Abstract
To identify genetic mechanisms involved in the interplay of risky sexual behaviors (RSBs) and alcohol dependence (AD), we conducted genome-wide gene-by-AD (GW-GxAD) analyses of RSB in 3924 alcohol-exposed and sexually experienced subjects. RSBs were defined as a score based on lifetime experiences of unprotected sex and multiple sexual partners. Diagnosis of lifetime AD was defined by DSM-IV criteria. To follow-up the genetic findings, functional magnetic resonance imaging analyses were conducted in an independent sample. A trans-population genome-wide significant signal was identified in LHPP (rs34997829; z=−5.573, p=2.51 × 10−8) in the GxAD analysis that also showed associations in the AD-stratified association analysis (AD z=−2.032 and non-AD z=4.903). The clinical relevance of the result was confirmed by the significant interaction between LHPP rs34997829 and AD with respect to self-reported sexually transmitted disease (STD; z=−2.809, p=4.97 × 10−3). The neuroimaging follow-up analysis of LHPP rs34997829 showed reduced power of the left superior frontal gyrus (t=−3.386, p=9.56 × 10−4) and increased power at the right amygdala (t=3.287, p=1.33 × 10−3) in the resting amplitude of low frequency fluctuations analysis; and reduced activation of the anterior cingulate region (t=−2.961, p=3.69 × 10−3) in the monetary incentive delay task. In conclusion, LHPP locus is associated to AD–RSB interaction; and with brain circuitries previously implicated in the inhibition of risky behavior and impulsiveness, emotional regulation, and impulse control/error monitoring. Thus, LHPP is a strong candidate to influence RSB and STD risk in the context of AD.
INTRODUCTION
Risky sexual behaviors (RSB) associated with alcohol misuse contribute substantially to the ~15 million new cases of sexually transmitted diseases (STD) diagnosed annually in the United States (Chaney et al, 2016; Corbin et al, 2016; Khan et al, 2012; Tran et al, 2016). Neurobiological investigations have provided insight into some of the alcohol-related mechanisms of brain disinhibition that may lead to RSB (Spanagel, 2009). However, there is very little available information on the molecular mechanisms by which alcohol-related behaviors contribute to RSB. Genome-wide analysis can be a powerful tool to identify molecular mechanisms of complex traits, including in the context of alcohol abuse (Polimanti et al, 2015). Although no large genomic investigation has been conducted on RSB, a recent genome-wide association study (GWAS) of age at first sexual intercourse indicated that the onset of sexual activity is genetically correlated to other reproductive and behavioral traits (Day et al, 2016). Our recent phenome-wide association study showed that risk alleles for alcohol-drinking behaviors are associated with reproductive health traits (Polimanti et al, 2016). These data suggest that alcohol use and abuse play a relevant role in RSB. The identification of loci associated with sexual behavior and, especially, RSB, could provide useful information to design preventive strategies to address the negative consequences of RSB. Moreover, preventive strategies and therapeutic approaches for subjects with alcohol use disorders could plausibly be developed based on an improved understanding of the molecular mechanisms that lead to alcohol-related RSB. Accordingly, genome-wide analyses of the interplay between alcohol behaviors and RSB are potentially of great clinical importance.
In the present study, we performed genome-wide gene-by-alcohol dependence (GW-GxAD) analyses in a multi-ethnic sample of 3924 subjects including African-Americans (AA) and European-Americans (EA). Our GWAS subjects were evaluated using the semi-structured assessment for drug dependence and alcoholism (SSADDA; Pierucci-Lagha et al, 2005), which yields DSM-IV diagnoses of lifetime alcohol and drug dependence, and other major psychiatric traits. A RSB score was calculated using questions from the SSADDA section on antisocial personality.
We identified LHPP rs34997829 in our GW-GxAD analysis. Since LHPP variants seem to be associated with multiple psychiatric phenotypes, including depression (Converge consortium, 2015) and the AD-RSB interaction we report here, we explored neuroimaging data to understand whether and which brain region activities are affected by LHPP variation. Specifically, we investigated resting amplitude of low frequency fluctuations (ALFF) signals and the monetary incentive delay (MID) task. Resting ALFF signals have been proposed to be relevant markers for spontaneous brain activity and signals originating in gray matter (Meda et al, 2015). The MID task has been used to quantify brain activation during prospect (A1), anticipation (A2), and outcome (OC) phases for reward/loss trials to investigate reward system function, impulsivity, and risky behaviors (Patel et al, 2013). MID participants see cues indicating that they may win or lose money (A1 phase), then wait for a variable anticipatory delay period (A2 phase), and finally respond to a rapidly presented target with a single button press to try to either win or avoid losing money (A3 phase).
