Abstract
Toll-like receptor 3 (TLR3) plays a key role in innate immunity by recognizing pathogenic, double-stranded RNAs. Thus, activation of TLR3 is a major factor in antiviral defense and tumor eradication. Although downregulation of TLR3 gene expression has been mainly reported in patients infected with hepatitis C virus (HCV), the influence of TLR3 genotype on the risk of HCV infection, HCV-related cirrhosis, and/or hepatocellular carcinoma (HCC) remains to be determined. Single-nucleotide polymorphisms (SNPs) within the TLR3 gene and their associations with HCV-related disease risk were investigated in a Saudi Arabian population in this study. Eight TLR3 SNPs were analyzed in 563 patients with HCV, which consisted of 437 patients with chronic HCV infections, 88 with HCV-induced liver cirrhosis, and 38 with HCC. A total of 599 healthy control subjects were recruited to the study. Among the eight TLR3 SNPs studied, the rs78726532 SNP was strongly associated with HCV infection when compared to that in healthy control subjects. The rs5743314 was also strongly associated with HCV-related liver disease progression (cirrhosis and HCC). In summary, these results indicate that distinct genetic variants of TLR3 SNPs are associated with HCV infection and HCV-mediated liver disease progression in the Saudi Arabian population.
1. Introduction
Hepatitis C virus (HCV) is an enveloped virus containing a positive-sense, single-stranded RNA genome that infects hepatocytes specifically through a noncytopathic process. It is a member of the genus Hepacivirus and belongs to the Flaviviridae family. HCV is a blood-borne pathogen and results in a significant public health problem, affecting approximately 180 million people globally [1, 2]. Owing to its ability to escape the host's defense mechanisms, HCV infection is considered persistent. Nearly 80% of all infected individuals develop chronic infection that persists for many years. Chronically infected individuals develop liver complications that include fibrosis and cirrhosis. Once cirrhosis has occurred, 3–5% of these individuals will develop hepatocellular carcinoma (HCC) [2, 3].
HCV-related diseases are treated with interferon-alpha (IFN-α) and ribavirin (RBV), antiviral agents that are used as the standard of care for patients with chronic HCV infection. However, this treatment regimen is effective in only 50–60% of patients with HCV [4]. Recently, there has been a major advancement in the treatment and management of HCV through the development of direct-acting anti-HCV drugs in addition to drug-induced activation of the host's cell-mediated immunity. Toll-like receptors (TLRs), a family of evolutionary conserved receptors that recognizes pathogens, have emerged as key regulators of both innate and adaptive immune responses. Lately, activation of immune cells through TLR agonists, which triggers interferon production, is being developed for therapy against HCV infection [5].
TLRs are a group of molecules that are essential for the innate immune response against infecting pathogens [6]. The effect of TLRs is mediated by sensing of molecules called pathogen-associated microbial patterns (PAMPs) [7]. In humans, there are 10 TLRs, which are expressed in different organs of the immune system [6]. In particular, TLR3 is encoded by a gene located on 4q35.1 and spans five exons. It is expressed intracellularly and recognizes double-stranded RNA (dsRNA) by responding with subsequent increases in interferon-α transcription, and its cellular effect is mediated through the recruitment of downstream signaling molecules such as retinoic acid-inducible gene 1 (RIG-I), melanoma differentiation-associated protein 5 (MDA5), and NACHTLRR-PYD-containing protein-3 (NALP3) [8]. Lately, TLR3-mediated antitumor activities inhibiting HCC development and progression have also been described [9–11].
Several studies have shown that genetic variations in the TLR3 gene are associated with susceptibility and/or resistance to numerous infectious and immune diseases [12], including acquired immune deficiency syndrome (AIDS) [13, 14], hepatitis B viral (HBV) infection [15], liver diseases in HCV-infected patients [16], predisposition to tick-borne encephalitis [17], human herpes simplex virus type 2 (HHV-2) infection [18], cutaneous candidiasis [19], autoimmunity [19], and type 1 diabetes mellitus [20].
In this study, we aimed to investigate the influence of TLR3 single-nucleotide polymorphisms (SNPs) on the susceptibility to HCV infection and HCV-related cirrhosis with or without HCC in a Saudi Arabian population.
2. Patients and Methods
2.1. Patients
The study protocol conformed to the 1975 Declaration of Helsinki and was approved by the Institutional Review Boards of King Khalid University Hospital (KKUH), Prince Sultan Military Medical City (PSMMC), and King Faisal Specialist Hospital and Research Center (KFSHRC). This study included 563 patients with HCV and 599 healthy control subjects. Participants in this study were recruited from the three tertiary care hospitals mentioned above in Riyadh, Saudi Arabia. The patients were divided into three groups: group 1 consisted of patients with chronic HCV (n = 437), group 2 consisted of patients with liver cirrhosis (LC) (n = 88), and group 3 consisted of patients with HCC (n = 38). Chronic HCV was diagnosed by the detection of anti-HCV antibodies and persistent presence of serum HCV RNA for more than 6 months, without any signs of liver complications. LC was diagnosed clinically by the detection of ascites, esophageal varices, and imaging findings on ultrasonography, transient elastography, computed tomography (CT), and magnetic resonance imaging (MRI) [21, 22]. HCC was confirmed on the basis of a pathological examination and/or elevation of blood alpha-fetoprotein (>400 ng/mL) in conjunction with CT, MRI, or ultrasonography scans.
Blood samples from healthy control individuals were obtained from blood donors in the participating hospitals and were HBs antigen (HBsAg) and HBe antigen (HBeAg) negative, while also lacking any serological markers for HCV, HBV, and HIV. Informed consent was obtained from all participants prior to enrollment in the study, and their demographic and clinical data were also recorded.
2.2. DNA Extraction and TLR3 SNP Genotyping
DNA was purified from blood using Gentra Puregene Blood kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. All samples were genotyped for the eight TLR3 SNPs via a PCR-based genotyping assay with two sets of specific primers designed using Primer3 v.0.4.0 (http://frodo.wi.mit.edu/primer3/) for TLR3 amplification: set 1 consisted of 5′-GCTGGAAAATCTCCAAGAGC-3′ and 5′-AGAGACCAAGCCAGCTAACC-3′; and set 2 consisted of 5′-GCGCTAAAAAGTGAAGAACTGG-3′ and 5′-GGGCTCTTGACCATCGTACT-3′. PCR reactions were performed on the Veriti 96-well thermal cycler (Applied Biosystems, California, USA) under the following conditions: 5 min initial denaturation at 95°C, followed by 40 cycles of 95°C for 1 min, 58°C for 45 s, 72°C for 1 min, and a 5 min final extension at 72°C. The amplified PCR products were analyzed by direct sequencing using the BigDye® Terminator v3.1 Cycle Sequencing Kit according to the manufacturer's instructions (BigDye Terminator v3.1 Cycle Sequencing Kit, Applera, Connecticut, USA). Sequencing products were purified using DyeEx spin column and were analyzed on the ABI 3700 DNA Analyzer (Applied Biosystems, California, USA).
