif, and only 12 out of the 23 amino acids characterized in both helices and the loop are conserved (Figure 2). Additionally our MEME analysis show that PCF-like proteins do not have an R domain (motif 11), nor a target sequence for miR319 (motif 14) (Supplementary Figure 1).
Expression of TCP-like homologs from Hypoxis decumbens and Cattleya trianae
In order to hypothesize functional roles for the Asparagales TCP-like homologs, the expression patterns of all homologs isolated from transcriptomic analysis in H. decumbens and C. trianae were evaluated (Figure 5). Although in both species we were able to dissect floral organs in preanthesis and young leaves, we were not able to find fruits of C. trianae, thus only young and old fruits of H. decumbens were included in the expression study.
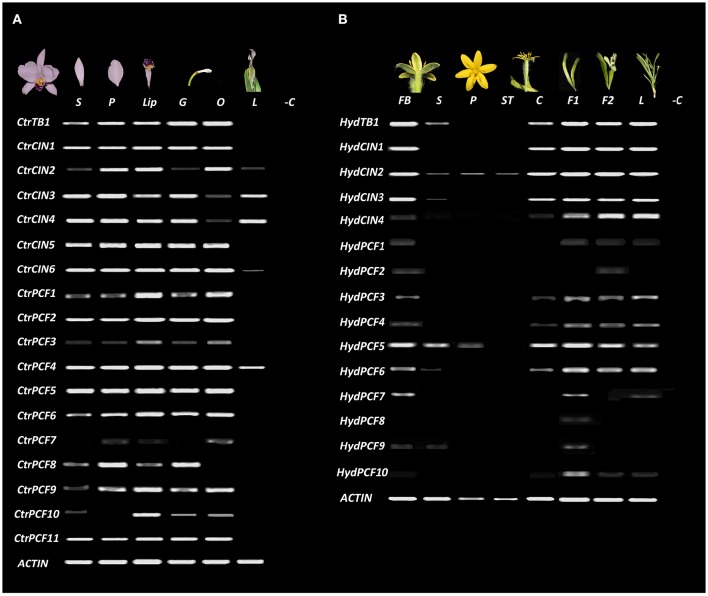
Figure 5.
TCP-like expression analysis in Asparagales. ACTIN was used as positive control. (A) Expression of all isolated TCP-like transcription factors in dissected floral organs and leaves of C. trianae (Orchidaceae). (B) Expression of all isolated TCP-like transcription factors in dissected floral organs, fruits and leaves of H. decumbens (Hypoxidaceae). BF, flower bud; C, carpels; F1, immature fruit; F2, mature fruit; G, gynostemium; L, Leaves; Lip, lip; O, Ovary; P, petals; S, sepals; ST, stamens. −C Indicates the amplification reaction of PCR without cDNA (negative control).
The CYC/TB1-like homologs have very different expression patterns in C. trianae and H. decumbens. HydTB1 is expressed in the floral bud, sepals, carpels, young and mature fruits and leaves, whereas CtrTB1 is expressed in all floral whorls and is not expressed in leaves (Figure 5). CIN-like homologs also exhibit different expression patterns in C. trianae and H. decumbens. HydCIN1, HydCIN2 and HydCIN3 are expressed in the floral bud, carpels, fruits and leaves. Only HydCIN2 expression is extended to sepals, petals and stamens at very low levels. Interestingly both HydCIN2 and HydCIN3 lack the miR319 binding site. HydCIN4 expression is restricted to fruits and leaves. CtrCIN1, CtrCIN2, CtrCIN5, and CtrCIN6 are expressed in sepals, petals, lip, gynostemium, and ovary, although, CtrCIN2 expression in sepals and gynostemium occurs at low levels. CtrCIN3 and CtrCIN4 have similar expression patterns to CtrCIN1, however, they are poorly expressed in the ovary and are the only CIN-like homologs that extend their expression to leaves.
Similar to CYC/TB1-like and CIN-like genes, the expression of PCF-like homologs varies dramatically between C. trianae and H. decumbens (Figure 5). CtrPCF1, CtrPCF2, CtrPCF3, CtrPCF4, CtrPCF5, CtrPCF6, CtrPCF9, and CtrPCF11 are expressed in all floral organs and the only copy with expression in leaves is CtrPCF4. CtrPCF3 has low expression in sepals and petals. CtrPCF7 is only expressed in petals and ovary. Finally, CtrPCF10 has expression in all floral whorls except in petals. In H. decumbens only HydPCF3, HydPCF4, HydPCF5, and HydPCF6 are expressed in the floral buds, carpels, fruits and leaves, although HydPCF3 and HydPCF4 have low expression in carpels. HydPCF5 is also expressed in the perianth with higher expression in sepals than in petals. HydPCF7 has restricted expression to the floral buds and the young fruits. HydPCF9 and HydPCF10 are only expressed in young fruits and the remaining copies (HydPCF1, HydPCF2, and HydPCF8) are only expressed at very low levels in the floral buds.
