Abstract
Background
The excessive and uncontrolled use of anthelmintics, e.g. ivermectin (IVM) for the treatment of livestock parasites has led to widespread resistance in gastrointestinal nematodes, such as Haemonchus contortus. There is an urgent need for better management of drug-use in nematode control and development of novel anthelmintics. Discovery and identification of anthelmintic resistance-associate molecules/markers can provide a basis for rational anthelmintics-use and development of novel drugs. Recent studies have shown that ivermectin resistance in H. contortus is likely to be multi-genic in nature except for several genes coding for IVM target and efflux pump. However, no other IVM resistance-associated genes were characterized by conventional methods or strategies. In the present study we adopted a new strategy, i.e. using genome-wide single nucleotide polymorphism (SNP) analysis based on 2b-RAD sequencing, for discovering SNPs markers across the genomes in both IVM susceptible and resistant isolates of H. contortus and identifying potential IVM resistance-associated genes.
Results
We discovered 2962 and 2667 SNPs within both susceptible and resistant strains of H. contortus, respectively. A relative lower and similar genetic variations were observed within both resistant and susceptible strains (average π values were equal to 0.1883 and 0.1953, respectively); whereas a high genetic variation was found across both strains (average π value was equal to 0.3899). A significant differentiation across 2b-RAD tags nucleotide sites was also observed between the two strains (average FST value was equal to 0.3076); the larger differences in average FST were observed at SNPs loci between coding and noncoding (including intronic) regions. Comparison between resistant and susceptible strains revealed that 208 SNPs loci exhibited significantly elevated FST values, 24 SNPs of those loci were located in the CDS regions of the nine genes and were likely to have signature of IVM directional selection. Seven of the nine candidate genes were predicted to code for some functional proteins such as potential IVM target and/or efflux pump proteins, component proteins of receptor complex in membrane on neuromuscular cells, and transcriptional regulation proteins. Those genes might be involved in resistance to IVM.
Conclusions
Our data suggest that candidate genes putatively associated with resistance to IVM in H. contortus may be identified by genome-wide SNP analysis using 2b-RAD sequencing.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-016-1959-6) contains supplementary material, which is available to authorized users.
Keywords: Candidate ivermectin resistance-associate genes, Haemonchus contortus, 2b-RAD sequencing, Genome-wide SNP analysis
Background
Parasite nematodes are major causes of morbidity in sheep and cattle. The infections can decrease production of meat and milk in those livestock animals. No vaccines are available for these diseases by far and major control measures rely on the use of anthelmintic drugs. The excessive and uncontrolled use of anthelmintics for the treatment of nematode diseases has led to widespread resistance in livestock nematodes [1]. Clearly, there is an urgent need for better management of drug-use in nematode control. To this end, development of molecular markers for anthelmintic resistance diagnosis is an attractive option for improvements in drug-use decisions [2]. In addition, identification of resistance markers can also help increase our understanding of mechanisms of drug effects, and provide the basis for development of novel anthemintics [2, 3].
Ivermectin (IVM) belongs to the macrocyclic lactone (ML) family of antiparasiticides. Introduced into the market in the early 1980s IVM has been widely used due to its broad-spectrum activity for the control of parasitic nematodes and ectoparasites in animals. As a consequence, IVM resistance has become widespread in nematodes of livestock [3]. Unfortunately, our understanding of the molecular mechanisms underlying resistance to IVM remains far from complete, despite some early indications that resistance to IVM may be due to specific polymorphisms in the drug target receptors, e.g. glutamate-gated chloride ion channel receptors (GluCIRs) [4, 5]. To date, there are no yet mutations identified that can explain the observed resistance to IVM in most field isolates of the parasitic nematode species [2]. In more recent years, studies on the likely genetic mechanisms of resistance suggest that ivermectin resistance in nematodes is multi-genic in nature [6, 7]. Therefore, it is important to discover novel genes associated with resistance to ivermectin for evaluating and testing most cases of field resistance in nematode species. One of the most important research priorities for anthelmintic resistance is to identify mutations in parasite genes that give rise to modification of drug target or nontarget-dependent development of resistance. These studies often examine a small number of loci, sometimes only one, and result in the missed detection for resistance-associated genes, and candidate gene studies are also based on prior assumptions about possible mechanisms of resistance. This situation has major limitations in identifying novel resistant genes and unsuspected mechanisms of resistance [2].
Recent progress resulting in genomic data available for some parasitic nematode species [8, 9], together with the advances of next generation sequencing (NGS) methods for genome-wide genetic marker discovery and genotyping, make it possible for researchers to screen out potential drug resistance genes in the whole genome scale [2, 10]. The principle of candidate gene discovery mentioned above is based on the genome-wide association study (GWAS), also known as whole genome association study (WGAS), which looks for associations between DNA sequence variants and phenotypes of interest. Conventional approach is to type thousands to hundreds of thousands of single nucleotide polymorphisms (SNPs), known as genetic markers, across the genome of interest, and compare the differences of these genetic markers between one case group and a control group, and identify regions/loci of genome/genes with variant genetic markers which are likely to be associated with traits like diseases or drug resistance [11].
2b-RADseq is a restriction site-associated DNA (RAD) sequencing based on sequencing the uniform fragments produced by type IIB restriction end nucleases, it produces high coverage of homologous SNP loci of fixed length, and provides a powerful method for genome-wide SNPs discovery and genomic studies at the population level [12, 13]. 2b-RAD is also well suited to identify genomic regions/loci under selection because of the uniform high density of markers across genomes [14]. It is a cost-effective method, and can be used in routine experimental laboratory. The aim of this study was to provide a preliminary “proof of principle” that candidate anthelmintic resistance-associated genes could be identified by identifying genome-wide signatures of drug selection. To this end, we first applied the 2b-RAD technique to discover thousands of SNPs in both susceptible and resistant strains of Haemonchus contortus to ivermectin, and investigated the patterns of genetic diversity and population differentiation across genome of the two strains using above SNPs markers; then we examined the variation in SNP allele frequencies which can be quantified by the statistic FST between the two strains, and identified candidate resistance-associated genes with signatures of anthelmintic selection by analyzing SNPs loci that exhibited significantly elevated FST values between the resistant and susceptible strains.
