Abstract
Vibrio cholerae converts glucose into either acid or the neutral end product acetoin and its survival in carbohydrate enriched media is linked to the nature of the byproducts produced. It has been demonstrated in this study that Escherichia coli strain isolated from the gut of healthy human volunteers and the commonly used probiotic E. coli Nissle strain that metabolize glucose to acidic byproducts drastically reduce the survival of V. cholerae strains irrespective of their glucose sensitivity and acetoin production status. Accordingly, E. coli glucose transport mutants that produce lower amounts of acidic metabolites had little effect on the survival of V. cholerae in cocultures. Thus, cross feeding of byproducts of glucose metabolism by heterologous bacteria modulates the survival of V. cholerae in glucose rich medium suggesting that composition of the gut microbiota could influence the outcome of V. cholerae infection especially when glucose based ORS is administered.
Electronic supplementary material
The online version of this article (doi:10.1186/s13099-016-0153-x) contains supplementary material, which is available to authorized users.
Keywords: V. cholerae survival, Cocultures, E. coli Nissle 1917, E. coli glucose transport mutants
Background
Successful infection by bacterial pathogens depends primarily upon a complex interplay between bacterial virulence factors and host responses and defense systems. Recently however, it is becoming apparent that presence of other microorganisms might profoundly influence the outcome of bacterial infections including that of Vibrio cholerae [1]. In this study we demonstrate that presence of heterologous organisms, with ability to produce acidic byproducts of glucose metabolism might modulate the survival of V. cholerae under glucose rich conditions.
All recorded cholera pandemics have been caused by strains of the O1 serogroup of V. cholerae that can be classified into two major biotypes, classical and El Tor. Although the classical and El Tor biotypes are closely related, several biochemical and genetic differences have been reported between the two biotypes including a unique difference in carbohydrate metabolism. In the presence of exogenous sugars the classical biotype strains (e.g. O395) produced organic acids resulting in a sharp decrease in media pH and drastic loss of viability. El Tor strains (e.g. N16961) have evolved to metabolize sugars to produce acetoin, a neutral fermentation end-product that did not inhibit bacterial growth [2]. It has been suggested that the ability to metabolize sugars without production of growth inhibitory acidic products might account for the evolutionary fitness of the V. cholerae El Tor biotype by virtue of which it displaced the classical biotype as a cause of epidemic cholera [2].
Here, we postulate that cross feeding of byproducts of glucose metabolism by heterologous bacteria might modulate the survival of V. cholerae in glucose rich growth medium. As proof of concept, V. cholerae classical and El Tor biotype strains were co-cultured in the presence of glucose with E.coli strains that produce acidic byproducts of glucose metabolism, and the effect of co-culturing on V. cholerae survival was determined. We observed a drastic loss of viability of V. cholerae strains irrespective of their acetoin production status in the co-culture with E. coli strains under carbohydrate rich condition. On the other hand, E. coli glucose transport mutants that produce lower amounts of acidic metabolites had little effect on the survival of V. cholerae in co-cultures.
Methods
Strains
All bacterial strains used for this study have been described in Additional file 1: Table S1. Bacterial cultures grown to the logarithmic phase in Luria broth (LB) were stored in glycerol (10% v/v) at −80 °C. When required, streptomycin was used at a concentration of 100 μg/ml.
Coculture studies
V. cholerae El Tor strain N16961 (SmR) or classical strain O395 (SmR) and E. coli strains mentioned in Table S1, were grown in LB medium up to mid-logarithmic phase. Cocultures (1:1) were set up in LB or LB containing 1% glucose (LBG). Monocultures were set up in LB and LBG in a similar manner as a control. At regular intervals aliquots of the cultures were removed and the number of V. cholerae cells was enumerated by serial dilution and plating on Luria agar containing streptomycin (100 μg/ml). At all time points, pH in the culture supernatants was measured. Statistical significance of the data has been calculated and expressed as ±SD in all experiments.