MATERIALS AND METHODS
Subjects and Diagnostic Procedures
The GWAS sample (2173 AAs and 1751 EAs) is constituted by subjects recruited for studies of the genetics of alcohol and drug dependencies funded by the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism (Gelernter et al, 2015; Gelernter et al, 2014a; 2014b; 2014c; Sherva et al, 2016). The sample consisted of small nuclear families originally collected for linkage studies, and unrelated individuals. Subjects were recruited at five US clinical sites: Yale University School of Medicine (APT Foundation, New Haven, CT, USA), the University of Connecticut Health Center (Farmington, CT, USA), the University of Pennsylvania Perelman School of Medicine (Philadelphia, PA, USA), the Medical University of South Carolina (Charleston, SC, USA), and McLean Hospital (Belmont, MA, USA). The institutional review board at each participating site approved the study and we obtained written informed consent from each participant. As reported above, RSBs were determined using questions from the SSADDA section on antisocial personality: I35B (‘Have you ever had sex with 10 different people within a single year?') and I37 (‘Have you more than once had unprotected sex (without a condom) with someone you believed could give you a disease, or when you had a disease that could be spread that way?'). On the basis of these two questions, we calculated an RSB score that ranged from 0 to 2 based on the number of affirmative responses. AD diagnosis was based on DSM-IV criteria (American Psychiatric Association, 1994). We included only subjects who reported having ever consumed more than 3 drinks in a 24-h period (ie, alcohol exposed) and reported sexual intercourse with at least 10 sexual partners (ie, sexually experienced). These inclusion criteria were selected on the basis of the SSADDA. The definitions of alcohol exposure and DSM-IV AD diagnosis are described in our previous GWAS of AD (Gelernter et al, 2014a). The definition of ‘sexually experienced' is derived from the criterion used in the SSADDA section on antisocial personality to identify the subjects to be screened for RSBs. To determine the STD status of the subjects, we used the response to the SSADDA item, ‘Has a doctor ever told you that you have (had): a sexually transmitted disease?'. Data from SSADDA interviews were also used to derive DSM-IV diagnosis of lifetime AD and other major psychiatric traits. Further details are available in our published GWAS of alcohol dependence in AAs and EAs (Gelernter et al, 2014a). Table 1 reports the characteristics of the sample analyzed here. We also used self-reported history of STD to evaluate the interaction between the loci identified and AD on STD status.
Table 1. Characteristics of Study Population.
| Ancestry-Sex | Age (SD) | RSB=0 (%) | RSB=1 (%) | RSB=2 (%) | STD (%) | Total |
|---|---|---|---|---|---|---|
| AD | ||||||
| AA-women | 40 (8) | 213 (47) | 151 (33) | 90 (20) | 151 (33) | 454 |
| AA-men | 43 (9) | 414 (38) | 466 (43) | 207 (19) | 328 (30) | 1087 |
| EA-women | 38 (11) | 196 (44) | 165 (37) | 83 (19) | 131 (30) | 444 |
| EA-men | 39 (11) | 465 (47) | 398 (40) | 129 (13) | 238 (24) | 992 |
| non-AD | ||||||
| AA-women | 41 (9) | 158 (68) | 52 (23) | 21 (9) | 43 (19) | 231 |
| AA-men | 43 (10) | 205 (51) | 160 (40) | 36 (9) | 76 (19) | 401 |
| EA-women | 38 (11) | 83 (58) | 41 (29) | 18 (13) | 33 (23) | 142 |
| EA-men | 39 (11) | 88 (51) | 69 (40) | 16 (9) | 36 (21) | 173 |
Genotyping and Imputation
We genotyped the samples using two different arrays. We used the Illumina HumanOmni1-Quad v1.0 microarray containing 988 306 autosomal SNPs to genotype 2660 subjects at the Center for Inherited Disease Research or the Yale Center for Genome Analysis; and the Illumina HumanCoreExome array to genotype 1264 additional subjects. Principal component (PC) analysis was conducted based on each genotyping array and for each ancestry group (AAs and EAs) using Eigensoft and SNPs that were common to the GWAS data sets and HapMap panel (after pruning the genome-wide SNPs for LD, r2>80%). Detailed information about the pre-imputation quality control pipeline is available in our published AD GWAS (Gelernter et al, 2014a). Imputation was performed using Impute2 software and the 1000 Genomes Phase 1 reference panel. After imputation, we included SNPs with minor allele frequency >5% and high imputation quality (certainty>0.9, info>0.8).