Methods
Collection of Haemonchus contortus
The susceptible strain of H. contortus was originally obtained from Australian and maintained for 2 years in sheep in Huazhong Agricultural University; a resistant strain of H. contortus to ivermectin was originally obtained from Moredern Institute of England and maintained for 6 years in sheep in Inner Mongolia Agricultural University. We first tested whether the above-mentioned strains were susceptible or resistant to IVM by using Larval Development Assay (LDA) [15–17]. Indigenous male goats at the age of 3 months were transported from pasture to pens and treated with ivermectin (at dosage of 0.4 mg/kg) and albendazole (at dosage of 30 mg/kg). Each goat was housed in a single pen, and had a free access to a commercial-concentrated-feeding-stuff and water. After 7 days, faecal samples were collected and examined by a modified McMaster technique. Twenty days later nematode egg counts of all animals were found to have negative values (mean faecal egg count of 0 eggs per gram) [18]. One goat was infected with approximately 7000 of the third-stage larvae of susceptible or resistant isolates of H. contortus, respectively. On day 30 post-infection, egg counts were examined, eggs were collected, and 100 eggs were used for LDA in each of both experimental and control groups. The results showed that the susceptible strain was very sensitive to ivermectin; whereas resistant strain had a lethal dose of 428.38 ng/ml (LD99) (see Additional file 1: Tables S1 and S2). The results of aforementioned assays indicated that the two strains could be used as the resources of worm samples in the following experiments. On day 15 after examination of LDA, the animals were slaughtered and adult worms were collected for further use.
Library construction and sequencing
2b-RAD libraries were prepared for H. contortus samples by following the protocol developed by Wang et al. [19]. Briefly, each of six genomic DNA samples (3 samples from each of the above-mentioned two strains of H. contortus) was extracted by phenol-chloroform method using GenElute Genomic DNA Miniprep Kit (Sigma-Aldrich, Shanghai, China); each sample contained a pool of four adult individuals. DNA from each sample was digested using BsaXI, then verified and separated on agarose gel. Next, library-specific adaptors and the digestion products were linked with T4 DNA ligase. Ligation products were amplified by PCR and the target band was excised from a 2% agarose gel. Finally, sample-specific barcodes were introduced by PCR with platform-specific barcode-bearing primers. PCR products were purified using QIA quick PCR purification kit and then pooled for sequencing using the Illumina HiseqXTen platform. All of the 2b-RAD sequences were archived in the NCBI SRA database (Assay ID: ss2137098375–2137102521).
Sequence data processing, SNPs discovery and identification of candidate resistance-associated genes
We used Stacks tool with default parameters described by Catchen et al. [20] to conduct the data processing the raw 2b-RAD sequences, discovery of SNPs, patterns of genetic diversity and population differentiation across genome of the two strains and identification of candidate resistance-associated genes. Briefly, billions of raw reads were filtered and cleaned, the data cleaned were aligned to a reference genome of H. contortus (ftp://ftp.sanger.ac.uk/pub/pathogens/Haemonchus/contortus) which was a version of draft assembly consisting of 67,687 contigs linked into 26,044 scaffolds of total length 370 Mb, and SNPs markers were identified across all of scaffolds of both strains, and density of SNPs was calculated across nucleotide sites for which sequence information was generated. We calculated population genomic statistics according to method described by Hohenlohe et al. [21] using Stacks tool, we measured the degree of polymorphism within populations with statistic nucleotide diversity π (equivalent to expected heterozygosity) and differentiation among populations with statistic fixation index FST. We calculated average values of FST across whole-genome using a kernel smoothing approach described by Hohenlohe et al. [21], and the feature of SNPs distribution was examined by comparison of average FST values of SNPs that were located in three different regions (CDS, intronic and noncoding regions) according to the approach described by Akey et al. [22]. We identified SNPs loci with signature of selection by selecting SNPs sites that exhibited significantly elevated FST values between the resistant and susceptible strains (Smoothed AMOVA FST > 0.4652 and P < 0.16). We identified candidate resistance-associated genes by using following criteria: (i) the genes contained above-mentioned SNPs loci with signature of selection; (ii) those SNPs were located in CDS region and were found in only resistant strain; and (iii) the genes had been annotated. Functional information and annotation of the genes were analyzed by interrogating WormBase.
Results
SNPs discovery and their features of distribution
To efficiently discover SNPs in both susceptible and resistant strains of H. contortus to ivermectin, we adopted 2b-RAD technique. After raw reads data were filtered and processed by using Stacks software, 2962 and 2667 SNPs were identified within both susceptible and resistant strains of H. contortus, respectively (Additional file 2: Table S3). To further determine whether above-mentioned SNPs are representatives of SNPs across the genome, we analyzed their features of distribution. As described by Laing et al. [8], draft assembly of H. contortus genome consisting of 67,687 contigs linked into 26,044 scaffolds with a total length of 370 Mb, we observed that a total number of 2,176,234 nucleotide sites for which sequence information was generated, could be aligned to 9258 scaffolds of total length 348.936639 Mb (Additional file 2: Table S3), hence the total number of nucleotide sites accounted for about 0.6% of the H. contortus genome (Table 1). In addition, almost half of the nucleotide sites were aligned to the top 1000 scaffolds which had almost half of length of the genome. It was the case for the distribution of SNPs identified across the nucleotide sites (Table 1). We also calculated the density of these SNPs. Since 4873 SNPs identified were distributed across these 2,176,234 nucleotide sites, therefore the density of SNPs across the nucleotide sites was about 1/447 bp (Table 1). This density is lower than that of SNPs across the genome described by Gilleard et al. [23]; these authors provided some data on the level of SNPs across the whole genome in several laboratory strains with the density of the SNPs of 1/202–1/283 bp. The low density of SNPs identified in this study may be due partly to the fact that number of worm samples sequenced is small. Overall, these findings suggest that the distribution of SNPs across the nucleotide sites is similar to that of SNPs across the genome.
Table 1.