Discussion
Most functional studies on TCP genes have focused on identifying their contribution to floral symmetry and plant architecture, as expected by the functions of the canonical CYC and DICH from A. majus and AtTCP1 from A. thaliana respectively (Luo et al., 1996, 1999; Costa et al., 2005). Studies on the evolution of TCP transcription factors have concentrated on core eudicots and particularly on CYC2 homologs (Damerval and Manuel, 2003; Howarth and Donoghue, 2006; Preston and Hileman, 2009; Martín-Trillo and Cubas, 2010; Mondragón-Palomino and Trontin, 2011; Sarvepalli and Nath, 2011; Danisman et al., 2012, 2013; Uberti-Manassero et al., 2012; Aguilar-Martínez and Sinha, 2013; Das Gupta et al., 2014; Lin et al., 2016). Our matrix includes sampling from the Phalaenopsis genome as well as all transcriptomes available for Asparagales. The phylogenetic analysis made with the full-length coding sequences, allowed us to identify a number of large scale as well as local TCP gene duplications and changes in protein sequences linked to these duplications. Moreover, this is the first large scale analysis of CIN-like and PCF-like genes. We are able to report a comparative expression pattern in two Asparagales species, representing the two floral groundplans in the order, and present hypotheses on the putative role of these genes in floral patterning in representative Asparagales.
Asparagales CYC/TB1 homologs are found predominantly as single copies and have divergent expression patterns in Hypoxis decumbens and Cattleya trianae
Our study detected a single copy CYC/TB1 gene in each of the species investigated in the Asparagales. The Asparagales CYC/TB1 homologs fall outside of the Poales (TB1/REP clades) or Commelinales identified local duplications, described before and recovered here (Doebley et al., 1995, 1997; Yuan et al., 2009; Mondragón-Palomino and Trontin, 2011). The tree topology suggests that independent duplications have occurred in CYC-like genes in the monocots (Yuan et al., 2009; Mondragón-Palomino and Trontin, 2011; Hileman, 2014). Species-specific duplications in Asparagales were found only in Phalaenopsis (Orchidaceae). Moreover, expression data of CYC/TB1 genes in Asparagales show remarkable differences between H. decumbens and C. trianae, even though sampled organs correspond to fairly well developed tissues. CtrTB1 is expressed homogeneously in all floral whorls while HydTB1 is expressed predominantly in carpels, fruits and leaves (Figure 5). Homogeneous expression of CYC-like genes in dorsal and ventral floral organs in C. trianae suggest that CtrTB1 is likely not playing important roles in maintenance of bilateral symmetry in orchid flowers (see also Horn et al., 2015). However, only pre-anthethic floral buds were sampled and earlier stages are needed to test whether CtrTB1 can be contributing to the establishment of bilateral symmetry in the flower primordia. Comparative expression studies of CYC/TB1-like genes in other Orchidaceae point to significant variations. For instance, while the O. italica homolog OitaTB1 is only expressed in leaves (De Paolo et al., 2015), two CYC/TB1 genes in P. equestris do exhibit differential dorsiventral expression in the floral bud suggesting species specific roles in bilateral symmetry establishment (Lin et al., 2016).
Expression of CYC/TB1 genes in Orchidaceae contrasts with studies in Commelinales, Zingiberales, and Alstroemeriaceae that show differential dorsiventral expression of CYC/TB1 genes and hence support convergent recruitment of CYC homologs in the acquisition of bilateral symmetry in different monocots (Bartlett and Specht, 2011; Preston and Hileman, 2012; Hoshino et al., 2014). Within core eudicots, only the CYC2/TCP1 clade members have been linked to shifts toward bilateral floral symmetry. This has been extensively documented for CYC and DICH in A. majus (recent paralogs within the CYC2/TCP1 clade) which are expressed in the dorsal regions of the floral meristems and negatively regulate cell proliferation (Luo et al., 1996, 1999). However, many other CYC2 orthologs have been shown to control bilateral symmetry in Asterales, Brassicales, Dipsacales, Fabales, Lamiales, and Malpighiales (Busch and Zachgo, 2007; Gao et al., 2008; Preston et al., 2009; Wang et al., 2010; Zhang et al., 2010, 2013; Howarth et al., 2011; Tähtiharju et al., 2012; Yang et al., 2012; Ma et al., 2016).
Recruitment of CYC2 homologs in bilateral symmetry is likely facilitated by conserved protein-protein interactions mediated by the LxxLL motif (Heery et al., 1997; Damerval and Manuel, 2003; Reeves and Olmstead, 2003; Howarth and Donoghue, 2006; Li et al., 2009; Preston et al., 2009; Tähtiharju et al., 2012; Parapunova et al., 2014; Ma et al., 2016). If so, it is possible that different CYC-like proteins can form specific homo- and heterodimers, or even have unique partners, and thus protein motifs can provide clues to protein affinity and functional specificity (Kosugi and Ohashi, 2002). Our MEME analysis rescues motifs 6, 9 and 12, shared only between P. equestris PETCP11715 and the A. majus AmCYC and AmDICH, which are not present in other orchid CYC/TB1 proteins (i.e., C. trianae and O. italica) and hence putatively involved in establishing early bilateral floral symmetry in some Orchidaceae species (Supplementary Figure 1). When full length CtrTB1 is compared to the three P. equestris CYC/TB1 proteins very little similarity is detected (PETCP11715–0.38; PETCP06750–0.22; PETCP06750–0.24); for instance, CtrTB1 lacks motif 11 (R-domain). Nevertheless, the TCP domain is extremely conserved (0.91, 0.80, and 0.84 respectively). These results suggest that besides the key bHLH amino acids that target conserved genes, there are likely important motifs in the flanking regions allowing unique interactions and downstream partners in each species.