Genetic Data Analysis
We performed the GW-GxAD analysis using the R package GWAF to fit a generalized estimating equations (GEE) model to adjust for correlations among related individuals (Chen and Yang, 2010). Specifically, we used the geepack.quant.int.batch.imputed function (ie, GEE model including a main effect (SNP) and the effect of the interaction term (the product of allelic dosage and a covariate for interaction)) from the GWAF package to test the interaction between the imputed allele dosage and DSM-IV AD diagnosis for the RSB score, after adjusting for DSM-IV CD, OD, and ND diagnoses, age, and the first three ancestry PCs. The analysis was performed stratifying the samples by genotyping array, ancestry, and sex, and the results were combined by meta-analysis using the program METAL (Willer et al, 2010). The METAL approach uses the direction of effect and p-value observed in each study to compute a signed Z-score. Z-scores for each allele are combined across samples in a weighted sum, with weights proportional to the square-root of the sample size for each study. We applied a genomic control correction to all input files and negligible inflation of meta-analyzed p-values was observed (Supplementary Figure 1).
Neuroimaging Analysis
Unrelated and alcohol-exposed EA subjects (age range=18–23) were selected from the Brain and Alcohol Research in College Students (BARCS) study (Dager et al, 2013). All subjects underwent a single 5-min run of resting state functional magnetic resonance imaging (fMRI) on a 3T scanner as described previously (Meda et al, 2015). Following pre-processing, ALFF images were computed by extracting power spectra via a fast Fourier transform, and computing the sum of amplitudes in the 0.01–0.08 Hz frequency band. The ALFF measure at each voxel represents the averaged square root of the power in the above frequency windows normalized by the mean within-brain ALFF value for that subject. Following this, we used the REX toolbox to extract the mean ALFF signal from all anatomical atlas labeling regions of interest (ROIs). Participants were scanned for one run of the MID task to measure fMRI activation during anticipatory and consummatory reward, and loss processing. In our published MID task version (Andrews et al, 2011; Patel et al, 2013), there are three periods: (1) reward prospect (measuring subjective ‘wanting' of the reward), occurring following the initial cue; (2) reward anticipation (measuring anticipation of reward/loss), occurring following participant response, and (3) outcome feedback delivery (measuring reward experience) between box offset and outcome information varies randomly from 3000 to 5000 ms. We assessed main effects of these conditions and then used selected (left and right) ROIs to extract primary eigenvariate values. The subjects were genotyped using one of two arrays, the Illumina HumanOmni1-Quad or the Illumina HumanOmni2.5. PC analysis was conducted to confirm the European ancestry of the sample and to exclude cryptic relatedness. The best-guess genotypes of LHPP rs34997829 were estimated considering a genotype probability threshold ⩾0.8. Fifty-four subjects with genotype probability<0.8 were excluded from further analysis. The final sample size was 128 individuals. Linear regression analysis was performed considering ALFF and MID task traits as phenotypes, and age, sex, and the first three ancestry PCs as covariates. The false discovery rate was applied to correct the results for multiple testing (Benjamini and Hochberg, 1995) and q values<0.1 were considered to be significant.
RESULTS
AD and Differences in RSB
AD subjects were significantly more likely to reply affirmatively to both RSB questions I35B (p<1.1 × 10−10) and I37 (p=1.9 × 10−7; Supplementary Figure 2); thus they had higher RSB scores (p=1.57 × 10−13; Supplementary Figure 3) than non-AD subjects.
GWAS and GW-GxAD Analysis of RSB
In the trans-population meta-analyses, we observed one GWS signal in the GW-GxAD analysis. We observed that rs34997829, a SNP located within the LHPP gene, was GWS in the GW-GxAD analysis (z=−5.573, p=2.51 × 10−8; Figure 1, Table 2), and nominally significant with opposite effect directions in AD-stratified association analysis of RSB (AD: z=−2.032, p=0.042; non-AD: z=4.903, p=9.44 × 10−7). Specifically, the imputed allele dosage of rs34997829 was positively associated with RSB in non-AD subjects and negatively associated in AD samples (Supplementary Figure 4). Considering population-specific results, the significant result for LHPP rs34997829 comes from nearly equal support in both the African and European ancestry groups (AA Cohen's D=−0.083; EA Cohen's D=−0.096). The allele frequencies of the LHPP rs34997829 imputed allele were 0.668, 0.812, and 0.732 in AAs, EAs, and in the trans-population meta-analysis, respectively. Since an independent LHPP variant (rs35936514; no LD with rs34997829, Supplementary Figure 5) was recently associated with major depressive disorder (MDD) (Converge consortium, 2015), we also evaluated whether MDD status affected the RSB LHPP finding. Adjusting the analysis for MDD status, no change was observed in our genome-wide significant result (z=−5.707, p=1.15 × 10−8). We also observed that LHPP rs34997829 was not associated with AD and MDD in our sample (AD p=0.419; MDD p=0.055).