Nucleotide sites and SNPs identified in scaffolds
| Scaffolds | Lengtha | Sitesb | IRc | LS | IR_LS | Density of SNPsd | Site contente |
|---|---|---|---|---|---|---|---|
| 1 to 1000 | 171,808,466 | 1,047,366 | 1381 | 1465 | 2454 | 1/427 | 0.61% |
| Others | 177,128,173 | 1,128,868 | 1286 | 1497 | 2419 | 1/467 | 0.64% |
| Total (9258) | 348,936,639 | 2,176,234 | 2667 | 2962 | 4873 | 1/447 | 0.62% |
aTotal length (bp) of genome
bThe total number of nucleotide sites for which sequence information was generated in at least one sample, after trimming restriction enzyme recognition sequence
cThe remaining columns give the number of single-nucleotide polymorphisms identified within each population. IR population is resistant strain of H. contortus, LS is susceptible strain of H. contortus and IR_LS is 2 populations combined
dDensity of SNPs = No. of IR_LS/No. of sites
eSite content = No. of sites/Total length (bp) of genome
Furthermore, we analyzed the feature of distribution of the SNPs identified indifferent functional regions (i.e. CDS, intronic and noncoding), and found that the larger differences in average FST were observed between coding and noncoding as well as intronic SNPs (Table 2), which is consistent with the finding described by Akey et al. [22] who observed a similar feature of distribution of FST in human genes. Taken together, the SNPs identified in this study can provide an excellent fractional representation of the total of SNPs across the entire genome of H. contortus and can be used in further analysis.
Table 2.
Average FST as a function of SNP category
| N | Average FST | SE | |
|---|---|---|---|
| CDS | 484 | 0.2700104 | 0.011941471 |
| Intro | 847 | 0.3163460 | 0.009877181 |
| Non-coding | 1472 | 0.3069602 | 0.007520325 |
Abbreviations: N the number of SNPs within each region; SE standard error
Genome-wide estimates of genetic diversity and population differentiation
To understand whether the susceptible and resistant strains have been subject to different selection pressures, we analyzed genetic diversity and population differentiation within or among the two strains using these SNPs across genome (Fig. 1). We identified lower and similar genetic variation within strains, the average genetic diversity π values were equal to 0.1883 and 0.1953 within the susceptible and resistant strains, respectively; and a significant genetic variation was observed across both strains, the average π value was 0.3899 (Table 3). This finding is in agreement with previous studies of genetic variation within and among susceptible and resistant populations in the field [24]. A significant differentiation across 2b-RAD tags nucleotide sites was also observed between the two strains (Fig. 1c), the average FST value was equal to 0.3076 (Table 3). These estimates indicated that the susceptible and resistant strains had been affected by different selection and provided a basis for further identification of a genome-wide signature of selection.
Fig. 1.
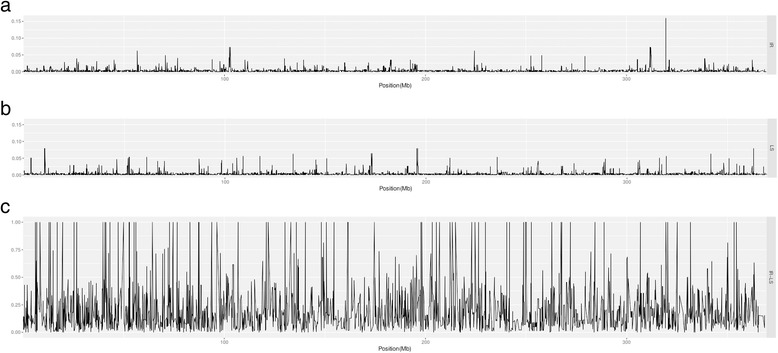
Patterns of genetic diversity and population differentiation distributed across the genome. a Genome-wide patterns of nucleotide diversity for resistant strain of H. contortus. b Genome-wide patterns of nucleotide diversity for susceptible strain of H. contortus. c Genome-wide differentiation among populations of susceptible and resistant strains of H. contortus
Table 3.
Pairwise nucleotide diversity and population differentiation among two H. contortus populations. Above the diagonal: the average nucleotide diversity (π) in each combined pair of the populations; along the diagonal: π within each single population; below the diagonal: average FST between the two populations
| IR | LS | |
|---|---|---|
| IR | 0.1883 | 0.3899043 |
| LS | 0.3076215 | 0.1953 |
Abbreviations: IR resistant H. contortus; LS susceptible H. contortus
Identification of candidate resistance-associated genes
To identify candidate genes that could be subject to IVM selection, we first selected those SNPs loci whose FST values were significantly greater than that of the genome-wide average 0.3076 (F ST > 0.4652 and P ≤ 0.16) (see Methods), and found that a total of 208 SNPs loci met this criterion (Additional file 3: Table S4), of which 24 loci were located in the CDS regions and the genes contained these loci had annotation information (Table 4). Then, among these 24 SNPs loci we selected those SNPs that were resistant strain-specific (i.e. only presented in resistant strain) for further analysis, we obtained nine SNPs loci that may have been affected by IVM selection (Table 4). In addition, we further analyzed the functional information of the genes that contained these nine SNPs loci by interrogating wormbase and relevant literatures, finally we identified seven candidate resistance-associated genes that encompassed eight SNPs loci (Table 5).
Table 4.