Conversely, the expression of HydTB1 in H. decumbens is indicative of exclusive roles in carpels, fruits and leaves. This data suggests that HydTB1 may have similar roles to other CYC genes that do not participate in establishing floral bilateral symmetry. Such is the case of the A. thaliana, AtTCP1, which promotes shoot growth and regulates leaf lamina size (Costa et al., 2005; Guo et al., 2010; Koyama et al., 2010b). To date less attention has been given to the putative role of CYC genes in the development of leaves, carpels and fruits, despite the fact that expression has been detected in these organs in other core eudicots. This is the case for CYC homologs in Citrullus, Gerbera, Gossypium, Lotus, Solanum, and some Papaveraceae that have been detected in seedlings, young leaves, and immature fruits (Damerval et al., 2007; Wang et al., 2010; Parapunova et al., 2014; Ma et al., 2016; Shi et al., 2016).
CIN-like genes have undergone numerous duplications in angiosperms, monocots and orchidaceae and show broad expression patterns in Cattleya trianae when compared to Hypoxis decumbens
Here we show the first comprehensive phylogenetic analysis of CIN-like genes. Our results point to the occurrence of at least one duplication event predating angiosperm diversification, at least one duplication occurring prior to the origin of the Asparagales and one specific duplication prior to the diversification of Orchidoideae and Epidendroideae (Figure 3, Supplementary Figure 2; Floyd and Bowman, 2007; Martín-Trillo and Cubas, 2010). By comparison to CYC genes, CIN genes functional characterization is restricted to model species only. The canonical CINCINNATA in A. majus has dual roles in limiting the growth of leaf margins while promoting epidermal cell differentiation in petals (Nath et al., 2003; Crawford et al., 2004). The cin mutant in A. majus, as well as the tcp4 mutant in A. thaliana, exhibit curly leaves as a result of excessive growth in leaf margins (Crawford et al., 2004; Koyama et al., 2010a). CIN regulates leaf shape through direct or indirect negative regulation of the boundary CUP-SHAPED COTYLEDON (CUC) genes, likely via the activation of ASSYMETRIC LEAVES 1 (AS1), miR164, INDOLE-3-ACETIC ACID3/SHORT HYPOCOTYL2 (IAA3/SHY2), and SMALL AUXIN UP RNA (SAUR) (Koyama et al., 2007, 2010a). CIN also controls leaf development through the regulation of cell proliferation by activating miR396, CYCLIN-DEPENDENT KINASE INHIBITOR/KIP RELATED PROTEIN 1 (ICK1/KRP1) and jasmonate biosynthesis (Schommer et al., 2014). Similarly, AtTCP4 regulates the transition between cell proliferation and differentiation, controlling cytokinin and auxin receptors (Efroni et al., 2013; Das Gupta et al., 2014). AtTCP4 also controls leaf senescence, maintains petal growth, and regulates early embryo development and seed viability, and finally it regulates jasmonic acid (JA) biosynthesis by the activation of LIPOXIGENASE2 (LOX2) (Schommer et al., 2008; Nag et al., 2009; Sarvepalli and Nath, 2011; Danisman et al., 2012). Leaf patterning is also controlled by CIN-like homologs in tomato and rice (Ori et al., 2007; Yang et al., 2013; Zhou et al., 2013; Ballester et al., 2015).
In addition to the roles of CIN genes in leaf patterning, other functions in carpel and fruit development have been identified. AtTCP2 and AtTCP3 are known to activate NGATHA genes that regulate carpel apical patterning. NGA orthologs from A. thaliana, rice, tomato and bean have conserved putative TCP binding site suggesting that this regulation is conserved in monocots and dicots (Ballester et al., 2015). Moreover, the tcp3 mutant has shorter siliques with a crinkled surface (Koyama et al., 2007; Ballester et al., 2015). Additionally, expression analyses in different Solanaceae species suggest an important contribution of these genes to fruit development and maturation, as they are downstream targets of key ripening regulators including RIPENING INHIBITOR (RIN), COLORLESS NON-RIPENING (CNR) and SlAP2a (Supplementary Figure 3; Crawford et al., 2004; Parapunova et al., 2014). More recently PeCIN8, a P. equestris CIN-like homolog, was shown to be broadly expressed and to have roles in leaf cell proliferation, determining the fruit final size and controlling proper embryo and ovule development (Lin et al., 2016). In summary, CIN-like genes are pleiotropic regulators of cell division and differentiation in leaves, petals, carpels, ovules, fruits and seeds across angiosperms.
This study identified significant changes in the helix I residues and the loop between Orchidaceae sequences and other Asparagales CIN-like proteins (Figure 2). For instance, a number of motifs involved in protein interaction, including the R domain characteristic of class II TCP proteins were only identified in the CIN1a clade, but are absent in all other paralogous clades (Supplementary Figure 1, Cubas et al., 1999; Damerval and Manuel, 2003). Here we have also identified conserved motifs like the miR319 binding site, previously reported in CIN-like homologs from O. italica (Nag et al., 2009; De Paolo et al., 2015), for all CIN-like sequences in Asparagales. This indicates, that Asparagales homologs are also regulated by miR319, similar to AtTCP4, and other CIN-like Arabidopsis paralogs, including AtTCP2/3/10 and 24 (Palatnik et al., 2003; Koyama et al., 2007; Schommer et al., 2008, 2012; Koyama et al., 2010a; Danisman et al., 2012).