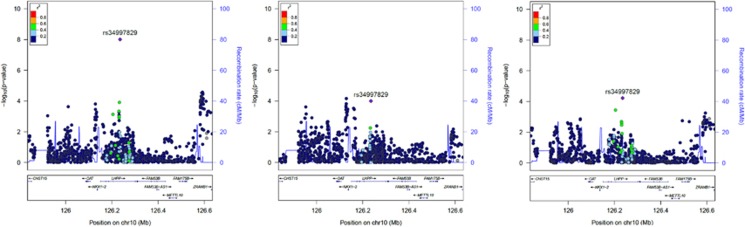
Figure 1.
Regional Manhattan plots of LHPP rs34997829 in the genome-wide gene-by-alcohol dependence (GW-GxAD) analyses of risky sexual behaviors (RSB) considering the trans-population meta-analysis (left), African-Americans (AAs; center), and European-Americans (EAs; right). There were no markers in either population (ie, African and European reference populations of the 1000 Genomes Project Phase 3) for which LHPP rs34997829 showed LD r2>0.6; therefore these figures appear comparatively sparse.
Table 2. LHPP rs34997829 Summary of GW-GxAD Results in Trans-Population Meta-Analysis, AAs, and EAs. AF: Imputed Allele Frequency; N: Sample Size.
| Meta-analysis | AF | N | Z score | P value |
|---|---|---|---|---|
| Trans-population | 0.732 | 3924 | −5.573 | 2.51 × 10−8 |
| AAs | 0.668 | 2173 | −3.888 | 1.01 × 10−4 |
| EAs | 0.812 | 1751 | −4.011 | 6.04 × 10−5 |
GxAD Analysis of STD Status
A self-reported history of STD was strongly associated with higher RSB scores in both AD and non-AD subjects (p<2.2 × 10−16 and p=2.2 × 10−11, respectively; Supplementary Figure 6) and with AD (p=0.024; Supplementary Figure 7). Performing a GxAD analysis of STD status, we observed a significant interaction of LHPP rs34997829 with AD on STD history in the trans-population meta-analysis (z=−2.809, p=4.97 × 10−3). The interaction for STD history was in the same direction as that observed for the RSB score: the variant is associated with STD in both AD and non-AD subjects but with opposite directions (AD z=−1.512; non-AD z=2.294). To verify the effect of RSB on STD results, we included RSB as a covariate in the model. No interaction was observed between LHPP and AD with respect to STD status after RSB adjustment (p=0.075).