SNPs loci that exhibit significantly elevated FST value, identified in CDS regions
| Chr | BPa | Smoothed AMOVA FST | Smoothed AMOVA FST P-value | Gene ID |
|---|---|---|---|---|
| Scaffold_11180 | 1238 | 1 | < 0.05 | cds:HCOI00022400.t1 |
| Scaffold_1311 | 4966 | 1 | < 0.05 | cds:HCOI00277600.t1 |
| Scaffold_1311 | 4972 | 1 | < 0.05 | cds:HCOI00277600.t1 |
| Scaffold_1409 | 28,964 | 1 | < 0.05 | cds:HCOI00335100.t1 |
| Scaffold_148 | 58,650 | 0.750886831 | < 0.05 | bcds:HCOI00506600.t3 |
| Scaffold_148 | 65,574 | 0.755702686 | < 0.05 | bcds:HCOI00506600.t3 |
| Scaffold_1483 | 47,819 | 1 | < 0.05 | bcds:HCOI00378500.t1 |
| Scaffold_1511 | 40,561 | 1 | < 0.05 | cds:HCOI00392800.t1 |
| Scaffold_1595 | 23,974 | 1 | < 0.05 | cds:HCOI00449900.t1 |
| Scaffold_207 | 42,678 | 1 | < 0.05 | cds:HCOI00808400.t1 |
| Scaffold_2990 | 2020 | 1 | < 0.05 | bcds:HCOI01034200.t1 |
| Scaffold_340 | 169,069 | 1 | < 0.05 | bcds:HCOI01315600.t1 |
| Scaffold_340 | 169,075 | 1 | < 0.05 | cds:HCOI01315600.t1 |
| Scaffold_3511 | 5425 | 1 | < 0.05 | bcds:HCOI01200700.t1 |
| Scaffold_3558 | 3252 | 1 | < 0.05 | cds:HCOI01210600.t1 |
| Scaffold_3989 | 17,378 | 1 | < 0.05 | cds:HCOI01318200.t3 |
| Scaffold_4175 | 4946 | 1 | < 0.05 | bcds:HCOI01355700.t1 |
| Scaffold_4362 | 13,965 | 1 | < 0.05 | bcds:HCOI00703000.t1 |
| Scaffold_4362 | 13,975 | 1 | < 0.05 | cds:HCOI00703000.t1 |
| Scaffold_4362 | 13,977 | 1 | < 0.05 | cds:HCOI00703000.t1 |
| Scaffold_645 | 29,323 | 0.784867758 | < 0.05 | cds:HCOI01894100.t1 |
| Scaffold_6841 | 192 | 1 | < 0.05 | cds:HCOI01753600.t1 |
| Scaffold_774 | 42,481 | 1 | < 0.05 | bcds:HCOI02035600.t2 |
| Scaffold_793 | 2897 | 0.465277778 | 0.16 | bcds:HCOI02054500.t1 |
aBase position of SNP loci located in a given scaffold
bThe SNPs that are resistant strain-specific and only present in a resistant strain
Table 5.
Candidate genes related to IVM resistance, identified within CDS regions that exhibit significantly elevated FST value
| Scaffold | BPa | Gene ID | Annotation |
|---|---|---|---|
| Scaffold_148 | 58,650 | cds:HCOI00506600.t3 | Low density lipoprotein-receptor domain containing protein |
| Scaffold_148 | 65,574 | cds:HCOI00506600.t3 | Low density lipoprotein-receptor domain containing protein |
| Scaffold_1483 | 47,819 | cds:HCOI00378500.t1 | 4Fe-4S ferredoxin and ABC transporter domain containing protein |
| Scaffold_340 | 169,069 | cds:HCOI01315600.t1 | RNA polymerase-associated protein RTF1 |
| Scaffold_3511 | 5425 | cds:HCOI01200700.t1 | L27-1 and PDZ and Src homology-3 and guanylate kinase domain containing protein |
| Scaffold_4362 | 13,965 | cds:HCOI00703000.t1 | AIR synthase related protein domain containing protein |
| Scaffold_774 | 42,481 | cds:HCOI02035600.t2 | WW Rsp5 WWP and FF domain containing protein |
| Scaffold_793 | 2897 | cds:HCOI02054500.t1 | Neurotransmitter-gatedion-channel ligand-binding |
aBase position of SNP loci located in a given scaffold
After analyzing functional information of relevant genes according to the Wormbase data and references, we selected those genes that might take part in changes in the drug receptor or modulation of drug concentration as candidate genes; they may code for some functional molecules such as potential IVM target and/or efflux pump proteins, component proteins of receptor complex in membrane on neuromuscular cells, and transcriptional regulation proteins. We found that the four molecules encoded by HCOI02054500, HCOI00378500, HCOI01200700 and HCOI02035600 genes, respectively, were likely to be a potential IVM target or efflux pump proteins. According to Wormbase (http://parasite.wormbase.org/Haemonchus_contortus_prjeb506/Info/Index) we found that the protein encoded by the HCOI02054500 gene was likely to have the function similar to that of GABA receptors; the protein encoded by the HCOI00378500 gene may participate transfer of energy and substrates; and the two proteins, encoded by the HCOI01200700 and HCOI02035600 genes may participate in the formation of receptor complex in membrane on the neurons and or muscle cells of H. contortus, respectively. We also found that the protein encoded by the HCOI00506600 gene may take part in lipid metabolism and influence the nematode P-gp activity. Meanwhile, we inferred that enzyme encoded by HCOI00703000 was likely to be a new class of ATP-binding proteins and may be related to the drug metabolism and detoxification. A recent study had indicated that some genes responsible for transcriptional regulation may also be involved in IVM resistance [25]; according to the annotation, we inferred that molecule encoded by the HCOI01315600 gene was likely to play a role in the regulation of transcription of ABC transporter genes in H. contortus.
As for the HCOI01034200 and HCOI01355700 genes which encode glycoside hydrolase domain containing protein and peptidase S9 domain containing protein, respectively, we were unable to provide evidence that the proteins encoded by the two genes are likely to be responsible for the resistance to IVM in H. contortus. Hence, we did not categorize them into our list of candidate IVM resistance-associate genes.
Discussion
Haemonchus contortus has shown a great ability to develop resistance to all the anthelmintic drug classes including macro cyclic (ML) lactones, e.g. IVM [3]. Previous studies have shown that changes in target site and drug efflux pathways, e.g. over expression of a number of P-glycoprotein genes may play roles in resistant isolates to IVM, but no definitive mechanism could explain the observed field resistance [2]. Recent investigations on the likely genetic mechanisms of resistance have indicated that multigenic basis and another mechanism may have contributed to ML resistance in H. contortus [7, 26, 27]. Therefore, discovery and identification of novel molecules responsible for ML resistance has caught more attention. Conventional strategy for discovery of candidate anthelmintic resistance-associated molecules in H. contortus is to draw on the experience of similar work in mammals and C. elegans worms [28–31]. In this present study, we adopted a new strategy for the first time, i.e. genome-wide scan based on SNPs analysis to identify other candidate genes or molecular markers associated with resistance to IVM in H. contortus.