2.3. Statistical Analysis
The genotypic and allelic distribution of TLR3 SNPs between the patients and healthy control groups were evaluated by Pearson's χ 2 test. The association between the SNPs and the disease groups was calculated under dominant and recessive genetic models and was expressed in terms of an odds ratio (OR) and their 95% confidence intervals (CI). A p value of less than 0.05 was considered statistically significant. Statistical analysis was performed using SPSS version 20.0 (SPSS Inc., Chicago, IL, USA). The SNPs were tested for Hardy–Weinberg equilibrium (HWE) and a p value of 0.01 was set for the analysis.
3. Results
3.1. Genotype and Allele Frequency Distributions of the TLR3 Polymorphisms Associated with HCV Infection
Polymorphisms of TLR3 gene were previously described as genetic factors associated with the susceptibility to HBV infection in a Saudi Arabian population [15]. Here, we analyzed the allelic frequency distribution of eight TLR3 SNPs in patients with HCV (n = 563) in comparison with that in healthy control subjects (n = 599). The baseline and clinical characteristics of the study subjects, HCV-infected patients, and healthy control subjects are presented in Table 1. Older age and gender were significantly linked to a higher risk for chronic HCV infection. Predictive indicators of the progression of liver disease such as body mass index (BMI), platelet count, and HCV viral load (ALT, ASP, and ALP) were also assessed in patients with HCV. End-stage liver disease was significantly correlated with a reduction in platelet count and increase in AST levels, whereas no significant differences were observed among other groups.
Table 1.
Baseline and clinical characteristics of HCV-infected patients and healthy control subjects.
| Variable | Chronic HCV (n = 437) | Liver cirrhosis (n = 88) | HCC (n = 38) | Healthy control (599) | p a value |
|---|---|---|---|---|---|
| Age (yrs.) | 43.23 ± 22.25 | 53.20 ± 19.79 | 63.23 ± 10.70 | 30.79 ± 8.90 | <0.0001 |
| Sex | |||||
| Male count (%) | 214 (49%) | 44 (50%) | 18 (47.4%) | 577 (96.3%) | <0.0001 |
| Female count (%) | 223 (51%) | 44 (50%) | 20 (52.6%) | 22 (3.7%) | |
| BMI^∗ | 29.2 (25.67–33.39) | 30.06 (26.22–33.06) | 25.99 (21.89–31.39) | 0.804 | |
| Platelet (per 109/L)∗ | 230 (178.75–282.25) | 163.5 (108.50–205.50) | 104.00 (51.50–139.00) | <0.0001 | |
| ALT∗∗ | 87.58 ± 119.76 | 93.18 ± 68.88 | 114.33 ± 31.78 | 0.738 | |
| AST∗∗ | 55.36 ± 41.87 | 77.28 ± 47.20 | 91.33 ± 24.58 | <0.0001 | |
| ALP∗∗ | 113.37 ± 78.91 | 133.66 ± 90.04 | 147.00 ± 96.70 | 0.072 | |
| HCV viral Load (log10)∗ | 6.04 (5.42–6.53) | 6.08 (5.57–6.45) | 5.98 (2.79–7.24) | 0.552 |
∗Values are expressed as median interquartile range (25th–75th). ∗∗Values are expressed as mean ± SD.
p a: nonparametric test and one-way ANOVA for continuous data and Chi square test for categorical data.
ALT: alanine aminotransferase; AST: aspartate aminotransferase; ALP: alkaline phosphatase.
Genotype frequencies and distributions of each TLR3 SNPs were in accordance with HWE in both the control and HCV-infected groups (Table 2).
Table 2.
Heterozygosity, HWE, and minor allele frequency of the TLR3 SNPs.
| Name | Position | ObsHET | PredHET | HWpval | MAF | Alleles |
|---|---|---|---|---|---|---|
| rs5743311 | 187000164 | 0.036 | 0.035 | 1 | 0.018 | G:A |
| rs5743312 | 187000256 | 0.255 | 0.267 | 0.3738 | 0.159 | C:T |
| rs1879026 | 187000321 | 0.295 | 0.288 | 0.7829 | 0.175 | G:T |
| rs111611328 | 187000364 | 0.006 | 0.01 | 0.021 | 0.005 | G:C |
| rs5743313 | 187000367 | 0.417 | 0.394 | 0.2523 | 0.269 | C:T |
| rs5743314 | 187000375 | 0.408 | 0.39 | 0.3682 | 0.265 | G:C |
| rs5743315 | 187000416 | 0.211 | 0.198 | 0.2656 | 0.112 | C:A |
| rs78726532 | 187000464 | 0.215 | 0.202 | 0.2266 | 0.114 | A:G |
We also confirmed the relevance of the TLR3 SNPs as markers for chronic HCV infection in patients compared to that in the noninfected healthy control subjects. The genotype distribution and allele frequency for TLR3 polymorphisms between patients with HCV and the control group are summarized in Table 3. Our results showed that the GG genotype of TLR3 rs78726532 was associated with the susceptibility to HCV infection when compared with healthy controls (OR = 0.264, 95% CI = 0.074–0.942, and p value = 0.027). No significant difference in the genotype and allele distributions of rs5743311, rs5743312, and rs1879026; rs5743313 and rs5743314; and rs5743315 and rs111611328 TLR3 SNPs was observed in patients with HCV when compared with that in healthy controls.
Table 3.
Genotypic distribution of TLR3 gene polymorphisms when all HCV-infected patients were compared to healthy control group.