Spotting assays
Survival of V. cholerae N16961 strain in cell free conditioned medium was assayed by spotting on LA plates containing streptomycin (100 μg/ml). Cell free conditioned medium was prepared from 12 h cultures of N16961 and E. coli strains grown in LB or LBG at 37 °C with aeration. Mid-logarithmic phase V. cholerae N16961 was resuspended in the conditioned medium and cultures were allowed to grow for 12 h at 37 °C with aeration. Survival was assayed by spotting dilutions (10−3–10−6) on LB agar plates containing streptomycin to select V. cholerae N16961 (SmR).
Results and discussion
To ascertain how the survival of V. cholerae is influenced by heterologous bacteria in glucose enriched medium, V. cholerae classical and El Tor biotypes strains were co-cultured with wild type E. coli strains (Additional file 1: Table S1) in the presence or absence of glucose. The E. coli strains produce largely acidic metabolites upon glucose fermentation [3–5]; it is therefore conceivable that the survival of V. cholerae may be affected in the co-culture with E. coli in LBG.
As reported earlier, V. cholerae classical biotype strain O395 exhibited severe growth defect in Luria broth medium supplemented with 1% glucose (LBG) since this biotype produces acidic byproducts of glucose metabolism. On the other hand, V. cholerae El Tor biotype strain N16961 that metabolizes glucose to acetoin, could grow normally in LBG [2]. However, when the strains were grown in co-cultures with E. coli 40 or E. coli Nissle, a drastic decline in cell count of both the V. cholerae strains was observed concomitant with acidification of the growth media presumably due to acidic glucose fermentation metabolites produced by E. coli (Fig. 1). Interestingly, it was noted that although N16961 is capable of producing acetoin in LBG medium, it failed to overcome the lethal effect of strong acidification caused by the E. coli strains. These results suggest that E. coli strains by virtue of their ability to metabolize glucose to acidic byproducts and cause acidification of the growth medium in LBG, affect survival of V. cholerae strains irrespective of the acetoin producing capability of the latter, when the E. coli and V. cholerae strains are grown together. To examine if acidification of the medium by acidic byproducts of glucose metabolism during growth of E. coli cultures in LBG was primarily responsible for the killing of V. cholerae strains in the co-cultures, the V. cholerae strains were next co-cultured with the E. coli MG1655 glucose transport mutants (Additional file 1: Table S1). In these co-cultures much lower acidification of the growth medium was observed and survival of the V. cholerae strains was much higher than that in co-cultures with the wild type E. coli strains (Fig. 2) strongly suggesting that glucose uptake and metabolism produced acidic byproducts and the resulting acidification of the media in wild type E. coli cultures resulted in severe decline in cell count of V. cholerae strains in LBG. Suspension of V. cholerae strain N16961 in conditioned medium prepared from 12 h cultures of E. coli resulted in loss of cell count indicating that cell–cell contact was not necessary for the decline in cell count of V. cholerae in the presence of the E. coli strains (Additional file 2: Fig. S1).
Fig. 1.