Our strategy is based on a basic principle of population genomics, i.e. under the condition of selective neutrality, FST statistic of population is determined by genetic drift, which will affect all loci across the genome in a similar fashion, but naturals election (e.g. under pressure of drug) is a locus-specific force that can cause systematic deviations in FST values for a selected gene and nearby genetic markers [32–34]. Therefore, for FST statistic we can calculate both a genome-wide average and outliers by population genomics methods, the genome-wide average provides a baseline value of neutral processes, and outliers from the background are likely to be the locus-specific signature of positive directional selection [22, 32]. It is noteworthy to mention that such strategy has been reported in several recent studies, e.g. Cheeseman et al. [23, 35] reported that they were able to identify genome regions underlying Artemisia in resistance in malaria by mapping genome-wide divergence (FST) between drug-resistant and drug-sensitive parasites . Tennessen et al. [36] also adopted a similar strategy to identify a region of the snail Biomphalaria glabrata genome that correlates with resistance to Schistosoma mansoni infection. So it is a feasible that this approach can be applied to our study.
In order to obtain genome-wide DNA markers, we first identified thousands of SNPs from 2b-RAD tag sequences for both resistant and sensitive to IVM worms. Relative lower density (about 7.56 SNPs per Mb) and genetic diversity of the SNPs were observed across the whole genome within both strains, this is due partly to the fact that the number of worm samples (similar to the term census population size) sequenced is small, this is consistent with the results of previous investigations indicating that the high levels of genetic diversity within H. contortus populations are largely due to their large census population sizes [24]. On the other hand, the factors that adult worms from each of the two strains were collected from a single host (known as infra population) may influence genetic diversity. We also observed a similar result within two field infra populations in Sichuan and Inner Mongolia China (unpublished data). In spite of the limitation mentioned above, some features of the population genetic structure of H. contortus can still be demonstrated by statistical analysis of these SNPs, e.g. in the present study, a similar level of genetic diversity was observed within both resistant and susceptible strains, consistent with previous studies showing that H. contortus field populations that are resistant to anthelmintic drugs seem to have a similar level of overall genetic diversity as susceptible populations [24]. In addition, Akey et al. [22] interrogated a high-density SNP map to analyze signatures of natural selection in the human genome, and found that the largest difference in average FST could be observed between coding and noncoding SNPs due to natural purifying selection (FST for coding region < FST for noncoding region). In spite of the low-density of SNPs obtained in the present study, we were also able to obtain a similar result. These results suggest that those SNPs from 2b-RAD tag sequences can provide an excellent fractional representation of the total of SNPs in the entire genome of H. contortus, and can be used to reveal the locus-specific signature of drug selection.
In fact, we found that 208 SNP loci exhibited significantly elevated FST value, of which 24 loci were located in CDS region, the others within intron and another noncoding region, those genes with significant variation in SNP allele frequencies should be considered as potential IVM resistance-associated genes. Among those genes, although our study failed to find known candidate IVM resistant genes such as Hco-glc (encoding IVM target glutamate-gated chloride channel receptor, GluClR) and Hco-pgp (encoding ABC transporter membrane protein P-glycoprotein for IVM efflux pumps) [6, 8, 37, 38], we were still able to identify some genes with functions of drug target and efflux pump similar to that of Hco-glc and Hco-pgp, for example, HCOI02054500 and HCOI00378500 genes. According to the annotation by Wormbase parasite (http://parasite.wormbase.org/Haemonchus_contortus_prjeb506/Info/Index), HCOI02054500 gene was predicted to encode an uncharacterized protein with a molecular function of extracellular ligand-gated ion channel activity, and to have a C. elegans or thologue lgc-36 which is an or tholog of members of the human GABR [Gamma-amino butyric acid (GABA) A receptors] family including GABRR1, suggesting that HCOI02054500 is likely to have the function similar to that of GABA receptors. Meanwhile, the HCOI00378500 gene was annotated as encoding 4Fe-4S ferredoxin and ABC transporter domain containing protein; this protein belongs to ATP-binding cassette sub-family E member 1 (ABCE1); it was predicted to have ATP binding activity and ATPase activity, and might be involved in nematode larval development and reproduction, suggesting that this protein is likely to mediate transfer of energy and substrates. As we have known, IVM is believed to act by opening glutamate-gated chloride channels and GABA-gated channels in nematodes neurons or muscle cells which leads to a permanent hyperpolarisation and an inhibitory paralysis of the cells. Early reports on the mechanism of IVM also indicated that changes in GluCl and GABA receptors may be implicated in ML resistance in laboratory-selected resistant H. contortus [2]. In addition, multidrug resistance ABC transporters are essential for many cellular processes that require the transport of substrates across cell membranes; and the over expression of those transporter genes can be rapidly and transiently induced following IVM and moxidectin treatment in H. contortus [39], which leads to therapy failure by decreasing drug concentration at the target. Some early molecular analyses also demonstrated that polymorphisms in H. contortus P-gp genes may have been associated with resistance to MLs [2]. All these data suggest that genetic changes in drug sites and drug efflux pump transporters of H. contortus may have been implicated in IVM resistance. Therefore, we speculate that the two novel genes identified in this study may have contributed to the IVM resistance in H. contortus, and can be selected as candidate IVM resistance-associated genes.
Among the candidate genes screened in this study, it is worth to note that the HCOI00703000 gene (http://parasite.wormbase.org/Haemonchus_contortus_prjeb506/Info/Index) is annotated to code for AIR synthase-related protein domain containing protein which probably contains ATP-binding site; this protein is believed to represent a new class of ATP-binding proteins. This kind of protein (enzyme) was reported to catalyze the conversion of formylglycinamide ribonucleotide (FGAM) and ATP to AIR, ADP and Pi, the fifth step in de novo purine biosynthesis. Moreover, previous studies have suggested that a pyrrolo [2,3-d] pyrimidine folate analog inhibits variety of human folate-requiring enzymes, including PurN and PurH, and purine biosynthesis pathway was thought as a chemotherapeutic target [40]. Whether the protein encoded by the HCOI00703000 gene in H. contortus is related to the drug metabolism and detoxification or even can be one of the drug targets, remain to be further validated.