| SNPs | Genotype/allele distribution | Healthy controls | HCV patients | OR (95% CI) | χ 2 | p value |
|---|---|---|---|---|---|---|
| n = 599 | n = 563 | |||||
| rs5743311 | AA | 1 (0.17%) | 0 (0%) | 0.360 (0.015–8.846) | 0.93 | 0.336 |
| AG | 10 (1.67%) | 18 (3.2%) | 1.942 (0.889–4.244) | 2.87 | 0.091 | |
| GG | 588 (98.16%) | 545 (96.8%) | Ref | |||
| A | 12 (1%) | 18 (1.6%) | 1.606 (0.770–3.348) | 1.62 | 0.203 | |
| G | 1186 (99%) | 1108 (98.4%) | ||||
| AA+AG.VS.GG | 1.765 (0.826–3.772) | 2.21 | 0.137 | |||
| AA.VS.AG+GG | 2.825 (0.115–69.479) | 0.94 | 0.332 | |||
|
| ||||||
| rs5743312 | TT | 18 (3.01%) | 16 (2.84%) | 0.959 (0.482–1.905) | 0.01 | 0.903 |
| CT | 141 (23.54%) | 139 (24.69%) | 1.063 (0.811–1.393) | 0.2 | 0.657 | |
| CC | 440 (73.46%) | 408 (72.47%) | Ref | |||
| T | 177 (14.77%) | 171 (15.19%) | 1.033 (0.822–1.297) | 0.08 | 0.781 | |
| C | 1021 (85.23%) | 955 (84.81%) | ||||
| TT+CT.VS.CC | 1.051 (0.811–1.362) | 0.14 | 0.705 | |||
| TT.VS.CT+CC | 1.059 (0.535–2.098) | 0.03 | 0.869 | |||
|
| ||||||
| rs1879026 | TT | 23 (3.84%) | 15 (2.66%) | 0.701 (0.361–1.363) | 1.11 | 0.292 |
| GT | 160 (26.71%) | 161 (28.6%) | 1.082 (0.835–1.401) | 0.35 | 0.552 | |
| GG | 416 (69.45%) | 387 (68.74%) | Ref | |||
| T | 206 (17.2%) | 191 (16.96%) | 0.984 (0.792–1.221) | 0.02 | 0.882 | |
| G | 992 (82.8%) | 935 (83.04%) | ||||
| TT+GT.VS.GG | 1.034 (0.806–1.326) | 0.07 | 0.793 | |||
| TT.VS.GT+GG | 1.459 (0.753–2.825) | 1.27 | 0.2602 | |||
|
| ||||||
| rs5743313 | TT | 42 (7.01%) | 37 (6.57%) | 0.952 (0.595–1.524) | 0.04 | 0.838 |
| CT | 250 (41.74%) | 242 (42.98%) | 1.046 (0.824–1.329) | 0.14 | 0.71 | |
| CC | 307 (51.25%) | 284 (50.44%) | Ref | |||
| T | 334 (27.88%) | 316 (28.06%) | 1.009 (0.842–1.210) | 0.01 | 0.921 | |
| C | 864 (72.12%) | 810 (71.94%) | ||||
| TT+CT.VS.CC | 1.033 (0.821–1.300) | 0.08 | 0.783 | |||
| TT.VS.CT+CC | 1.072 (0.678–1.694) | 0.09 | 0.766 | |||
|
| ||||||
| rs5743314 | CC | 44 (7.35%) | 37 (6.57%) | 0.897 (0.563–1.429) | 0.21 | 0.646 |
| GC | 249 (41.57%) | 239 (42.45%) | 1.023 (0.805–1.301) | 0.04 | 0.8501 | |
| GG | 306 (51.09%) | 287 (50.98%) | Ref | |||
| C | 337 (28.13%) | 313 (27.8%) | 0.984 (0.821–1.179) | 0.03 | 0.858 | |
| G | 861 (71.87%) | 813 (72.2%) | ||||
| CC+GC.VS.GG | 1.004 (0.798–1.264) | 0 | 0.971 | |||
| CC.VS.GC+GG | 1.127 (0.716–1.773) | 0.27 | 0.605 | |||
|
| ||||||
| rs5743315 | AA | 9 (1.5%) | 3 (0.53%) | 0.352 (0.095–1.309) | 2.65 | 0.104 |
| AC | 123 (20.53%) | 118 (20.96%) | 1.014 (0.763–1.347) | 0.01 | 0.926 | |
| CC | 467 (77.96%) | 442 (78.51%) | Ref | |||
| A | 141 (11.77%) | 124 (11.01%) | 0.928 (0.718–1.199) | 0.33 | 0.566 | |
| C | 1057 (88.23%) | 1002 (88.99%) | ||||
| AA+AC.VS.CC | 0.969 (0.733–1.280) | 0.05 | 0.822 | |||
| AA.VS.AC+CC | 2.847 (0.767–10.572) | 2.67 | 0.102 | |||
|
| ||||||
| rs111611328 | CC | 3 (0.5%) | 1 (0.18%) | 0.353 (0.037–3.404) | 0.89 | 0.346 |
| GC | 6 (1%) | 5 (0.89%) | 0.883 (0.268–2.909) | 0.04 | 0.837 | |
| GG | 590 (98.5%) | 557 (98.93%) | Ref | |||
| C | 12 (1%) | 7 (0.62%) | 0.618 (0.243–1.576) | 1.03 | 0.309 | |
| G | 1186 (99%) | 1119 (99.38%) | ||||
| CC+GC.VS.GG | 0.706 (0.250–1.997) | 0.43 | 0.509 | |||
| CC.VS.GC+GG | 2.829 (0.293–27.275) | 0.88 | 0.347 | |||
|
| ||||||
| rs78726532 | GG | 12 (2%) | 3 (0.53%) | 0.264 (0.074–0.942) | 4.84 | 0.027 |
| AG | 120 (20.03%) | 118 (20.96%) | 1.039 (0.781–1.382) | 0.07 | 0.793 | |
| AA | 467 (77.96%) | 442 (78.51%) | Ref | |||
| G | 144 (12.02%) | 124 (11.01%) | 0.906 (0.702–1.169) | 0.58 | 0.447 | |
| A | 1054 (87.98%) | 1002 (88.99%) | ||||
| GG+AG.VS.AA | 0.969 (0.733–1.280) | 0.05 | 0.822 | |||
| GG.VS.AG+AA | 3.816 (1.071–13.594) | 4.92 | 0.026 | |||
Note: risk alleles are marked in bold letters.
3.2. Genotype and Allele Frequency Distributions of the TLR3 Polymorphisms Associated with HCV-Related Liver Diseases
Genotype and allele distributions were also determined in patients with HCV that were developing cirrhosis, a late-stage liver disease marked by inflammation. Among the eight TLR3 polymorphisms, only two SNPs (rs5743313 and rs5743314) showed a significant association with the risk for cirrhosis in patients with HCV when compared with patients with chronic HCV infections (Table 4). The rs5743313 SNP was dominantly associated with liver cirrhosis (OR = 1.605, CI = 1.009–2.554, and p value = 0.045). In addition, the rs5743314 GC genotype was significantly correlated with the risk for cirrhosis (OR = 1.702, 95% CI = 1.050–2.759, and p value = 0.029) when compared with HCV patients (Table 4). The percentage of the rs5743314 C allele among the patients diagnosed with liver cirrhosis (34.6%) was higher than that of patients with chronic HCV (OR = 1.468, 95% CI = 1.040–2.072, and p value = 0.03), and the C allele was found to be dominantly associated (OR = 1.730, 95% CI = 1.085–2.757, and p value = 0.0204), suggesting that the C allele may contribute to HCV progression. No significant difference in the genotype and allele distributions of the other SNPs was observed between patients with HCV-related liver cirrhosis and those with chronic HCV (Table 4).
Table 4.
Genotypic distribution of TLR3 gene polymorphisms when chronic HCV patients were compared to patients diagnosed with liver cirrhosis.