Neuroimaging Follow-Up Analysis of LHPP rs34997829
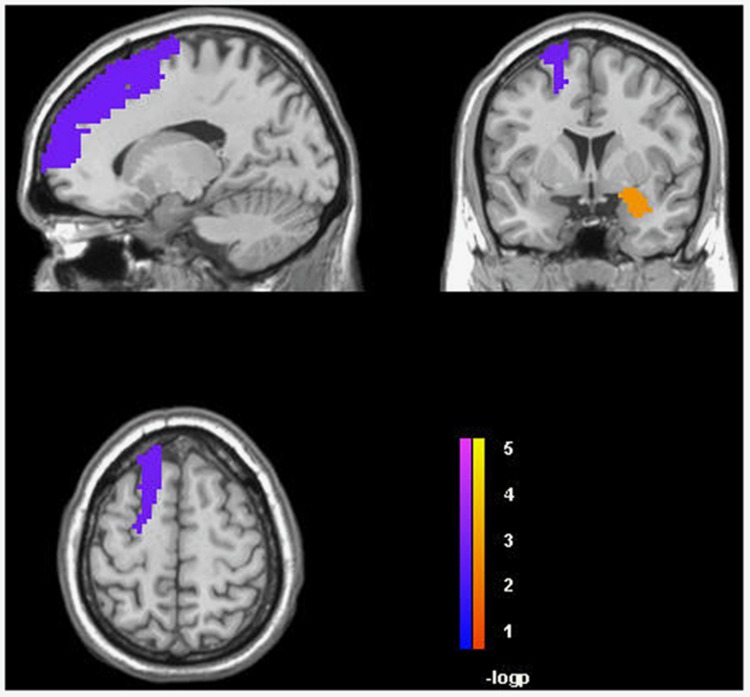
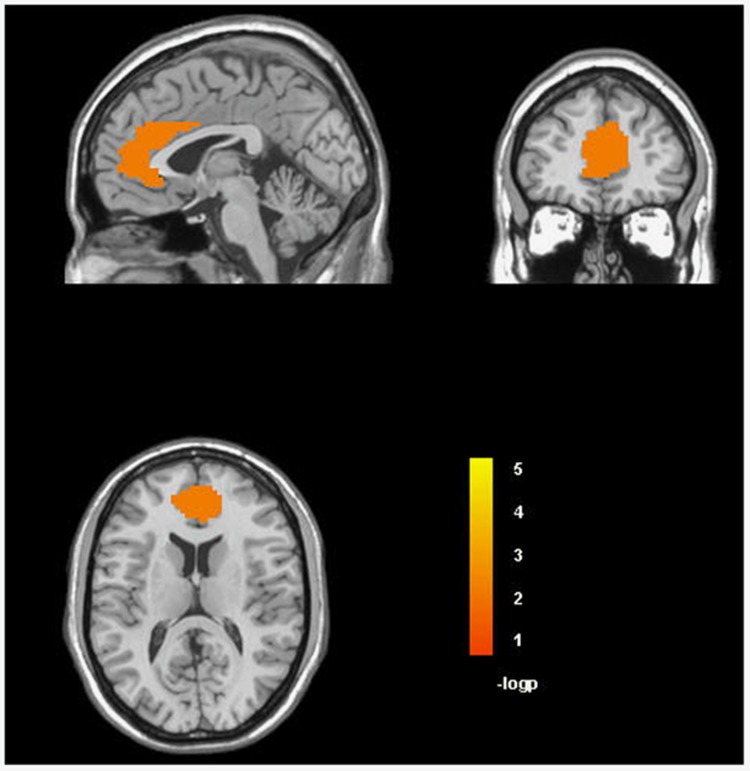
One hundred twenty-eight unrelated EA subjects were included in a follow-up fMRI analysis of LHPP rs34997829, the significant locus in the trans-population meta-analysis, confirmed by the GxAD analysis of STD status. In that analysis, we investigated both the resting ALFF signals in all ROIs prescribed using the automatic anatomical labeling atlas; and activation during reward and loss processing in selected ROIs (ie, anterior cingulate, Brodmann area 10, Brodmann area 11, insula, nucleus accumbens, para-hippocampal gyrus, ventral tegmental area, amygdala, caudate, and hippocampus) using the MID Task. We observed that the C allele of LHPP rs34997829 (ie, the imputed risk allele in our genome-wide investigation) was associated with reduced power of the left superior frontal gyrus (t=−3.386, p=9.56 × 10−4, q=0.076; Figure 2) and increased power of the right amygdala (t=3.287, p=1.33 × 10−3, q=0.076; Figure 2) in the ALFF analysis. In the MID task, LHPP rs34997829*C was associated with reduced activation (A1 L5 phase: lose $5 during the prospect phase) of the anterior cingulate (t=−2.961, p=3.69 × 10−3, q=0.06; Figure 3).
Figure 2.
Association of LHPP rs34997829 with the power of left superior frontal gyrus and the right amygdala in the amplitude of low frequency fluctuations (ALFF) analysis (false discovery rate (FDR) q<0.1). A full color version of this figure is available at the Neuropsychopharmacology journal online.
Figure 3.
Association of LHPP rs34997829 with the activation of anterior cingulate region in the monetary incentive delay (MID) task (false discovery rate (FDR) q<0.1). A full color version of this figure is available at the Neuropsychopharmacology journal online.
DISCUSSION
To our knowledge, this is the first reported genome-wide analysis of RSB in the context of AD. Our multi-ethnic genome-wide investigation yielded evidence for effects of the LHPP locus and AD, as well as the interaction of the two, on an ordinal measure of RSB that takes into account experiences of unprotected sex and multiple sexual partners.