It is also interesting to noting that the HCOI00506600 gene (http://parasite.wormbase.org/Haemonchus_contortus_prjeb506/Info/Index), coding for low density lipoprotein-receptor domain containing protein, has significantly shown higher genetic diversity, and may participate in the regulation of lipid metabolism which could lead to the drug resistance via multi-drug resistance (MDR) proteins (e.g. P-gp) [41]. In facts, the P-gps have been localized to the biological membranes and the membrane environment has been shown to modulate their activity. The membrane environment is mainly composed of lipids. Low density lipoprotein-receptor can mediate the endocytosis of cholesterol-rich LDL, and change membrane environment [42]. According to the studies by Riou et al. [43], changes in the cholesterol content in H. contortus eggs induced changes in benzimidazoles and ivermectin anthelmintic resistance; cholesterol depletion gave increased resistance and cholesterol loading gave decreased resistance; and the effect is likely to be correlated with changes in the function of membrane P-glycoprotein [44]. Above experimental evidence indirectly suggests that polymorphism of the HCOI00506600 gene may influence the nematode P-gp activity via modulating lipid composition of membrane and this gene may be involved in IVM resistance in H. contortus. Hence, this gene should be selected as a potential IVM resistance-associated gene.
We also found that the HCOI01200700 gene, encoding L27-1 and PDZ and Srchomology-3 and guanylate kinase domain containing protein, was shown to have significant change of SNP sites. According to the annotation (http://parasite.wormbase.org/Haemonchus_contortus_prjeb506/Info/Index), this gene has a C. elegans orthologue dlg-1 that encodes a membrane-associated guanylate kinases (MAGUK) protein [45]. They are a super family of proteins, have emerged as a key element in the organization of protein complexes in specialized membrane regions. These proteins are characterized by the presence of multiple protein-protein interaction domains including PDZ and SH3 domains. They are located either on the pre- and/or post-synaptic sides of synapses or at cell-cell adhesion sites of epithelial cells. MAGUK proteins can interact with glutamate receptors and various ionic channels, they have ability to form protein-protein interactions with cytoskeleton proteins, microtubule/actin-based machinery and molecules involved in signal transduction [46]. Meanwhile, we found that the HCOI02035600 gene probably has similar function to HCOI01200700 gene (http://parasite.wormbase.org/Haemonchus_contortus_prjeb506/Info/Index), this gene, coding for WW- Rsp5- WWP- and FF domain-containing protein, was predicted to be required for embryonic viability and for normally high rates of postembryonic growth. The WW domain has been originally discovered as a short conserved region in a number of unrelated proteins, e.g. dystrophin, amultidomain cytoskeletal protein that is thought to have multiple functions including involvement in membrane stability, transduction of contractile forces to the extracellular environment and organization of membrane specialization. Mutations in the dystrophin gene lead to muscular dystrophy of Duchenne or Becker type [47]. Therefore, we inferred that proteins encoded by HCOI01200700 and HCOI02035600 genes may participate in the formation of receptor complex in membrane on the neurons and/or muscle cells of H. contortus, and these receptors may be anthelmintic targets or efflux pumps, suggesting that both HCOI01200700 and HCOI02035600 genes may indirectly be implicated in resistance to IVM.
A recent study has indicated that some ABC transporter genes were shown to have significant increase in transcription following 3 h exposure to both IVM and LEV in the resistant H. contortus isolate, suggesting that some genes responsible for transcriptional regulation may also be involved in IVM resistance [25]. But up to now, no transcriptional regulation genes that may be related to IVM resistance were identified. In the present study we found that the HCOI01315600 gene (http://parasite.wormbase.org/Haemonchus_contortus_prjeb506/Info/Index) was likely to be one of this kind of genes. This gene was annotated as coding for RNA polymerase-associated protein RTF1 and likely to play a role in regulation of transcription. Tenney et al. [48] showed that Drosophila Rtf1 (dRtf1) protein was required for proper gene expression and development, and also participated in histone methylation and Notch signaling; these transcriptional regulation functions of Rtf1 via the Paf1 complex are highly conserved among eukaryotes [48]. We inferred that the functions of H. contortusRtf1 protein was likely to be similar to that of dRtf1and may be indirectly implicated in resistance to IVM by increasing the transcription levels of ABC transporter genes in resistant strains, a prediction that remains to be further confirmed experimentally.
Conclusions
In conclusion, our data suggest that candidate genes putatively associated with resistance to IVM in H. contortus may be identified by genome-wide SNP analysis using 2b-RAD sequencing. Seven candidate genes were predicted to code for some functional molecules such as potential IVM target and/or efflux pump proteins, component proteins of receptor complex in membrane on neuromuscular cells, and transcriptional regulation proteins; and might be involved in resistance to IVM via the mechanisms of changes in the drug receptor or modulation of drug concentration in H. contortus. These findings provide not only an indirect evidence for multigenic model of resistance but also a theoretical basis for further experimental validation of these novel IVM resistance-associated proteins.
Acknowledgements
Not applicable.
Funding
This study was supported by the Special Fund for Agro-scientific Research in the Public Interest, China (Grant no. 201303037), the National Key Basic Research Program (973 program) of China (Grant No. 2015CB150303), the Fund on Sci & Tech Innovation Program of the Chinese Academy of Agricultural Sciences (Grant No. SVRICAAS001) and the National Natural Science Foundation of China (Grant No. 31660710).
Availability of data and materials
The data supporting the findings of this study are included within the article and its additional files; and SNPs identified in this study can be found in the NCBI SRA database (Assay ID: ss2137098375–ss2137102521).
Authors’ contributions
XF and XY conceived and designed the experiments; XL, XS and CG performed the experiments; CG, XS and MA participated animal model and sample collection; XL wrote the manuscript; XF, XY and MH critically revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
This study was approved by the Animal Ethics Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Permit No. shvri-sh-0886). All goats were handled in strict accordance with good animal practice according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
Abbreviations
- ABCE1
ATP-binding cassette sub-family E member 1
- CDS
Coding sequence
- FGAM
Formylglycinamide ribonucleotide
- GABA
Gamma-amino butyric acid
- glc
Glutamate-gated chloride
- GluCIRs
Glutamate-gated chloride ion channel receptors
- GWAS
Genome-wide association study
- Hco
Haemonchus contortus
- IR
Ivermectin resistant
- IVM
Ivermectin
- LDA
Larval development assay
- LS
Ivermectin susceptible
- MAGUK
Membrane-associated guanylate kinases
- MDR
Multi-drug resistance
- ML
Macrocyclic lactone
- NGS
Next generation sequencing
- P-gp
P-glycoprotein
- RAD
Restriction site-associated DNA
- SNP
Single nucleotide polymorphism
- WGAS
Whole genome association study
Additional files
Larval development assay to determine ivermectin resistance in the susceptible strain of H. contortus. Table S2. Larval development assay to determine ivermectin resistance in the resistant strain of H. contortus. (DOCX 16 kb)
Nucleotide sites and SNPs identified on each scaffold of both susceptible and resistant strains of H. contortus. (XLSX 345 kb)
A complete list of 208 SNPs loci exhibits significantly elevated FST value. (XLSX 16 kb)
Contributor Information
Xiaoping Luo, Email: luoxpnmg@163.com.