| SNPs | Genotype/allele distribution | Chronic HCV | Liver cirrhosis | OR (95% C.I.) | χ 2 | p value |
|---|---|---|---|---|---|---|
| n = 437 | n = 88 | |||||
| rs5743311 | AA | 0 (0%) | 0 (0%) | 4.829 (0.095–244.994) | NaN | 1.00 |
| AG | 15 (3.43%) | 1 (1.14%) | 0.323 (0.042–2.480) | 1.31 | 0.253 | |
| GG | 422 (96.57%) | 87 (98.86%) | Ref | |||
| A | 15 (1.72%) | 1 (0.57%) | 0.327 (0.043–2.494) | 1.29 | 0.515 | |
| G | 859 (98.28%) | 175 (99.43%) | ||||
| AA+AG.VS.GG | 0.323 (0.042–2.480) | 1.31 | 0.253 | |||
| AA.VS.AG+GG | 0.202 (0.004–10.263) | NaN | 1.00 | |||
|
| ||||||
| rs5743312 | TT | 15 (3.43%) | 1 (1.14%) | 0.301 (0.039–2.321) | 1.49 | 0.223 |
| CT | 110 (25.17%) | 18 (20.45%) | 0.740 (0.422–1.299) | 1.11 | 0.292 | |
| CC | 312 (71.4%) | 69 (78.41%) | Ref | |||
| T | 140 (16.02%) | 20 (11.36%) | 0.672 (0.408–1.108) | 2.46 | 0.117 | |
| C | 734 (83.98%) | 156 (88.64%) | ||||
| TT+CT.VS.CC | 0.687 (0.397–1.190) | 1.81 | 0.178 | |||
| TT.VS.CT+CC | 3.092 (0.403–23.720) | 1.31 | 0.253 | |||
|
| ||||||
| rs1879026 | TT | 13 (2.97%) | 2 (2.27%) | 0.698 (0.154–3.169) | 0.22 | 0.639 |
| GT | 129 (29.52%) | 21 (23.86%) | 0.739 (0.433–1.260) | 1.24 | 0.265 | |
| GG | 295 (67.51%) | 65 (73.86%) | Ref | |||
| T | 155 (17.73%) | 25 (14.2%) | 0.768 (0.486–1.213) | 1.29 | 0.257 | |
| G | 719 (82.27%) | 151 (85.8%) | ||||
| TT+GT.VS.GG | 0.735 (0.439–1.231) | 1.37 | 0.241 | |||
| TT.VS.GT+GG | 1.318 (0.292–5.948) | 0.13 | 0.718 | |||
|
| ||||||
| rs5743313 | TT | 28 (6.41%) | 8 (9.09%) | 1.825 (0.772–4.317) | 1.92 | 0.165 |
| CT | 179 (40.96%) | 44 (50%) | 1.570 (0.970–2.543) | 3.40 | 0.065 | |
| CC | 230 (52.63%) | 36 (40.91%) | Ref | |||
| T | 235 (26.89%) | 60 (34.09%) | 1.406 (0.995–1.987) | 3.76 | 0.052 | |
| C | 639 (73.11%) | 116 (65.91%) | ||||
| TT+CT.VS.CC | 1.605 (1.009–2.554) | 4.03 | 0.045 | |||
| TT.VS.CT+CC | 0.685 (0.301–1.557) | 0.83 | 0.363 | |||
|
| ||||||
| rs5743314 | CC | 28 (6.41%) | 8 (9.09%) | 1.902 (0.803–4.505) | 2.19 | 0.138 |
| GC | 176 (40.27%) | 45 (51.14%) | 1.702 (1.050–2.759) | 4.72 | 0.029 | |
| GG | 233 (53.32%) | 35 (39.77%) | Ref | |||
| C | 232 (26.54%) | 61 (34.66%) | 1.468 (1.040–2.072) | 4.79 | 0.03 | |
| G | 642 (73.46%) | 115 (65.34%) | ||||
| CC+GC.VS.GG | 1.730 (1.085–2.757) | 5.38 | 0.0204 | |||
| CC.VS.GC+GG | 0.685 (0.301–1.557) | 0.83 | 0.363 | |||
|
| ||||||
| rs5743315 | AA | 3 (0.69%) | 0 (0%) | 0.694 (0.035–13.585) | 0.61 | 0.434 |
| AC | 92 (21.05%) | 18 (20.45%) | 0.956 (0.542–1.685) | 0.02 | 0.876 | |
| CC | 342 (78.26%) | 70 (79.55%) | Ref | |||
| A | 98 (11.21%) | 18 (10.23%) | 0.902 (0.530–1.534) | 0.14 | 0.703 | |
| C | 776 (88.79%) | 158 (89.77%) | ||||
| AA+AC.VS.CC | 0.926 (0.526–1.630) | 0.07 | 0.789 | |||
| AA.VS.AC+CC | 1.426 (0.073–27.846) | 0.61 | 0.436 | |||
|
| ||||||
| rs111611328 | CC | 0 (0%) | 0 (0%) | 5.023 (0.099–254.873) | NaN | 1.00 |
| GC | 3 (0.69%) | 2 (2.27%) | 3.364 (0.554–20.436) | 1.95 | 0.162 | |
| GG | 434 (99.31%) | 86 (97.73%) | Ref | |||
| C | 3 (0.34%) | 2 (1.14%) | 3.337 (0.554–20.120) | 1.94 | 0.468 | |
| G | 871 (99.66%) | 174 (98.86%) | ||||
| CC+GC.VS.GG | 3.364 (0.554–20.436) | 1.95 | 0.126 | |||
| CC.VS.GC+GG | 0.202 (0.004–10.263) | NaN | 1.00 | |||
|
| ||||||
| rs78726532 | GG | 3 (0.69%) | 0 (0%) | 0.671 (0.034–13.130) | 0.63 | 0.426 |
| AG | 94 (21.51%) | 16 (18.18%) | 0.804 (1.447–1.447) | 0.53 | 0.466 | |
| AA | 340 (77.8%) | 72 (81.82%) | Ref | |||
| G | 100 (11.44%) | 16 (9.09%) | 0.774 (0.445–1.348) | 0.82 | 0.364 | |
| A | 774 (88.56%) | 160 (90.91%) | ||||
| GG+AG.VS.AA | 0.779 (0.433–1.401) | 0.7 | 0.403 | |||
| GG.VS.AG+AA | 1.426 (0.073–27.846) | 0.61 | 0.436 | |||
To assess the influence of TLR3 polymorphisms on the risk of progression of end-stage liver diseases (liver cirrhosis and HCC), the genotype and allelic distributions were analyzed between patients with chronic HCV and patients diagnosed with liver cirrhosis and developing HCC. The TLR3 SNP rs5743314 that was previously described to be associated with risk for cirrhosis was also found to be associated with the risk for chronic HCV-related end-stage liver disease progression (rs5743314 GC genotype: OR = 1.545, 95% CI = 1.022–2.334, and p value = 0.0383), also the dominant model of rs5743314 associated with HCV liver disease progression (OR = 1.523, 95% CI = 1.021–2.271, and p value = 0.0385) (Table 5). The CT genotype and the dominant model TT+CT versus CC of rs5743313 were only slightly statistically significantly associated with liver cirrhosis and HCC (p value = 0.054 and 0.053, resp.). Similarly, the C allele and the recessive model CC.VS.GC+GG of rs111611328 were slightly statistically associated when compared to those in patients with chronic HCV with cirrhotic and HCC individuals (OR = 4.683, 95% CI = 1.041–21.062, and p value = 0.088; OR = 0.096, 95% CI = 0.004–2.362, and p value = 0.062; resp.) (Table 5). No significant difference in the genotype and allele distributions of the other SNPs was observed between patients with chronic HCV and those with HCV diagnosed with liver cirrhosis and HCC.
Table 5.