In trans-population meta-analysis (N=3924), the only GWS finding was rs34997829 at LHPP. Specifically, LHPP rs34997829 was associated with RSB in opposite directions with respect to DSM-IV AD diagnosis (ie, crossing-interaction). This interaction scenario has been observed previously in psychiatric traits and disorders, and it is mainly detectable through genome-wide analysis (Dick, 2011; Murcray et al, 2009). However, since the association observed in AD subjects is close to the nominal significance threshold, our results may alternatively indicate that LHPP rs34997829 affects RSB in non-AD subjects only. The LHPP SNP rs34997829, according to ENCODE data (available at https://www.encodeproject.org/), is located in an experimentally validated regulatory intronic element involved in distal transcriptional regulation (ENCODE assay: Chromatin Interaction Paired-End Tags) and RNA binding protein mediated regulation (ENCODE assay: RNA IP Sequencing). A linkage and sequencing analysis on families with a strong history of MDD identified disease-segregating SNPs in LHPP that interacted with the HTR1A -1019G allele (Neff et al, 2009). A recent whole-genome sequencing analysis from the CONVERGE Consortium yielded statistically strong and consistent results, identifying a LHPP variant (rs35936514) associated with MDD in 10 640 Han Chinese women (Converge consortium, 2015). The depression-associated variant (ie, LHPP rs35936514) shows less variability in African and European populations than in Asians (minor allele frequency: Africa 18% Asia 28% and Europe 7% from AFR, EAS, and EUR reference populations, respectively, of 1000 Genomes Project Phase 3 (1000 Genomes Project Consortium et al, 2015). Our previous GWAS of cocaine dependence identified a GWS variant in FAM53B (rs2629540), located 190 kb from LHPP rs34997829 (Gelernter et al, 2014c), and this finding is also supported by a recent mouse-model study (Dickson et al, 2015). These three variants (ie, LHPP rs34997829, LHPP rs35936514, and FAM53B rs2629540) are not in linkage disequilibrium (r2<0.1) in the reference populations of 1000 Genomes Project Phase 3 and thus they represent independent signals (Supplementary Figure 5). In addition, we demonstrated that in our study the LHPP-AD interaction with respect RSB is not attributable to MDD. Although the existence of multiple independent GWS signals related to behavioral phenotypes in the same chromosomal region could be a chance finding, an alternate hypothesis is that they affect the same locus or mechanism involved in different behavioral phenotypes. Besides its GWS interaction with AD in relation to RSB, LHPP rs34997829 demonstrated significant interaction with AD in relation to STD status (p=4.97 × 10−3). This supports the clinical importance of the finding, and suggests that RSB associated with LHPP rs34997829 may mediate the risk of STD in the context of AD. In the fMRI follow-up investigation, the ALFF analysis showed that LHPP rs34997829 is associated in the inhibitory/excitatory mechanisms of frontal and subcortical regions (ie, superior frontal gyrus and amygdala, respectively) that govern emotional regulation (Banks et al, 2007; Ochsner et al, 2012). Resting ALFF analysis provide relevant data regarding spontaneous brain activity and signals originating in gray matter (Meda et al, 2015). Accordingly, although the significant association between LHPP rs34997829 and inhibitory/excitatory mechanisms of frontal and subcortical regions suggest the involvement of this locus in emotional regulation, further investigations are needed to understand how LHPP gene variation interacts with alcohol-related behaviors in contributing to the brain activities underlying RSBs. In the MID task, which is directly relevant to reward-driven impulsivity (Andrews et al, 2011; Patel et al, 2013), LHPP rs34997829 was associated with reduced activity in the anterior cingulate, a key region for impulse control/error monitoring/decisional choice (Carter et al, 1998; Hoerst et al, 2010). A previous fMRI MID task study observed reduced anterior cingulate activity during the A1 phase in drug users compared with controls, correlating it to impulsivity-compulsivity factors (Patel et al, 2013) similar to LHPP rs34997829.
As noted above, LHPP has recently been confirmed as a genetic risk locus for MDD, based on a whole genome sequencing study in a Chinese population (Converge consortium, 2015). A consistent literature has reported that RSBs are more frequent among patients with depression and other psychiatric disorders than in the general population (Ramrakha et al, 2000; Tull and Gratz, 2013). The molecular mechanisms at the basis of psychiatric diseases may include pathogenic pathways that cause the increased RSB observed in psychiatric patients. Subjects with AD also present increased RSB (Chaney et al, 2016; Corbin et al, 2016; Khan et al, 2012; Tran et al, 2016). We hypothesize that AD affects the same molecular mechanisms responsible for RSB in psychiatric patients. Accordingly, the identification of LHPP, a risk locus for MDD, as an AD-interactive locus in relation to RSB seems to support our hypothesis.