Xiaona Shi, Email: sxn0729@163.com.
Chunxiu Yuan, Email: yuanchx@shvri.ac.cn.
Min Ai, Email: aimy1991@foxmail.com.
Cheng Ge, Email: wodebaiyulan@163.com.
Min Hu, Email: mhu@mail.hzau.edu.cn.
Xingang Feng, Email: xingangf62@aliyun.com.
Xiaoye Yang, Email: xiaoyeyang122@sohu.com.
References
- 1.Kaplan RM, Vidyashankar AN. An inconvenient truth: global warming and anthelmintic resistance. Vet Parasitol. 2012;186(1–2):70–78. doi: 10.1016/j.vetpar.2011.11.048. [DOI] [PubMed] [Google Scholar]
- 2.Kotze AC, Hunt PW, Skuce P, von Samson-Himmelstjerna G, Martin RJ, Sager H, et al. Recent advances in candidate-gene and whole-genome approaches to the discovery of anthelmintic resistance markers and the description of drug/receptor interactions. Int J Parasitol Drugs Drug Resist. 2014;4(3):164–184. doi: 10.1016/j.ijpddr.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotze AC, Prichard RK. Anthelmintic resistance in Haemonchus contortus: History, mechanisms and diagnosis. Adv Parasitol. 2016;93:397–428. [DOI] [PubMed]
- 4.Blackhall WJ, Pouliot JF, Prichard RK, Beech RN. Haemonchus contortus: selection at a glutamate-gated chloride channel gene in ivermectin-and moxidectin-selected strains. Exp Parasitol. 1998;90(1):42–48. doi: 10.1006/expr.1998.4316. [DOI] [PubMed] [Google Scholar]
- 5.Njue AI, Hayashi J, Kinne L, Feng XP, Prichard RK. Mutations in the extracellular domains of glutamate-gated chloride channel alpha3 and beta subunits from ivermectin-resistant Cooperia oncophora affect agonist sensitivity. J Neurochem. 2004;89(5):1137–47. [DOI] [PubMed]
- 6.Lespine A, Ménez C, Bourguinat C, Prichard RK. P-glycoproteins and other multidrug resistance transporters in the pharmacology of anthelmintics: prospects for reversing transport-dependent anthelmintic resistance. Int J Parasitol Drugs Drug Resist. 2012;2:58–75. doi: 10.1016/j.ijpddr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redman E, Sargison N, Whitelaw F, Jackson F, Morrison A, Bartley DJ, et al. Introgression of ivermectin resistance genes into a susceptible Haemonchus contortus strain by multiple backcrossing. Plos Pathog. 2012;8(2):e1002534. [DOI] [PMC free article] [PubMed]
- 8.Laing R, Kikuchi T, Martinelli A, Tsai IJ, Beech RN, Redman E, et al. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14(8):R88. [DOI] [PMC free article] [PubMed]
- 9.Schwarz EM, Korhonen PK, Campbell BE, Young ND, Jex AR, Jabbar A, et al. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14(8):R89. [DOI] [PMC free article] [PubMed]
- 10.Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12(7):499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 11.Casto AM, Feldman MW. Genome-wide association study SNPs in the human genome diversity project populations: does selection affect unlinked SNPs with shared trait associations? Plos Genet. 2011;7(1):e1001266. doi: 10.1371/journal.pgen.1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Meyer E, McKay JK, Matz MV. 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat Methods. 2012;9(8):808–810. doi: 10.1038/nmeth.2023. [DOI] [PubMed] [Google Scholar]
- 13.Seetharam AS, Stuart GW. Whole genome phylogeny for 21 Drosophila species using predicted 2b-RAD fragments. Peer J. 2013;1:e226. [DOI] [PMC free article] [PubMed]
- 14.Pecoraro C, Babbucci M, Villamor A, Franch R, Papetti C, Leroy B, et al. Methodological assessment of 2b-RAD genotyping technique for population structure inferences in yellowfin tuna (Thunnus albacares). Mar Genomics. 2016;25:43–8. [DOI] [PubMed]
- 15.Gopal RM, Pomroy WE, West DM. Resistance of field isolates of Trichostrongylus colubriformis and Ostertagia circumcincta to ivermectin. Int J Parasitol. 1999;29(5):781–6. [DOI] [PubMed]
- 16.Dolinská M, Königová A, Várady M. Is the micro-agar larval development test reliable enough to detect ivermectin resistance? Parasitol Res. 2012;111(5):2201–2204. doi: 10.1007/s00436-012-2944-4. [DOI] [PubMed] [Google Scholar]
- 17.Zawadzki JL, Kotze AC, Fritz JA, Johnson NM, Hemsworth JE, Hines BM, et al. Silencing of essential genes by RNA interference in Haemonchus contortus. Parasitology. 2012;139(5):613–29. [DOI] [PubMed]
- 18.Waruiru RM. Efficacy of closantel, albendazole and levamisole on an ivermectin resistant strain of Haemonchus contortus in sheep. Vet Parasitol. 1997;73(1):65–71. [DOI] [PubMed]
- 19.Wang S, Meyer E, McKay JK, Matz MV. Nature methods. 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat Methods. 2012;9:808–810. doi: 10.1038/nmeth.2023. [DOI] [PubMed] [Google Scholar]
- 20.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohenlohe PA, Bassham SS, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. Plos Genetics. 2010;6(2):e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12(12):1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheeseman IH, Mcdew-White M, Phyo AP, Sriprawat K, Nosten F, Anderson TJ. Pooled sequencing and rare variant association tests for identifying the determinants of emerging drug resistance in malaria parasites. Mol Biol Evol. 2015;32(4):1080–1090. doi: 10.1093/molbev/msu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilleard JS, Redman E. Genetic diversity and population structure of Haemonchus contortus. Adv Parasitol. 2016;93:31–68. [DOI] [PubMed]