Genotypic distribution of TLR3 gene polymorphisms when chronic HCV patients were compared to patients diagnosed with liver cirrhosis and HCC.
| SNPs | Genotype/allele distribution | Chronic HCV | Cirrhosis + HCC | OR (95% C.I.) | χ 2 | p value |
|---|---|---|---|---|---|---|
| n = 437 | n = 126 | |||||
| rs5743311 | AA | 0 (0%) | 0 (0%) | 3.421 (0.068–173.296) | NaN | 1.00 |
| AG | 15 (3.43%) | 3 (2.38%) | 0.686 (0.195–2.409) | 0.35 | 0.554 | |
| GG | 422 (96.57%) | 123 (97.62%) | Ref | |||
| A | 15 (1.72%) | 3 (1.19%) | 0.690 (0.198–2.402) | 0.34 | 0.779 | |
| G | 859 (98.28%) | 249 (98.81%) | ||||
| AA+AG.VS.GG | 0.686 (0.195–2.409) | 0.35 | 0.554 | |||
| AA.VS.AG+GG | 0.289 (0.006–14.645) | NaN | 1.00 | |||
|
| ||||||
| rs5743312 | TT | 15 (3.43%) | 1 (0.79%) | 0.217 (0.028–1.662) | 2.61 | 0.106 |
| CT | 110 (25.17%) | 29 (23.02%) | 0.857 (0.536–1.369) | 0.42 | 0.517 | |
| CC | 312 (71.4%) | 96 (76.19%) | Ref | |||
| T | 140 (16.02%) | 31 (12.3%) | 0.735 (0.485–1.116) | 2.1 | 0.147 | |
| C | 734 (83.98%) | 221 (87.7%) | ||||
| TT+CT.VS.CC | 0.780 (0.493–1.235) | 1.13 | 0.288 | |||
| TT.VS.CT+CC | 4.43 (0.581–33.968) | 2.47 | 0.116 | |||
|
| ||||||
| rs1879026 | TT | 13 (2.97%) | 2 (1.59%) | 0.493 (0.109–2.226) | 0.88 | 0.348 |
| GT | 129 (29.52%) | 32 (25.4%) | 0.795 (0.506–1.250) | 0.99 | 0.321 | |
| GG | 295 (67.51%) | 92 (73.02%) | Ref | |||
| T | 155 (17.73%) | 36 (14.29%) | 0.773 (0.522–1.146) | 1.65 | 0.198 | |
| G | 719 (82.27%) | 216 (85.71%) | ||||
| TT+GT.VS.GG | 0.768 (0.494–1.194) | 1.38 | 0.239 | |||
| TT.VS.GT+GG | 1.901 (0.423–8.537) | 0.73 | 0.394 | |||
|
| ||||||
| rs5743313 | TT | 28 (6.41%) | 9 (7.14%) | 1.369 (0.611–3.069) | 0.59 | 0.444 |
| CT | 179 (40.96%) | 63 (50%) | 1.499 (0.992–2.265) | 3.72 | 0.054 | |
| CC | 230 (52.63%) | 54 (42.86%) | Ref | |||
| T | 235 (26.89%) | 81 (32.14%) | 1.288 (0.951–1.745) | 2.68 | 0.102 | |
| C | 639 (73.11%) | 171 (67.86%) | ||||
| TT+CT.VS.CC | 1.481 (0.993–2.209) | 3.74 | 0.053 | |||
| TT.VS.CT+CC | 0.890 (0.409–1.939) | 0.09 | 0.769 | |||
|
| ||||||
| rs5743314 | CC | 28 (6.41%) | 9 (7.14%) | 1.387 (0.619–3.109) | 0.64 | 0.425 |
| GC | 176 (40.27%) | 63 (50%) | 1.545 (1.022–2.334) | 4.29 | 0.0383 | |
| GG | 233 (53.32%) | 54 (42.86%) | Ref | |||
| C | 232 (26.54%) | 81 (32.14%) | 1.311 (0.967–1.777) | 3.05 | 0.0805 | |
| G | 642 (73.46%) | 171 (67.86%) | ||||
| CC+GC.VS.GG | 1.523 (1.021–2.271) | 4.28 | 0.0385 | |||
| CC.VS.GC+GG | 0.890 (0.409–1.939) | 0.09 | 0.769 | |||
|
| ||||||
| rs5743315 | AA | 3 (0.69%) | 0 (0%) | 0.487 (0.025–9.504) | 0.88 | 0.349 |
| AC | 92 (21.05%) | 26 (20.63%) | 0.967 (0.593–1.576) | 0.02 | 0.891 | |
| CC | 342 (78.26%) | 100 (79.37%) | Ref | |||
| A | 98 (11.21%) | 26 (10.32%) | 0.911 (0.577–1.439) | 0.16 | 0.689 | |
| C | 776 (88.79%) | 226 (89.68%) | ||||
| AA+AC.VS.CC | 0.936 (0.575–1.524) | 0.07 | 0.7903 | |||
| AA.VS.AC+CC | 2.038 (0.105–39.716) | 0.87 | 0.351 | |||
|
| ||||||
| rs111611328 | CC | 0 (0%) | 1 (0.79%) | 10.555 (0.427–260.710) | 3.51 | 0.061 |
| GC | 3 (0.69%) | 2 (1.59%) | 2.352 (0.389–14.236) | 0.92 | 0.337 | |
| GG | 434 (99.31%) | 123 (97.62%) | Ref | |||
| C | 3 (0.34%) | 4 (1.59%) | 4.683 (1.041–21.062) | 4.9 | 0.088 | |
| G | 871 (99.66%) | 248 (98.41%) | ||||
| CC+GC.VS.GG | 3.528 (0.703–17.701) | 2.66 | 0.103 | |||
| CC.VS.GC+GG | 0.096 (0.004–2.362) | 3.47 | 0.062 | |||
|
| ||||||
| rs78726532 | GG | 3 (0.69%) | 0 (0%) | 0.475 (0.024–9.263) | 0.90 | 0.343 |
| AG | 94 (21.51%) | 24 (19.05%) | 0.851 (0.516–1.403) | 0.4 | 0.526 | |
| AA | 340 (77.8%) | 102 (80.95%) | Ref | |||
| G | 100 (11.44%) | 24 (9.52%) | 0.815 (0.510–1.303) | 0.73 | 0.392 | |
| A | 774 (88.56%) | 228 (90.48%) | ||||
| GG+AG.VS.AA | 0.825 (0.501–1.358) | 0.57 | 0.448 | |||
| GG.VS.AG+AA | 2.038 (0.105–39.716) | 0.87 | 0.351 | |||
3.3. Haplotype Analyses
The haplotype combinations for three TLR3 SNPs (rs1879026, rs5743313, and rs5743314) and their genotypic distribution within the two comparative studies (patients with HCV diagnosed with cirrhosis compared to patients with chronic HCV) were established to determine the linkage disequilibrium (LD) pattern and the frequency of the haplotypes. The frequency of the haplotype GTC (freq. = 0.27), which includes the risk allele C for rs5743314, was statistically associated with the risk of progression to end-stage HCV-related liver diseases (p value = 0.0248), despite the absence of any correlation with the two other TLR3 SNPs (rs1879026 and rs5743313) with risk for HCV infection or end-stage liver disease (Table 6). Similarly, haplotype analysis was performed between patients with chronic HCV and patients diagnosed with liver cirrhosis and HCC, revealing that one out of three haplotypes was close to becoming significant, which includes the three TLR3 SNPs rs1879026-G, rs5743313-T, and rs5743314-C, with a p value = 0.0632 (Table 7, Figure 1).