In summary, we performed the first genome-wide analyses of RSB and RSB-AD interplay and our finding indicated potential mechanisms related to mental illness. We also observed that LHPP allele related to RSB-AD interplay can be associated with the interplay between AD and STD, linking behavior genes to the predisposition to STDs. LHPP rs34997829 was then evaluated in a neuroimaging paradigm, and that analysis provided supported for biological function of the risk variant. To our knowledge, no other large studies are publically available to replicate our current findings. For such a replication to be accomplished, it would be necessary to locate a large sample with DSM-IV AD diagnosis status and, ideally, a more sophisticated tool to assess RSB. Nevertheless, the LHPP rs34997829 result is supported by multiple findings, including trans-population evidence, involvement in RSB clinical consequences, and supportive neuroimaging data. The genetics of RSB is an under-investigated field. Our current data not only present novel findings, but also shed an innovative light on this topic. Molecular data can provide a basis to develop important preventive treatments, especially for high-risk categories like people with substance use disorders. Indeed, the LHPP protein product should be a promising target to develop drug therapies since its enzymatic function could be modulated by inhibitors or stimulators. Such pharmacological therapies might have multiple preventive effects with respect to depressive conditions and/or RSBs, reducing the consequences of these psychological traits.
FUNDING AND DISCLOSURE
This study was supported by National Institutes of Health grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, P50 AA012870, R01 AA016599, RC1 AA019036, the Connecticut and Crescenz Philadelphia VA MIRECCs, and a NARSAD Young Investigator Award (to RP) from the Brain & Behavior Research Foundation. Dr. Kranzler has been an advisory board member, consultant, or CME speaker for Indivior, Lundbeck, and Otsuka. He is also a member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, which is supported by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Pfizer, and XenoPort.The other authors reported no biomedical financial interests or potential conflicts of interest.
Acknowledgments
We appreciate the work in recruitment and assessment provided at Yale University School of Medicine and the APT Foundation by James Poling, PhD; at McLean Hospital by Roger Weiss, M.D.; at the Medical University of South Carolina by Kathleen Brady, MD, PhD and Raymond F. Anton, MD; and at the University of Pennsylvania by David Oslin, MD. Genotyping services for a part of our GWAS study were provided by the Center for Inherited Disease Research (CIDR) and Yale University Center for Genome Analysis. CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). We are grateful to Ann Marie Lacobelle, Catherine Aldi and Christa Robinson for their excellent technical assistance, to the SSADDA interviewers, led by Yari Nunez and Michelle Slivinsky, who devoted substantial time and effort to phenotype the study sample and to John Farrell and Alexan Mardigan for database management assistance.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp).
Supplementary Material
References
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM et al,1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (1994) Diagnostic and Statistical Manual of Mental Disorders 4th Ed. American Psychiatric Press: Washington, DC, USA. [Google Scholar]
- Andrews MM, Meda SA, Thomas AD, Potenza MN, Krystal JH, Worhunsky P et al (2011). Individuals family history positive for alcoholism show functional magnetic resonance imaging differences in reward sensitivity that are related to impulsivity factors. Biol Psychiatry 69: 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks SJ, Eddy KT, Angstadt M, Nathan PJ, Phan KL (2007). Amygdala-frontal connectivity during emotion regulation. Soc Cogn Affect Neurosci 2: 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 57: 289–300. [Google Scholar]
- Carter CS, Braver TS, Barch DM, Botvinick MM, Noll D, Cohen JD (1998). Anterior cingulate cortex, error detection, and the online monitoring of performance. Science 280: 747–749. [DOI] [PubMed] [Google Scholar]
- Chaney BH, Vail-Smith K, Martin RJ, Cremeens-Matthews J (2016). Alcohol use, risky sexual behavior, and condom possession among bar patrons. Addict Behav 60: 32–36. [DOI] [PubMed] [Google Scholar]