In comparison with reported functional data from model eudicots and monocots, the CIN-like differential expression observed in C. trianae and H. decumbens homologs points to three testable hypotheses. (1) While in H. decumbens all CIN-like genes are expressed in young leaves and may play roles in leaf development, only two of the six paralogs identified in C. trianae (CtrCIN3/4) are likely involved in cell division and differentiation during leaf development, similar to the P. equestris PeCIN8 (Lin et al., 2016). (2) While CIN-like genes are likely playing key roles in both perianth and fertile organs development and growth in C. trianae, their contribution to perianth development and growth is less clear in H. decumbens, perhaps only with HydCIN2 involved in cell proliferation in the perianth. (3) In both species CIN-like genes are strongly expressed in carpels and fruits, suggesting that their role in carpel patterning, ovule development as well as fruit maturation is likely conserved in Asparagales. Nevertheless, mRNA expression data for all CIN-like genes must be interpreted with caution given the putative conserved miR139 postranscriptional regulation.
PCF-like genes are extensively duplicated and have overlapping expression patterns with CIN-like genes in asparagales
Our results on the evolution of PCF-like genes points to numerous duplication events within Asparagales in comparison to the CYC-like and the CIN-like clades. In addition, the topology recovered suggests that PCF-like gene duplications in monocots are independent from the one that occurred in core eudicots (Figure 4, Supplementary Figure 2). Functional data available for PCF-like genes suggest redundancy with CIN-like genes (Aguilar-Martínez and Sinha, 2013; Danisman et al., 2013). For instance, both PCF-like and CIN-like genes control leaf development through regulation of LOX2. However, while AtTCP20 (PCF-like) inhibits, AtTCP4 (CIN-like) induces the expression of LOX2 (Danisman et al., 2012). PCF-like genes are also involved in the regulation of circadian clock genes, that is the case of CCA1 Hiking Expedition CHE (AtTCP21) (Pruneda-Paz et al., 2009; Giraud et al., 2010). In addition dimers formed between AtTCP15 and other TCP-like proteins (AtTCP2, AtTCP3, AtTCP11) are known to regulate circadian cycles, cell proliferation in floral organs and leaves, and to promote seed germination (Koyama et al., 2007; Kieffer et al., 2011; Resentini et al., 2015). PCF-like genes are also expressed in ovule, seed and fruit development in Solanum lycopersicum, Solanum tuberosum, O. sativa and in P. equestris, suggesting that they mediate cell proliferation in the carpel to fruit transition in both eudicots and monocots (Supplementary Figure 3; Kosugi and Ohashi, 1997; Yao et al., 2007; Parapunova et al., 2014; Lin et al., 2016).
Expression detected here of PCF-like homologs in C. trianae and H. decumbens show broad expression of most paralogs in C. trianae (except CtrPCF7) contrasting with a restricted expression of most paralogs in H. decumbens (except HydPCF5; Figure 5). Such expression patterns are in accordance with all putative functions identified in other monocots and in core eudicots including, but not restricted to, cell proliferation control in leaves, carpels, ovules, fruits and seeds (Lin et al., 2016). All data available point to pleiotropic roles of TCP-like genes with a high degree of redundancy among paralogs. Interestingly, when compared to CYC-like and CIN-like genes, the PCF-like gene HydPCF5 in H. decumbens is more likely to be playing cell proliferation roles in the perianth.
In conclusion, the Asparagales, unlike core eudicot model plants, have a reduction in the number of CYC-like homologs and an increase of CIN-like and PCF-like copies. Characteristic key amino acids in the bHLH domain, flanking motifs and binding sites are for the most part conserved in Asparagales CIN-like and PCF like proteins suggesting similar conserved mechanisms of post-transcriptional regulation and interacting partners. The most notorious exception to this is the lack of a miR319 binding site in HydCIN2/3. Nevertheless, the CYC-like proteins in Asparagales seem to be poorly expressed and have undergone important shifts in protein domains, suggesting changes in regulation as well as in protein-protein interactions. The orchid included in this study has similar numbers of TCP copies when compared to Asparagales with radially symmetrical flowers, and only the Epidendrioideae and Orchidoideae seem to have an additional CIN paralog lacking in other Orchidaceae and Asparagales. Expression data suggests different roles of TCP-like genes in C. trianae and H. decumbens, pointing to: (1) a possible decoupling of TB1 homologs from bilateral symmetry in C. trianae; (2) conserved roles of CIN-like and PCF-like genes in the control of cell proliferation in carpels, ovules and fruits in both species; and (3) preferential leaf expression of CIN-like and PCF-like genes in H. decumbens and perianth expression in C. trianae. Here we have performed the first large scale analysis of TCP genes in Asparagales which will provide a platform for in depth comparative expression analyses as well as much needed functional studies of these genes in emerging model orchids.
Author contributions
YM and NP planned and designed the research. YM and NP conducted fieldwork. YM, JA, and NP performed experiments. YM, JA, and NP analyzed the data and wrote and approved the final version of the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the recognition obtained by the Premio Fomento a la Investigación, Alcaldia de Medellín, 2016. We thank the OneKP repository staff (University of Alberta), the Orchidstra staff (Academia Sinica) and Orchidbase staff (National Cheng Kung University) for facilitating access to the online databases. We thank Favio González (Universidad Nacional de Colombia) and Mariana Mondragón-Palomino (Universität Regensburg) for their comments to the manuscript and to preliminary results presented in meetings, respectively. We thank Ricardo Callejas (Universidad de Antioquia) for allowing us to use office and laboratory space and Cecilia Zumajo-Cardona (The New York Botanical Garden) for laboratory assistance. We thank Victor Acosta, Francisco Villegas and the staff at Vivero Sol Rojo for maintaining cultivated plant material of C. trianae. Finally we thank two reviewers for their insightful comments on the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00009/full#supplementary-material
Conserved motifs in and Orchidaceae and non-Orchidaceae Asparagales TCP-like proteins. Model core eudicots and monocots used as reference include Arabidopsis thaliana, Antirrhinum majus, and Oryza sativa. Motifs 1, 2, and 3 correspond to the conserved TCP domain. Motif 11 indicates the characteristic R domain in Class II TCP-like genes. Motif 14 corresponds to the miR319 binding site in CIN-like genes.