Table 6.
Haplotype frequencies of TLR3 SNPs in chronic HCV-infected patients compared to patients with liver cirrhosis.
| Haplotype | Freq. | Liver cirrhosis, chronic HCV ratio counts | Liver cirrhosis, chronic HCV frequencies | Chi square | p value | ||
|---|---|---|---|---|---|---|---|
| rs1879026 | rs5743313 | rs5743314 | |||||
| G | C | G | 0.547 | 90.4 : 85.6, 483.8 : 390.2 | 0.513, 0.554 | 0.953 | 0.329 |
| G | T | C | 0.27 | 59.6 : 116.4, 224.1 : 649.9 | 0.339, 0.256 | 5.035 | 0.0248 |
| T | C | G | 0.167 | 24.6 : 151.4, 151.1 : 722.9 | 0.140, 0.173 | 1.144 | 0.2847 |
Table 7.
Haplotype frequencies of TLR3 SNPs in chronic HCV-infected patients compared to patients with cirrhosis + HCC infected patients.
| Haplotype | Freq. | Liver cirrhosis + HCC, chronic HCV ratio counts | Liver cirrhosis + HCC, chronic HCV frequencies | Chi Square | p value | ||
|---|---|---|---|---|---|---|---|
| rs1879026 | rs5743313 | rs5743314 | |||||
| G | C | G | 0.549 | 134.5 : 117.5, 483.7 : 390.3 | 0.534, 0.553 | 0.311 | 0.577 |
| G | T | C | 0.27 | 79.5 : 172.5, 224.2 : 649.8 | 0.315, 0.257 | 3.452 | 0.0632 |
| T | C | G | 0.166 | 35.5 : 216.5, 151.2 : 722.8 | 0.141, 0.173 | 1.458 | 0.2273 |
Figure 1.
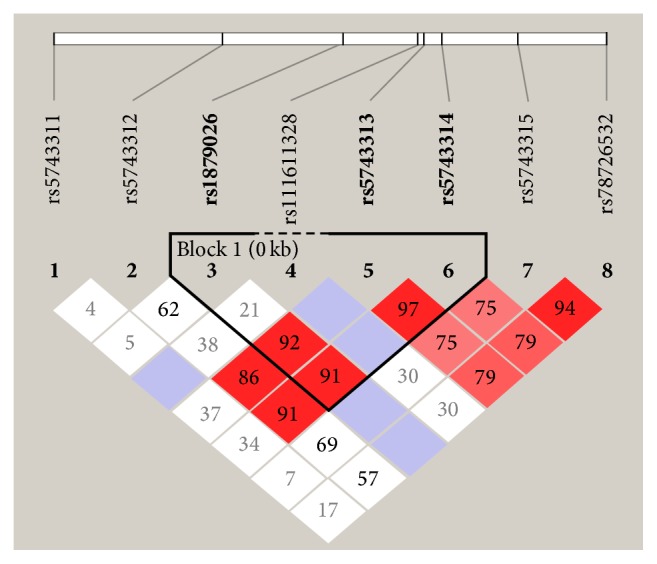
LD plot of SNPs in the TLR3 gene region. The plot was generated by Haploview 4.2. Numbers in squares show LD between the SNPs.
4. Discussion
Over the past 25 years, HCV has been a major causative factor in parenteral-acquired hepatitis and has received considerable attention owing to its ability to evade the host's defense mechanisms and a lack of a protective vaccine [23]. HCV persistence leads to chronic and progressive end-stage liver diseases, including HCC, which often necessitates liver transplantations for the patient, thus making HCV infections an important disease burden. Therefore, there is an urgent need for predictive genetic tools such as those based on SNP profiling (referred as personalized medicine), to characterize patients with susceptibility to HCV infection and HCV-mediated liver diseases. This will allow for better predictions in HCV infection progression rates and improvements in the outcome of standard therapies involving pegylated IFN-α combined with ribavirin [24]. TLR3, a sensor of the host immune system activated by viral dsRNA and responsible for the production of IFN, has been demonstrated to play an essential role during HCV infection and clearance, as well as in the severity of liver diseases. Thus, SNP genotyping of the TLR3 gene might be a good candidate approach to predicting the progression of HCV infections. Here, we investigated the influence of genetic variants within TLR3 and determined the degree of association with HCV infection and HCV-related liver damage that results in cirrhosis and HCC. Among the eight TLR3 SNPs genotyped, rs78726532 was strongly associated with HCV infection when compared to healthy control subjects. A protective role of the rs78726532 GG genotype for HCV infection was also observed. Three other TLR3 SNPs (rs5743313, rs5743314, and rs111611328) were also associated with HCV-related end-stage liver disease progression (liver cirrhosis and HCC).
Initially, low levels of TLR3 gene expression were detected in various liver cells, including nonparenchymal cells (i.e., Kupffer cells, sinusoidal endothelial cells, dendritic cells, stellate cells, and biliary epithelial cells) and hepatic lymphocytes [25–28]. TLR3 is mainly located on the endosome-lysosome membrane but can also be expressed on the plasma membrane of some cells. The host's immune defense is initiated with the presence of a byproduct of the replicative cycle of many single-stranded RNA viruses that could interact with the dsRNA-sensing receptor, TLR3. TLR3 activation results in the stimulation of interferon-regulatory factor- (IRF-3-) dependent type I IFN responses, NF-kB-dependent proinflammatory cytokine production, and stimulation of IFN genes (ISGs) that suppress HCV replication [29]. During early stages of HCV infection, TLR3 expression is upregulated by IFN, indicating a positive feedback regulation. Thus, TLR3 signaling may have pleiotropic functions and is involved in inflammation leading to HCV clearance during the course of acute liver injury. Herein, a protective role of the TLR3 rs78726532 GG genotype for HCV infection was observed, indicating a robust expression and function of the TLR3 receptor, which is an important determinant for viral clearance [30]. In a previous study assessing the function of TLR3 in macrophages of patients with chronic HCV, the intronic TLR3 rs1316816 SNP was found to be associated with high TLR3 expression and HCV clearance [31]. In our study, monitoring the level of expression of TLR3 in patients harboring the TLR3 rs78726532 SNP would be of interest.
Among the eight TLR3 SNPs genotyped, rs78726532 was strongly associated with HCV infection when compared to that in their healthy counterparts. Multiple SNPs positioned in the TLR3 gene targeted to assess the risk of HCV infection have yielded inconsistent results. Indeed, Sá et al., 2015, demonstrated that the rs5743305 and rs3775291 SNPs were not associated with a risk for HCV infection [32]. This was inconsistent with a study by Medhi et al., 2011, that analyzed polymorphisms at the promoter region of the TLR3 gene [33], as well as a recent meta-analysis that concluded TLR3 gene polymorphisms (mainly rs3775291) were associated with a risk for HCV infection [34]. This inconsistency may be explained by the involvement of genetic factors due to racial and ethnic differences, as well as controversies in regard to whether TLR3 expression is upregulated [35, 36] or downregulated [37, 38] in peripheral blood mononuclear cells from patients with chronic HCV. Furthermore, a growing body of evidence suggests that TLR3 signaling is inhibited or diminished during HCV infection, a mechanism that may contribute to HCV modulation of the host immune system leading to chronic infection. In addition, it has been shown that HCV can modulate p53 function, which has been demonstrated to activate TLR3 transcription by binding to the p53 consensus site in the TLR3 promoter [39]. A deeper understanding of the molecular and regulatory system that results in TLR3 downregulation is crucial to an improvement of the TLR3-mediated innate immunity as a therapeutic approach.