- Chen MH, Yang Q (2010). GWAF: an R package for genome-wide association analyses with family data. Bioinformatics 26: 580–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converge consortium (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523: 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin WR, Scott CJ, Treat TA (2016). Sociosexual attitudes, sociosexual behaviors, and alcohol use. J Stud Alcohol Drugs 77: 629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dager AD, Anderson BM, Stevens MC, Pulido C, Rosen R, Jiantonio-Kelly RE et al (2013). Influence of alcohol use and family history of alcoholism on neural response to alcohol cues in college drinkers. Alcohol Clin Exp Res 37(Suppl 1): E161–E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Helgason H, Chasman DI, Rose LM, Loh PR, Scott RA et al (2016). Physical and neurobehavioral determinants of reproductive onset and success. Nat Genet 48: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM (2011). Gene-environment interaction in psychological traits and disorders. Annu Rev Clin Psychol 7: 383–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson PE, Miller MM, Calton MA, Bubier JA, Cook MN, Goldowitz D et al (2015). Systems genetics of intravenous cocaine self-administration in the BXD recombinant inbred mouse panel. Psychopharmacology (Berl) 233: 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R et al (2015). Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry 77: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH et al (2014. a). Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 19: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao HY et al (2014. b). Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry 76: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR et al (2014. c). Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry 19: 717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerst M, Weber-Fahr W, Tunc-Skarka N, Ruf M, Bohus M, Schmahl C et al (2010). Correlation of glutamate levels in the anterior cingulate cortex with self-reported impulsivity in patients with borderline personality disorder and healthy controls. Arch Gen Psychiatry 67: 946–954. [DOI] [PubMed] [Google Scholar]
- Khan MR, Berger AT, Wells BE, Cleland CM (2012). Longitudinal associations between adolescent alcohol use and adulthood sexual risk behavior and sexually transmitted infection in the United States: assessment of differences by race. Am J Public Health 102: 867–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Wang Z, Ivleva EI, Poudyal G, Keshavan MS, Tamminga CA et al (2015). Frequency-specific neural signatures of spontaneous low-frequency resting state fluctuations in psychosis: evidence from bipolar-schizophrenia network on intermediate phenotypes (B-SNIP) Consortium. Schizophr Bull 41: 1336–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcray CE, Lewinger JP, Gauderman WJ (2009). Gene-environment interaction in genome-wide association studies. Am J Epidemiol 169: 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff CD, Abkevich V, Packer JC, Chen Y, Potter J, Riley R et al (2009). Evidence for HTR1A and LHPP as interacting genetic risk factors in major depression. Mol Psychiatry 14: 621–630. [DOI] [PubMed] [Google Scholar]
- Ochsner KN, Silvers JA, Buhle JT (2012). Functional imaging studies of emotion regulation: a synthetic review and evolving model of the cognitive control of emotion. Ann N Y Acad Sci 1251: E1–E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KT, Stevens MC, Meda SA, Muska C, Thomas AD, Potenza MN et al (2013). Robust changes in reward circuitry during reward loss in current and former cocaine users during performance of a monetary incentive delay task. Biol Psychiatry 74: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A et al (2005). Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA). Drug Alcohol Depend 80: 303–312. [DOI] [PubMed] [Google Scholar]
- Polimanti R, Kranzler HR, Gelernter J (2016). Phenome-wide association study for alcohol and nicotine risk alleles in 26,394 Women. Neuropsychopharmacology (e-pub ahead of print; doi:10.1038/npp.2016.72). [DOI] [PMC free article] [PubMed]
- Polimanti R, Zhang H, Smith AH, Zhao H, Farrer LA, Kranzler HR et al (2015). Genome-wide association study of body mass index in subjects with alcohol dependence. Addict Biol (e-pub ahead of print; doi: 10.1111/adb.12317). [DOI] [PMC free article] [PubMed]
- Ramrakha S, Caspi A, Dickson N, Moffitt TE, Paul C (2000). Psychiatric disorders and risky sexual behaviour in young adulthood: cross sectional study in birth cohort. BMJ 321: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A et al (2016). Genome-wide association study of cannabis dependence severity, novel risk variants, and shared genetic risks. JAMA Psychiatry 73: 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R (2009). Alcoholism: a systems approach from molecular physiology to addictive behavior. Physiol Rev 89: 649–705. [DOI] [PubMed] [Google Scholar]
- Tran BX, Nguyen LH, Nguyen CT, Phan HT, Latkin CA (2016). Alcohol abuse increases the risk of HIV infection and diminishes health status of clients attending HIV testing services in Vietnam. Harm Reduct J 13: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tull MT, Gratz KL (2013). Major depression and risky sexual behavior among substance dependent patients: the moderating roles of distress tolerance and gender. Cognit Ther Res 37: 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.