- 25.Raza A, Kopp SR, Bagnall NH, Jabbar A, Kotze AC. Effects of in vitro exposure to ivermectin and levamisole on the expression patterns of ABC transporters in Haemonchus contortus larvae. Int J Parasitol Drugs Drug Resist. 2016;6(2):103–15. [DOI] [PMC free article] [PubMed]
- 26.Ashraf S, Beech RN, Hancock MA, Prichard RK. Ivermectin binds to Haemonchus contortus tubulins and promotes stability of microtubules. Int J Parasitol. 2015;45(9–10):647–54. [DOI] [PubMed]
- 27.Urdaneta-Marquez L, Bae SH, Janukavicius P, Beech R, Dent J, Prichard R. A dyf-7 haplotype causes sensory neuron defects and is associated with macrocyclic lactone resistance worldwide in the nematode parasite Haemonchus contortus. Int J Parasitol. 2014;44(14):1063–71. [DOI] [PubMed]
- 28.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 29.Gilleard JS. The use of Caenorhabditis elegans in parasitic nematode research. Parasitology. 2004;128 Suppl 1:S49–70. [DOI] [PubMed]
- 30.Glendinning SK, Buckingham SD, Sattelle DB, Wonnacott S, Wolstenholme AJ. Glutamate-gated chloride channels of Haemonchus contortus restore drug sensitivity to ivermectin resistant Caenorhabditis elegans. Plos One. 2011;6(7):e22390. [DOI] [PMC free article] [PubMed]
- 31.Bürglin TR, Lobos E, Blaxter ML. Caenorhabditis elegans as a model for parasitic nematodes. Int J Parasitol. 1998;28(3):395–411. doi: 10.1016/S0020-7519(97)00208-7. [DOI] [PubMed] [Google Scholar]
- 32.Hohenlohe PA, Catchen J, Cresko WA. Population genomic analysis of model and nonmodel organisms using sequenced RAD tags. Methods Mol Biol. 2012;888:235–260. doi: 10.1007/978-1-61779-870-2_14. [DOI] [PubMed] [Google Scholar]
- 33.Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59(11):2312–2323. doi: 10.1554/05-273.1. [DOI] [PubMed] [Google Scholar]
- 34.Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol. 2005;14(3):671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 35.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336(6077):79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tennessen JA, Bonner KM, Bollmann SR, Johnstun JA, Yeh JY, Marine M, et al. Genome-wide scan and test of candidate genes in the snail Biomphalaria glabrata reveal new locus influencing resistance to Schistosoma mansoni. Plos Negl Trop Dis. 2015;9(9):e0004077. [DOI] [PMC free article] [PubMed]
- 37.Mccavera S, Rogers AT, Yates DM, Woods DJ, Wolstenholme AJ. An ivermectin-sensitive glutamate-gated chloride channel from the parasitic nematode Haemonchus contortus. Mol Pharmacol. 2009;75(6):1347–55. [DOI] [PMC free article] [PubMed]
- 38.Harder A. The biochemistry of Haemonchus contortus and other parasitic nematodes. Adv Parasitol. 2016;93:69–94. [DOI] [PubMed]
- 39.Prichard RK, Roulet A. ABC transporters and beta-tubulin in macrocyclic lactone resistance: prospects for marker development. Parasitology. 2007;134(Pt 8):1123–1132. doi: 10.1017/S0031182007000091. [DOI] [PubMed] [Google Scholar]
- 40.Mueller EJ, Oh S, Kavalerchik E, Kappock TJ, Meyer E, Li C, et al. Investigation of the ATP binding site of Escherichia coli aminoimidazole ribonucleotide synthetase using affinity labeling and site-directed mutagenesis. Biochemistry. 1999;38(31):9831–9839. doi: 10.1021/bi990638r. [DOI] [PubMed] [Google Scholar]
- 41.Riou M, Guégnard F, Sizaret PY, Le Vern Y, Kerboeuf D. Drug resistance is affected by colocalization of P-glycoproteins in raft-like structures unexpected in eggshells of the nematode Haemonchus contortus. Biochem Cell Biol. 2010;88(3):459–67. [DOI] [PubMed]
- 42.Eckford PD, Sharom FJ. Interaction of the P-glycoprotein multidrug efflux pump with cholesterol: effects on ATPase activity, drug binding and transport. Biochemistry. 2008;47(51):13686–13693. doi: 10.1021/bi801409r. [DOI] [PubMed] [Google Scholar]
- 43.Riou M, Koch C, Kerboeuf D. Increased resistance to anthelmintics of Haemonchus contortus eggs associated with changes in membrane fluidity of eggshells during embryonation. Parasitol Res. 2005;95(4):266–72. [DOI] [PubMed]
- 44.Riou M, Guégnard F, Le Vern Y, Kerboeuf D. Modulation of the multidrug resistance (MDR) system in the nematode Haemonchus contortus by changing cholesterol content: effects on resistance to anthelmintics. J Antimicrob Chemother. 2003;52(2):180–7. [DOI] [PubMed]
- 45.Chen K, Featherstone DE. Discs-large (DLG) is clustered by presynaptic innervation and regulates postsynaptic glutamate receptor subunit composition in Drosophila. BMC Biol. 2005;3:1. [DOI] [PMC free article] [PubMed]
- 46.Montgomery JM, Zamorano PL, Garner CC. MAGUKs in synapse assembly and function: an emerging view. Cell Mol Life Sci. 2004;61(7–8):911–929. doi: 10.1007/s00018-003-3364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haenggi T, Fritschy JM. Role of dystrophin and utrophin for assembly and function of the dystrophin glycoprotein complex in non-muscle tissue. Cell Mol Life Sci. 2006;63(14):1614–1631. doi: 10.1007/s00018-005-5461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, et al. Drosophila Rtf1 functions in histone methylation, gene expression, and notch signaling. Proc Natl Acad Sci USA. 2006;103(32):11970–4. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are included within the article and its additional files; and SNPs identified in this study can be found in the NCBI SRA database (Assay ID: ss2137098375–ss2137102521).