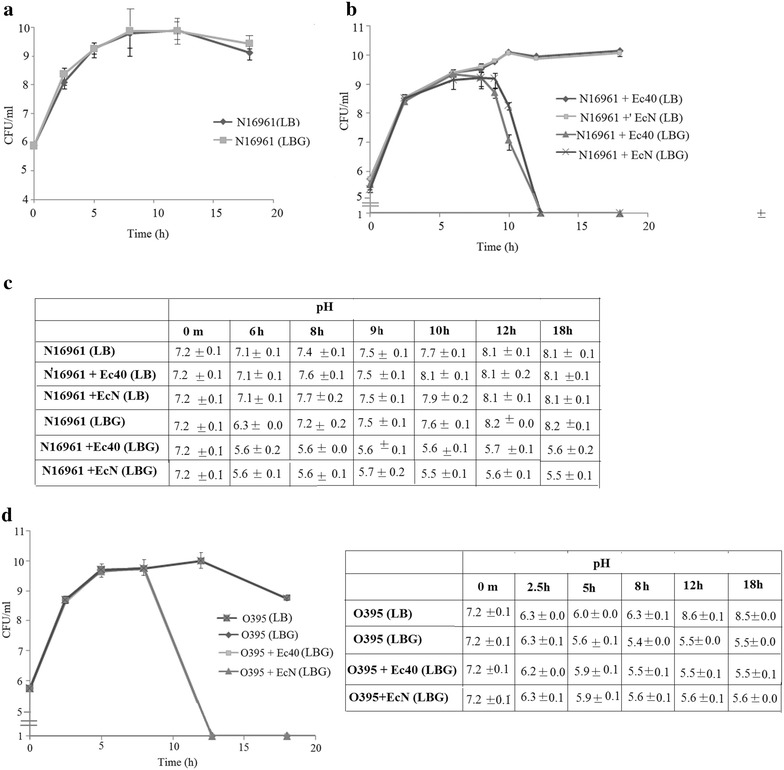
Cocultures of V. cholerae with E. coli strains.CFU of V. cholerae El Tor strain N16961 (SmR) was enumerated in individual cultures (a) or in cocultures (1:1) with E. coli 40 (Ec40) or E. coli Nissle (EcN) (b) at different time points. At all time points, pH of the culture supernatants was measured (c). CFU of V. cholerae classical strain O395 (SmR) in mono- and co-cultures was similarly assayed and pH of the culture supernatants was measured (d)
Fig. 2.
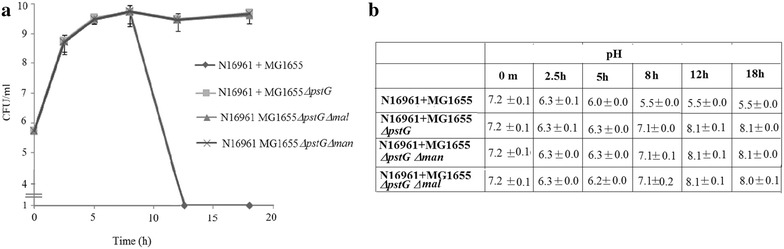
Cocultures of V. cholerae with E. coli WT and glucose transport mutants. V. cholerae N16961 (SmR) was grown together with E. coli MG1655 or glucose transport mutants (1:1 ratio) in LB medium containing glucose (1%). At regular intervals CFU of V. cholerae cells was determined (a) and pH of the culture supernatants was measured (b)
Based on its interaction with host, E. coli strains can be categorized broadly into non pathogenic commensal and pathogenic groups. The pathogenic groups can be further divided into intestinal pathogenic and extra intestinal pathogenic strains [6, 7]. Interestingly, the commensal and probiotic E. coli EcN strain has been exploited clinically to ameliorate the burden of ulcerative colitis and Crohn’s disease [8–10]. Other than clinical use, EcN as well as other non pathogenic E. coli strains have been evaluated for their potential as delivery vehicles [11], in reducing intestinal colonization of Salmonella typhimurium [12] and in cancer therapy [13]. Though non pathogenic commensal and probiotic E. coli strains have tremendous potential for therapeutic use, it must be borne in mind that efficacy of such strains as probiotics is strongly dependent on the composition of intestinal microbiota and immune status of host [14].
Conclusion
In essence, this study suggests that preponderance of bacterial strains that metabolize glucose to acidic compounds in the gut might hinder V. cholerae survival. The result is particularly important as glucose based oral rehydration therapy is currently highly recommended during cholera infections. Thus, selected E. coli strains isolated from healthy human gut and commonly used probiotic E. coli Nissle that metabolize glucose to acidic byproducts could be used as probiotics together with ORT for the control of cholera.
Authors’ contribution
SRC conceived the idea and initiated the work, SRC and RC designed the experiments, CS, ME and SA performed CFU analysis, CS and ME performed pH kinetics, ME and PD performed spotting experiment, SRC and RC wrote the manuscript, All the authors gave their editorial input. All authors read and approved the final manuscript.
Acknowledgements
We gratefully acknowledge Prof. Rudolf von Bünau Ardeypharm GmbH. Herdecke, Germany for the generous gift of Escherichia coli Nissle 1917 and Prof. Katja Bettenbrock for generously providing the E. coli MG1655 strains.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study. Please contact author for data requests.