In this present study, we showed that three other TLR3 SNPs (rs5743313, rs5743314, and rs111611328) were strongly associated with HCV-related end-stage liver disease progression (liver cirrhosis and HCC). However, several studies have suggested that TLR3 may contribute to resistance against HCV infection, even though TLR3 appears to have no role in disease advancement after a chronic infection is established [16, 40]. TLR3 polymorphisms (rs1879026 and rs3775290) have been described to be associated with a risk for HBV-related liver diseases in a Chinese population [41], whereas the same nine SNPs of our study were not correlated with any susceptibility to HBV-related liver diseases in the Saudi Arabian population [15]. Therefore, the impact of TLR3 polymorphisms may act in an ethnic- and viral-specific manner. At the cellular and molecular level, there is a growing body of evidence indicating that TLR3 plays a role in cirrhosis pathogenesis and hepatocarcinogenesis [42, 43]. For example, it has been documented that dsRNA activates TLR3 which subsequently results in NK cell accumulation and activation leading to liver inflammation. Such process could contribute to cirrhosis and HCC if left untreated [44–46]. Previous studies using a rat model showed that activated TLR3 could inhibit HCC development and progression by inhibiting cell invasion and inducing apoptosis in cancer cells [9, 10]. Therefore, the strong association of three TLR3 SNPs (rs5743313, rs5743314, and rs111611328) with HCV-related end-stage liver disease progression (cirrhosis and HCC) indicates an impairment of TLR3 function in the prevention of excessive inflammation and control of liver regeneration.
5. Conclusion
In conclusion, we showed that distinct TLR3 polymorphisms were associated with susceptibility to HCV infection and to HCV-related end-stage liver disease progression. These findings may act as indicators for differential TLR3 structures distinctively involved in immunity and carcinogenesis. A better understanding of these distinguishable structural and functional features will be helpful for developing therapeutic applications against TLR3-mediated HCV infections and TLR3-mediated, HCV-related liver diseases.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through Research Group no. RGP-018.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Muir A. J. The natural history of hepatitis C viral infection. Seminars in Gastrointestinal Disease. 2000;11(2):54–61. [PubMed] [Google Scholar]
- 2.Shepard C. W., Finelli L., Alter M. J. Global epidemiology of hepatitis C virus infection. Lancet Infectious Diseases. 2005;5(9):558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 3.Lavanchy D. The global burden of hepatitis C. Liver International. 2009;29(supplement 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison J. G., Lawitz E. J., Shiffman M. L., et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. New England Journal of Medicine. 2009;361(6):580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 5.Mifsud E. J., Tan A. C. L., Jackson D. C. TLR agonists as modulators of the innate immune response and their potential as agents against infectious disease. Frontiers in Immunology. 2014;5, article 79 doi: 10.3389/fimmu.2014.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nature Immunology. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto M., Funami K., Oshiumi H., Seya T. Toll-like receptor 3: a link between toll-like receptor, interferon and viruses. Microbiology and Immunology. 2004;48(3):147–154. doi: 10.1111/j.1348-0421.2004.tb03500.x. [DOI] [PubMed] [Google Scholar]
- 9.Yuan M.-M., Xu Y.-Y., Chen L., Li X.-Y., Qin J., Shen Y. TLR3 expression correlates with apoptosis, proliferation and angiogenesis in hepatocellular carcinoma and predicts prognosis. BMC Cancer. 2015;15(1, article 245) doi: 10.1186/s12885-015-1262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y.-Y., Chen L., Zhou I.-M., Wu Y.-Y., Zhu Y.-Y. Inhibitory effect of dsRNA TLR3 agonist in a rat hepatocellular carcinoma model. Molecular Medicine Reports. 2013;8(4):1037–1042. doi: 10.3892/mmr.2013.1646. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z., Chen L., Zhu Y., et al. Double-stranded RNA-induced TLR3 activation inhibits angiogenesis and triggers apoptosis of human hepatocellular carcinoma cells. Oncology Reports. 2012;27(2):396–402. doi: 10.3892/or.2011.1538. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y.-T., Verma A., Hodgkinson C. P. Toll-like receptors and human disease: lessons from single nucleotide polymorphisms. Current Genomics. 2012;13(8):633–645. doi: 10.2174/138920212803759712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sironi M., Biasin M., Cagliani R., et al. A common polymorphism in TLR3 confers natural resistance to HIV-1 infection. Journal of Immunology. 2012;188(2):818–823. doi: 10.4049/jimmunol.1102179. [DOI] [PubMed] [Google Scholar]
- 14.Huik K., Avi R., Pauskar M., et al. Association between TLR3 rs3775291 and resistance to HIV among highly exposed Caucasian intravenous drug users. Infection, Genetics and Evolution. 2013;20:78–82. doi: 10.1016/j.meegid.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Qahtani A., Al-Ahdal M., Abdo A., et al. Toll-like receptor 3 polymorphism and its association with hepatitis B virus infection in Saudi Arabian patients. Journal of Medical Virology. 2012;84(9):1353–1359. doi: 10.1002/jmv.23271. [DOI] [PubMed] [Google Scholar]
- 16.Askar E., Bregadze R., Mertens J., et al. TLR3 gene polymorphisms and liver disease manifestations in chronic hepatitis C. Journal of Medical Virology. 2009;81(7):1204–1211. doi: 10.1002/jmv.21491. [DOI] [PubMed] [Google Scholar]
- 17.Barkhash A. V., Voevoda M. I., Romaschenko A. G. Association of single nucleotide polymorphism rs3775291 in the coding region of the TLR3 gene with predisposition to tick-borne encephalitis in a Russian population. Antiviral Research. 2013;99(2):136–138. doi: 10.1016/j.antiviral.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Svensson A., Tunbäck P., Nordström I., Padyukov L., Eriksson K. Polymorphisms in Toll-like receptor 3 confer natural resistance to human herpes simplex virus type 2 infection. Journal of General Virology. 2012;93(8):1717–1724. doi: 10.1099/vir.0.042572-0. [DOI] [PubMed] [Google Scholar]
- 19.Nahum A., Dadi H., Bates A., Roifman C. M. The L412F variant of Toll-like receptor 3 (TLR3) is associated with cutaneous candidiasis, increased susceptibility to cytomegalovirus, and autoimmunity. Journal of Allergy and Clinical Immunology. 2011;127(2):528–531. doi: 10.1016/j.jaci.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 20.Assmann T. S., De Almeida Brondani L., Bauer A. C., Canani L. H., Crispim D. Polymorphisms in the TLR3 gene are associated with risk for type 1 diabetes mellitus. European Journal of Endocrinology. 2014;170(4):519–527. doi: 10.1530/EJE-13-0963. [DOI] [PubMed] [Google Scholar]