ML analysis of TCP-like genes with extended Solanaceae sampling. ML phylogenetic analysis of TCP-like genes with reduced sampling including only model organisms like Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum, Solanum tuberosum, two Orchidaceae species, Cattleya trianae, Orchis italica, one non-Orchidaceae Asparagales, Hypoxis decumbens and the early diverging angiosperm Amborella trichopoda. Yellow stars indicate large scale duplication events. TCP major clades are labeled to the right. Branch and taxa colors correspond to those in the conventions. BS values ≥ 50 are shown.
Expression patterns of TCP-like homologs in Arabidopsis thaliana (Brassicaceae), Solanum lycopersicum and Solanum tuberosum (Solanaceae). (A) Arabidopsis thaliana AthTCP24 (CIN-like). (B) Arabidopsis thaliana AthTCP20 (PCF-like). (C) Solanum lycopersicum SlyTCP24 (CIN-like). (D) Solanum lycopersicum SlyTCP11 (PCF-like). (E) Solanum tuberosum StuTCP2 (CIN-like). (F) Solanum tuberosum StuTCP11 (PCF-like). Expression patterns show here were selected for the genes that showed the broader, most comprehensive expression patterns after comparing expression for all paralogs, and are used to summarize putative expression patterns for each gene clade. Expression levels vary in intensity between different copies of CIN-like and PCF-like. Taken from the eFP browser, last accessed Sep 5/2016 (http://bar.utoronto.ca/efp_tomato/cgi-bin/efpWeb.cgi and http://bar.utoronto.ca/efp_potato/cgi-bin/efpWeb.cgi).
List of sequences used in this study.
Primers used for TCP-like gene expression analyses.
References
- Aguilar-Martínez J. A., Sinha N. (2013). Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 4:406. 10.3389/fpls.2013.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J., Rocheta M., Galego L. (1997). Genetic control of flower shape in Antirrhinum majus. Development 124, 1387–1392. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester P., Navarrete-Gómez M., Carbonero P., Oñate-Sánchez L., Ferrándiz C. (2015). Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol. Plant 155, 21–32. 10.1111/ppl.12327 [DOI] [PubMed] [Google Scholar]
- Bartlett M. E., Specht C. D. (2011). Changes in expression pattern of the TEOSINTE BRANCHED1- like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am. J. Bot. 98, 227–243. 10.3732/ajb.1000246 [DOI] [PubMed] [Google Scholar]
- Broholm S. (2009). The Role of MADS and TCP Transcription Factors in Gerbera Hybrida Flower Development. Available online at: https://helda.helsinki.fi/handle/10138/22335
- Busch A., Zachgo S. (2007). Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc. Natl. Acad. Sci. U.S.A. 104, 16714–16719. 10.1073/pnas.0705338104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase M. W., Cameron K. M., Freudenstein J. V., Pridgeon A. M., Salazar G., van den Berg C., et al. (2015). An updated classification of Orchidaceae. Bot. J. Linn. Soc. 177, 151–174. 10.1111/boj.12234 [DOI] [Google Scholar]
- Chase M. W., Christenhusz M. J. M., Fay M. F., Byng J. W., Judd W. S., Soltis D. E., et al. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20. 10.1111/boj.12385 [DOI] [Google Scholar]
- Chase M. W., Reveal J. L., Fay M. F. (2009). A subfamilial classification for the expanded asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae. Bot. J. Linn. Soc. 161, 132–136. 10.1111/j.1095-8339.2009.00999.x [DOI] [Google Scholar]
- Chen S., Kim D. K., Chase M. W., Kim J. H. (2013). Networks in a large-scale phylogenetic analysis: reconstructing evolutionary history of Asparagales (Lilianae) based on four plastid genes. PLoS ONE 8:e59472. 10.1371/journal.pone.0059472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citerne H. L., Le Guilloux M., Sannier J., Nadot S., Damerval C. (2013). Combining phylogenetic and syntenic analyses for understanding the evolution of TCP ECE genes in eudicots. PLoS ONE 8:e74803. 10.1371/journal.pone.0074803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley S. B., Carpenter R., Copsey L., Coen E. (2005). Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc. Natl. Acad. Sci. U.S.A. 102, 5068–5073. 10.1073/pnas.0501340102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M. M. R., Fox S., Hanna A. I., Baxter C., Coen E. (2005). Evolution of regulatory interactions controlling floral asymmetry. Development 132, 5093–5101. 10.1242/dev.02085 [DOI] [PubMed] [Google Scholar]
- Crawford B. C. W., Nath U., Carpenter R., Coen E. S. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135, 244–253. 10.1104/pp.103.036368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. 10.1046/j.1365-313X.1999.00444.x [DOI] [PubMed] [Google Scholar]
- Damerval C., Citerne H., Le Guilloux M., Domenichini S., Dutheil J., De Craene L. R., et al. (2013). Asymmetric morphogenetic cues along the transverse plane: shift from disymmetry to zygomorphy in the flower of fumarioideae. Am. J. Bot. 100, 391–402. 10.3732/ajb.1200376 [DOI] [PubMed] [Google Scholar]
- Damerval C., Le Guilloux M., Jager M., Charon C. (2007). Diversity and evolution of CYCLOIDEA-like TCP genes in relation to flower development in Papaveraceae. Plant Physiol. 143, 759–772. 10.1104/pp.106.090324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C., Manuel M. (2003). Independent evolution of Cycloidea-like sequences in several angiosperm taxa. Comptes Rendus Palevol 2, 241–250. 10.1016/S1631-0683(03)00031-9 [DOI] [Google Scholar]