Funding
This work is supported by the CSIR program on Human Microbiome (BSC-0119). CS received research fellowship from CSIR, ME and PDD from UGC and SA from CSIR network project on infectious disease (BSC 210). Funder has no role in the design of study and collection, analysis and interpretation of data and in writing the manuscript.
Additional files
Additional file 1: Table S1. List of strains.
Additional file 2: Figure S1. Survival of V. cholerae N16961 strain in cell free conditioned medium prepared from 12 h cultures of N16961 and E. coli strains grown in LB (A) or LBG (B). Survival was assayed by spotting dilutions (10−3 to 10−6) on LB agar plates containing streptomycin (100 μg/ml) different to select V. cholerae N16961 (SmR).
Footnotes
Chirantana Sengupta and Manjula Ekka contributed equally to this work
Contributor Information
Chirantana Sengupta, Email: chirantanasengupta@gmail.com.
Manjula Ekka, Email: manjulaekka@imtech.res.in.
Saurabh Arora, Email: saurabh.arora@imtech.res.in.
Prashant D. Dhaware, Email: prashant1@imtech.res.in
Rukhsana Chowdhury, Email: rukhsana@iicb.res.in.
Saumya Raychaudhuri, Email: saumya@rocketmail.com, Email: saumya@imtech.res.in.
References
- 1.Hsiao A, Ahmed A, Subramanian S, Griffin N, Drewry L, Petri W, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoon S, Mekalanos J. 2,3-Butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect Immun. 2006;74:6547–6556. doi: 10.1128/IAI.00695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock A, Sawers G. Escherichia coli and Salmonella: cellular and molecular biology, 2nd edition. In: Neidhardt FC, editor. Fermentation. Washington: ASM Press; 1996. pp. 262–282. [Google Scholar]
- 4.Cruz Ramos H, Hoffmann T, Marino M, Nedjari H, Presecan-Siedel E, Dreesen O, et al. Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. J Bacteriol. 2000;182:3072–3080. doi: 10.1128/JB.182.11.3072-3080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinsiek S, Bettenbrock K. Glucose transport in Escherichia coli mutant strains with defects in sugar transport systems. J Bacteriol. 2012;194:5897–5908. doi: 10.1128/JB.01502-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichial coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 7.Dobrindt U. (Patho-) genomics of Escherichia coli. Int J Med Micro. 2005;295:357–371. doi: 10.1016/j.ijmm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Kruis W, Fric P, Pokromieks J, Lukas M, Fixa B, Kascak M, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS ONE. 2007;12:1–9. doi: 10.1371/journal.pone.0001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1012–1018. doi: 10.1002/ibd.20377. [DOI] [PubMed] [Google Scholar]
- 11.Duan F, Curtis KL, March JC. Secretion of insulinotropic proteins by commensal bacteria: rewiring the gut to treat diabetes. Appl Environ Microbiol. 2008;74:7437–7438. doi: 10.1128/AEM.01019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deriu E, Liu ZJ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, et al. Probiotic bacteria reduces Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson CJ, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2005;355(4):1–9. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 14.Gronbach K, Eberle U, Muller M, Olschlager TA, Dobrindt U, Leithauser F, et al. Safety of probiotic Escherichia coli strain Nissle 1917 depends on intestinal microbiota and adaptive immunity of the host. Infect Immun. 2010;78:3036–3046. doi: 10.1128/IAI.00218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dureja C, Mahajan S, Raychaudhuri S. Phylogenetic distribution and prevalence of genes encoding class I integrons and CTX-M-15 extended-spectrum β-lactamases in Escherichia coli isolates from healthy humans in Chandigarh, India. PLoS ONE. 2014;9:e112551. doi: 10.1371/journal.pone.0112551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raychaudhuri S, Jain V, Dongre M. Identification of a constitutively active variant of LuxO that affects production of HA/protease and biofilm development in a non-O1, non-O139 Vibrio cholerae O110. Gene. 2006;369:126–133. doi: 10.1016/j.gene.2005.10.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study. Please contact author for data requests.