- 21.Foucher J., Chanteloup E., Vergniol J., et al. Diagnosis of cirrhosis by transient elastography (FibroScan): a prospective study. Gut. 2006;55(3):403–408. doi: 10.1136/gut.2005.069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halfon P., Munteanu M., Poynard T. FibroTest-ActiTest as a non-invasive marker of liver fibrosis. Gastroenterologie Clinique et Biologique. 2008;32(6):22–39. doi: 10.1016/S0399-8320(08)73991-5. [DOI] [PubMed] [Google Scholar]
- 23.Keyvani H., Fazlalipour M., Monavari S. H. R., Mollaie H. R. Hepatitis C virus—proteins, diagnosis, treatment and new approaches for vaccine development. Asian Pacific Journal of Cancer Prevention. 2012;13(12):5917–5935. doi: 10.7314/apjcp.2012.13.12.5917. [DOI] [PubMed] [Google Scholar]
- 24.Kim K. H., Jang B. K., Chung W. J., et al. Peginterferon alpha and ribavirin combination therapy in patients with hepatitis C virus-related liver cirrhosis. The Korean Journal of Hepatology. 2011;17(3):220–225. doi: 10.3350/kjhep.2011.17.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorner M., Brandt S., Tinguely M., et al. Plasma cell toll-like receptor (TLR) expression differs from that of B cells, and plasma cell TLR triggering enhances immunoglobulin production. Immunology. 2009;128(4):573–579. doi: 10.1111/j.1365-2567.2009.03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzio M., Bosisio D., Polentarutti N., et al. Differential expression and regulation of toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. Journal of Immunology. 2000;164(11):5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- 27.Wilson C. L., Mann J., Walsh M., et al. Quiescent hepatic stellate cells functionally contribute to the hepatic innate immune response via TLR3. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0083391.e83391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarember K. A., Godowski P. J. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. Journal of Immunology. 2002;168(2):554–561. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 29.Wang N., Liang Y., Devaraj S., Wang J., Lemon S. M., Li K. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. Journal of Virology. 2009;83(19):9824–9834. doi: 10.1128/JVI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Firdaus R., Biswas A., Saha K., et al. Modulation of TLR 3, 7 and 8 expressions in HCV genotype 3 infected individuals: potential correlations of pathogenesis and spontaneous clearance. BioMed Research International. 2014;2014:7. doi: 10.1155/2014/491064.491064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian F., Bolen C. R., Jing C., et al. Impaired toll-like receptor 3-mediated immune responses from macrophages of patients chronically infected with hepatitis C virus. Clinical and Vaccine Immunology. 2013;20(2):146–155. doi: 10.1128/CVI.00530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sá K. S., Pires-Neto O. D., Santana B. B., et al. Toll-like receptor 3 gene polymorphisms are not associated with the risk of hepatitis B and hepatitis C virus infection. Revista da Sociedade Brasileira de Medicina Tropical. 2015;48(2):136–142. doi: 10.1590/0037-8682-0008-2015. [DOI] [PubMed] [Google Scholar]
- 33.Medhi S., Deka M., Deka P., et al. Promoter region polymorphism & expression profile of toll like receptor-3 (TLR-3) gene in chronic hepatitis C virus (HCV) patients from India. Indian Journal of Medical Research. 2011;134(8):200–207. [PMC free article] [PubMed] [Google Scholar]
- 34.Geng P., Song L., An H., Huang J., Li S., Zeng X. Toll-like receptor 3 is associated with the risk of HCV infection and HBV-Related Diseases. Medicine. 2016;95(21):p. e2302. doi: 10.1097/md.0000000000002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Q., Graham C. S., Mangoni E. D., Koziel M. J. Differential expression of toll-like receptor mRNA in treatment non-responders and sustained virologic responders at baseline in patients with chronic hepatitis C. Liver International. 2006;26(9):1100–1110. doi: 10.1111/j.1478-3231.2006.01357.x. [DOI] [PubMed] [Google Scholar]
- 36.Sato K., Ishikawa T., Okumura A., et al. Expression of Toll-like receptors in chronic hepatitis C virus infection. Journal of Gastroenterology and Hepatology. 2007;22(10):1627–1632. doi: 10.1111/j.1440-1746.2006.04783.x. [DOI] [PubMed] [Google Scholar]
- 37.Mohammed K. I., Adel L. A., Ali-Eldin F. A., Eladawy S. Expression of Toll like receptors 3 & 7 in peripheral blood from patients with chronic hepatitis C virus infection and their correlation with interferon-alpha. The Egyptian Journal of Immunology. 2013;20(1):13–22. [PubMed] [Google Scholar]
- 38.Yang Q., Fu S., Wang J. Hepatitis C virus infection decreases the expression of Toll-like receptors 3 and 7 via upregulation of miR-758. Archives of Virology. 2014;159(11):2997–3003. doi: 10.1007/s00705-014-2167-3. [DOI] [PubMed] [Google Scholar]
- 39.Taura M., Eguma A., Suico M. A., et al. p53 regulates toll-like receptor 3 expression and function in human epithelial cell lines. Molecular and Cellular Biology. 2008;28(21):6557–6567. doi: 10.1128/MCB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin S., Gao B. Toll-like receptor 3 in liver diseases. Gastroenterology Research and Practice. 2010;2010:6. doi: 10.1155/2010/750904.750904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X., Li H., Wang J., et al. Genetic polymorphisms in Toll-like receptor 3 gene are associated with the risk of hepatitis B virus-related liver diseases in a Chinese population. Gene. 2015;569(2):218–224. doi: 10.1016/j.gene.2015.05.054. [DOI] [PubMed] [Google Scholar]
- 42.Sun L., Dai J., Hu W., Wang J. Expression of toll-like receptors in hepatic cirrhosis and hepatocellular carcinoma. Genetics and Molecular Research. 2016;15(2) doi: 10.4238/gmr.15027419. [DOI] [PubMed] [Google Scholar]
- 43.Takii Y., Nakamura M., Ito M., et al. Enhanced expression of type I interferon and toll-like receptor-3 in primary biliary cirrhosis. Laboratory Investigation. 2005;85(7):908–920. doi: 10.1038/labinvest.3700285. [DOI] [PubMed] [Google Scholar]
- 44.Gardner T. R., Chen Q., Jin Y., Ajuebor M. N. Toll-like receptor 3 ligand dampens liver inflammation by stimulating Vα14 invariant natural killer T cells to negatively regulate γδT cells. The American Journal of Pathology. 2010;176(4):1779–1789. doi: 10.2353/ajpath.2010.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zampino R., Marrone A., Restivo L., et al. Chronic HCV infection and inflammation: clinical impact on hepatic and extra-hepatic manifestations. World Journal of Hepatology. 2013;5(10):528–540. doi: 10.4254/wjh.v5.i10.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J., Xu J., Zhang W., Wei H., Tian Z. TLR3 ligand-induced accumulation of activated splenic natural killer cells into liver. Cellular & Molecular Immunology. 2005;2(6):449–453. [PubMed] [Google Scholar]