- Danisman S., van der Wal F., Dhondt S., Waites R., de Folter S., Bimbo A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. 10.1104/pp.112.200303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S., Van Dijk A. D. J., Bimbo A., van der Wal F., Hennig L., De Folter S., et al. (2013). Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64, 5673–5685. 10.1093/jxb/ert337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta M., Aggarwal P., Nath U. (2014). CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 204, 901–912. 10.1111/nph.12963 [DOI] [PubMed] [Google Scholar]
- De Paolo S., Gaudio L., Aceto S. (2015). Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica. Sci. Rep. 5:16265. 10.1038/srep16265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Gustus C. (1995). teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Efroni I., Han S. K., Kim H. J., Wu M. F., Steiner E., Birnbaum K. D., et al. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24, 438–445. 10.1016/j.devcel.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress P. K. (2016). Development and evolution of extreme synorganization in angiosperm flowers and diversity: a comparison of Apocynaceae and Orchidaceae. Ann. Bot. 117, 749–767. 10.1093/aob/mcv119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S. K., Bowman J. L. (2007). The ancestral developmental tool kit of land plants. Int. J. Plant Sci. Spec. Issue Discern. Homol. Gene Expr. 168, 1–35. 10.1086/509079 [DOI] [Google Scholar]
- Galego L., Almeida J. (2002). Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev. 16, 880–891. 10.1101/gad.221002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Tao J. H., Yan D., Wang Y. Z., Li Z. Y. (2008). Expression differentiation of CYC-like floral symmetry genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae). Dev. Genes Evol. 218, 341–351. 10.1007/s00427-008-0227-y [DOI] [PubMed] [Google Scholar]
- Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C. P., et al. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934. 10.1105/tpc.110.074518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish T. J., Zuluaga A., Marques I., Lam V. K. Y., Gomez M. S., Iles W. J. D., et al. (2016). Phylogenomics and historical biogeography of the monocot order Liliales: out of Australia and through Antarctica. Cladistics 32, 581–605. 10.1111/cla.12153 [DOI] [PubMed] [Google Scholar]
- Gong Y.-B., Huang S.-Q. (2009). Floral symmetry: pollinator-mediated stabilizing selection on flower size in bilateral species. Proc. R. Soc. B Biol. Sci. 276, 4013–4020. 10.1098/rspb.2009.1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E. B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22, 1161–1173. 10.1105/tpc.109.069203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery D. M., Kalkhoven E., Hoare S., Parker M. G. (1997). A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387, 733–736. 10.1038/42750 [DOI] [PubMed] [Google Scholar]
- Hileman L. C. (2014). Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 369, 1–10. 10.1098/rstb.2013.0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman L. C., Baum D. A. (2003). Why do paralogs persist? Molecular evolution of CYCLOIDEA and related floral symmetry genes in Antirrhineae (Veronicaceae). Mol. Biol. Evol. 20, 591–600. 10.1093/molbev/msg063 [DOI] [PubMed] [Google Scholar]
- Horn S., Pabón-Mora N., Theuß V. S., Busch A., Zachgo S. (2015). Analysis of the CYC/TB1 class of TCP transcription factors in basal angiosperms and magnoliids. Plant J. 81, 559–571. 10.1111/tpj.12750 [DOI] [PubMed] [Google Scholar]
- Hoshino Y., Igarashi T., Ohshima M., Shinoda K., Murata N., Kanno A., et al. (2014). Characterization of CYCLOIDEA-like genes in controlling floral zygomorphy in the monocotyledon Alstroemeria. Sci. Hortic. (Amsterdam: ). 169, 6–13. 10.1016/j.scienta.2014.01.046 [DOI] [Google Scholar]
- Howarth D. G., Donoghue M. J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U.S.A. 103, 9101–9106. 10.1073/pnas.0602827103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth D. G., Martins T., Chimney E., Donoghue M. J. (2011). Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann. Bot. 107, 1521–1532. 10.1093/aob/mcr049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. 10.1111/j.1365-313X.2011.04674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocyan A. (2007). The discovery of polyandry in Curculigo (Hypoxidaceae): implications for androecium evolution of asparagoid monocotyledons. Ann. Bot. 100, 241–248. 10.1093/aob/mcm091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. 10.1105/tpc.9.9.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. 10.1046/j.1365-313X.2002.01294.x [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484. 10.1105/tpc.106.044792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010a). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588. 10.1105/tpc.110.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Sato F., Ohme-Takagi M. (2010b). A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci. Biotechnol. Biochem. 74, 2145–2147. 10.1271/bbb.100442 [DOI] [PubMed] [Google Scholar]
- Li S., Lauri A., Ziemann M., Busch A., Bhave M., Zachgo S. (2009). Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21, 429–441. 10.1105/tpc.108.064477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. F., Chen Y. Y., Hsiao Y. Y., Shen C. Y., Hsu J. L., Yeh C. M., et al. (2016). Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 67, 5051–5066. 10.1093/jxb/erw273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99, 367–376. 10.1016/S0092-8674(00)81523-8 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. 10.1038/383794a0 [DOI] [PubMed] [Google Scholar]
- Ma J., Liu F., Wang Q., Wang K., Jones D. C., Zhang B. (2016). Comprehensive analysis of TCP transcription factors and their expression during cotton (Gossypium arboreum) fiber early development. Sci. Rep. 6:21535. 10.1038/srep21535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. 10.1016/j.tplants.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees, in Gateway Computing Environments Workshop, GCE (San Diego, CA: ). [Google Scholar]
- Mondragón-Palomino M. (2013). Perspectives on MADS-box expression during orchid flower evolution and development. Front. Plant Sci. 4:377. 10.3389/fpls.2013.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Theißen G. (2009). Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann. Bot. 104, 583–594. 10.1093/aob/mcn258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Trontin C. (2011). High time for a roll call: gene duplication and phylogenetic relationships of TCP-like genes in monocots. Ann. Bot. 107, 1533–1544. 10.1093/aob/mcr059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539. 10.1073/pnas.0908718106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B. C. W., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Am. Assoc. Adv. Sci. 299, 1404–1407. 10.1126/science.1079354 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. 10.1007/s00239-006-0174-z [DOI] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791. 10.1038/ng2036 [DOI] [PubMed] [Google Scholar]
- Pabón-Mora N., González F. (2008). Floral ontogeny of Telipogon spp. (Orchidaceae) and insights on the perianth symmetry in the family. Int. J. Plant Sci. 169, 1159–1173. 10.1086/591982 [DOI] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Parapunova V., Busscher M., Busscher-Lange J., Lammers M., Karlova R., Bovy A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14:157. 10.1186/1471-2229-14-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Preston J. C., Hileman L. C. (2009). Developmental genetics of floral symmetry evolution. Trends Plant Sci. 14, 147–154. 10.1016/j.tplants.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Preston J. C., Hileman L. C. (2012). Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. Evodevo 3:6. 10.1186/2041-9139-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J. C., Kost M., A.„ Hileman L. C. (2009). Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytol. 182, 751–762. 10.1111/j.1469-8137.2009.02794.x [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J. L., Breton G., Para A., Kay S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485. 10.1126/science.1167206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimundo J., Sobral R., Bailey P., Azevedo H., Galego L., Almeida J., et al. (2013). A subcellular tug of war involving three MYB-like proteins underlies a molecular antagonism in Antirrhinum flower asymmetry. Plant J. 75, 527–538. 10.1111/tpj.12225 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2014). FigTree: Tree Figure Drawing Tool.
- Reeves P. A, Olmstead R. G. (2003). Evolution of the TCP gene family in asteridae: cladistic and network approaches to understanding regulatory gene family diversification and its impact on morphological evolution. Mol. Biol. Evol. 20, 1997–2009. 10.1093/molbev/msg211 [DOI] [PubMed] [Google Scholar]
- Resentini F., Felipo-Benavent A., Colombo L., Blázquez M. A., Alabadí D., Masiero S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8, 482–485. 10.1016/j.molp.2014.11.018 [DOI] [PubMed] [Google Scholar]
- Rudall P. J. (2002). Unique floral structures and iterative evolutionary themes in asparagales: insights from a morphological cladistic analysis. Bot. Rev. 68, 488–509. 10.1663/0006-8101(2002)068[0488:UFSAIE [DOI] [Google Scholar]
- Rudall P. J., Bateman R. M. (2002). Roles of synorganisation, zygomorphy and heterotopy in floral evolution: the gynostemium and labellum of orchids and other lilioid monocots. Biol. Rev. Camb. Philos. Soc. 77, 403–441. 10.1017/S1464793102005936 [DOI] [PubMed] [Google Scholar]
- Rudall P. J., Bateman R. M. (2004). Evolution of zygomorphy in monocot flowers: iterative patterns and developmental constraints. New Phytol. 162, 25–44. 10.1111/j.1469-8137.2004.01032.x [DOI] [Google Scholar]
- Rudall P. J., Perl C. D., Bateman R. M. (2013). Organ homologies in orchid flowers re-interpreted using the Musk Orchid as a model. PeerJ 1, 1–23. 10.7717/peerj.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K., Nath U. (2011). Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67, 595–607. 10.1111/j.1365-313X.2011.04616.x [DOI] [PubMed] [Google Scholar]
- Schommer C., Bresso E. G., Spinelli S. V., Palatnik J. F. (2012). MicroRNAs in plant development and stress responses. Screen 15, 29–47. 10.1007/978-3-642-27384-1 [DOI] [Google Scholar]
- Schommer C., Debernardi J. M., Bresso E. G., Rodriguez R. E., Palatnik J. F. (2014). Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 7, 1533–1544. 10.1093/mp/ssu084 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P., Guy K. M., Wu W., Fang B., Yang J., Zhang M., et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 16:85. 10.1186/s12870-016-0765-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson M. G. (2006). Plant Systematics. San Diego, CA: Elsevier; Academic Press. [Google Scholar]
- Su C. L., Chen W. C., Lee A. Y., Chen C. Y., Chang Y. C. A., Chao Y. T., et al. (2013). A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. PLoS ONE 8:e80462. 10.1371/journal.pone.0080462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tähtiharju S., Rijpkema A. S., Vetterli A., Albert V. A., Teeri T. H., Elomaa P. (2012). Evolution and diversification of the CYC/TB1 gene family in asteraceae-a comparative study in Gerbera (mutisieae) and sunflower (heliantheae). Mol. Biol. Evol. 29, 1155–1166. 10.1093/molbev/msr283 [DOI] [PubMed] [Google Scholar]
- Tsai W. C., Fu C. H., Hsiao Y. Y., Huang Y. M., Chen L. J., Wang M., et al. (2013). OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 54, 1–8. 10.1093/pcp/pcs187 [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero N. G., Lucero L. E., Viola I. L., Vegetti A. C., Gonzalez D. H. (2012). The class i protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63, 809–823. 10.1093/jxb/err305 [DOI] [PubMed] [Google Scholar]
- Vieira C. P., Vieira J., Charlesworth D. (1999). Evolution of the cycloidea gene family in Antirrhinum and Misopates. Mol. Biol. Evol. 16, 1474–1483. 10.1093/oxfordjournals.molbev.a026059 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Y., Luo D. (2010). LjCYC genes constitute floral dorsoventral asymmetry in Lotus japonicus. J. Integr. Plant Biol. 52, 959–970. 10.1111/j.1744-7909.2010.00926.x [DOI] [PubMed] [Google Scholar]
- Wilfinger W. W., Mackey K., Chomczynski P. (1997). NanoDrop and design are registered trademarks of NanoDrop Technologies 260/280 and 260/230 Ratios NanoDrop® ND-1000 and ND-8000 8-Sample Spectrophotometers. Biotechniques 22, 474–481.9067025 [Google Scholar]
- Yang C., Li D., Mao D., Liu X., Ji C., Li X., et al. (2013). Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativaL.). Plant Cell Environ. 36, 2207–2218. 10.1111/pce.12130 [DOI] [PubMed] [Google Scholar]
- Yang X., Pang H. B., Liu B. L., Qiu Z. J., Gao Q., Wei L., et al. (2012). Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24, 1834–1847. 10.1105/tpc.112.099457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Ma H., Wang J., Zhang D. (2007). Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 49, 885–897. 10.1111/j.1744-7909.2007.00509.x [DOI] [Google Scholar]
- Yuan Z., Gao S., Xue D.-W., Luo D., Li L.-T., Ding S.-Y., et al. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149, 235–244. 10.1104/pp.108.128231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kramer E. M., Davis C. C. (2010). Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc. Natl. Acad. Sci. U.S.A. 107, 6388–6393. 10.1073/pnas.0910155107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Steinmann V. W., Nikolov L., Kramer E. M., Davis C. C. (2013). Divergent genetic mechanisms underlie reversals to radial floral symmetry from diverse zygomorphic flowered ancestors. Front. Plant Sci. 4:302. 10.3389/fpls.2013.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Li D., Li Z., Hu Q., Yang C., Zhu L., et al. (2013). Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161, 1375–1391. 10.1104/pp.112.208702 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conserved motifs in and Orchidaceae and non-Orchidaceae Asparagales TCP-like proteins. Model core eudicots and monocots used as reference include Arabidopsis thaliana, Antirrhinum majus, and Oryza sativa. Motifs 1, 2, and 3 correspond to the conserved TCP domain. Motif 11 indicates the characteristic R domain in Class II TCP-like genes. Motif 14 corresponds to the miR319 binding site in CIN-like genes.
ML analysis of TCP-like genes with extended Solanaceae sampling. ML phylogenetic analysis of TCP-like genes with reduced sampling including only model organisms like Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum, Solanum tuberosum, two Orchidaceae species, Cattleya trianae, Orchis italica, one non-Orchidaceae Asparagales, Hypoxis decumbens and the early diverging angiosperm Amborella trichopoda. Yellow stars indicate large scale duplication events. TCP major clades are labeled to the right. Branch and taxa colors correspond to those in the conventions. BS values ≥ 50 are shown.
Expression patterns of TCP-like homologs in Arabidopsis thaliana (Brassicaceae), Solanum lycopersicum and Solanum tuberosum (Solanaceae). (A) Arabidopsis thaliana AthTCP24 (CIN-like). (B) Arabidopsis thaliana AthTCP20 (PCF-like). (C) Solanum lycopersicum SlyTCP24 (CIN-like). (D) Solanum lycopersicum SlyTCP11 (PCF-like). (E) Solanum tuberosum StuTCP2 (CIN-like). (F) Solanum tuberosum StuTCP11 (PCF-like). Expression patterns show here were selected for the genes that showed the broader, most comprehensive expression patterns after comparing expression for all paralogs, and are used to summarize putative expression patterns for each gene clade. Expression levels vary in intensity between different copies of CIN-like and PCF-like. Taken from the eFP browser, last accessed Sep 5/2016 (http://bar.utoronto.ca/efp_tomato/cgi-bin/efpWeb.cgi and http://bar.utoronto.ca/efp_potato/cgi-bin/efpWeb.cgi).
List of sequences used in this study.
Primers used for TCP-like gene expression analyses.