Abstract
Antagonists of growth hormone-releasing hormone (GHRH) inhibit the growth of various human cancers by multiple mechanisms, which include direct effects on tumor cells through the splice variants (SV) of the GHRH receptor. Our findings suggest that the tumoral protein encoded by SV 1 (SV1) is a likely functional receptor. The aim of this study was to develop a polyclonal antiserum against a polypeptide analog of segment 1-25 of the putative SV1 receptor protein. Rabbits were immunized with [Ala-23]SV1 (1-25)-Tyr-26-Cys-27-NH2 as a hapten, conjugated to BSA or keyhole limpet hemocyanin. The antisera thus generated were evaluated by RIA for binding to the radiolabeled hapten. The specificity and sensitivity of the antisera were studied on xenografts of RL and HT human non-Hodgkin's lymphomas. The sera raised against keyhole limpet hemocyanin-SV1 hapten, showed binding values of 50-75% at a 1:56,000 dilution. In Western blot analyses, the purified polyclonal antibody recognized a specific signal with a molecular mass of ≈40 kDa in RL and HT lymphomas. This band corresponds to the estimated molecular mass of the GHRH receptor isoform encoded by SV1. RT-PCR and ligand binding studies also revealed the expression of SV1 and the presence of high-affinity binding sites for GHRH on RL and HT tumors. Because the antiserum developed recognizes the tumoral GHRH receptor protein encoded by SV1, it should be of value in various investigations.
Keywords: splice variant, growth hormone-releasing hormone receptor, polyclonal antibody, non-Hodgkin's lymphoma
Antagonists of growth hormone-releasing hormone (GHRH) inhibit the growth of experimental human cancer cell lines xenografted into nude mice or cultured in vitro and are being developed for cancer therapy (1-4). To design still more potent antagonists, we have to fully understand their mechanism of action. GHRH antagonists suppress tumor growth through indirect and direct pathways. The indirect mechanism operates through a suppression of the growth hormone release from the pituitary and the resulting inhibition of the production of insulin-like growth factor I in the liver (1-11). However, the principal action of GHRH antagonists is probably exerted directly on tumors and appears to be mediated by specific receptors for GHRH antagonists on cancer cells (1-9, 11-13). Although the mRNA for GHRH and immunologically active GHRH were demonstrated in various human tumor cells, the mRNA for human pituitary GHRH receptor (GHRH-R) has not been detected on these tumor cells or any of the other cancer models (11, 14, 15).
Because of the structural similarities between GHRH and vasoactive intestinal peptide (VIP), the receptors for VIP could be a target for the GHRH antagonists, but GHRH antagonists inhibit the proliferation of MiaPaCa-2 human pancreatic tumor cells, which do not express the receptors for VIP (13). Moreover, in LNCaP human prostatic carcinoma cells, which are positive for the VIP receptors, GHRH antagonists inhibit tumor growth more powerfully than the antagonists of VIP (13).
These and other findings (1-15) suggested the involvement of specific receptors for GHRH antagonists on human cancers. We then reported the expression of four splice variants (SVs) of the full-length human GHRH-R in normal tissues and certain cancer cell lines on the basis of sequence analyses of cDNA encoding these receptors (16, 17). The deduced amino acid sequence of one of these SVs, called SV1, shows a close similarity to that of the full-length GHRH. However, the first 89 aa in the extracellular domain at the N terminus of the pituitary GHRH receptor are replaced in SV1 by a different, 25-aa polypeptide chain (16). Other studies show that, although the N-terminal extracellular domain plays an important role in the interaction between GHRH and the pituitary GHRH-R, the replacement of this domain with the N terminus of the receptor for VIP or secretin does not lead to the complete loss of function of the receptor (18). Unlike the other three SVs, the SV1 has all seven trans-membrane domains and the whole third intracellular loop, the latter being critical for the interaction with G proteins (16, 19-21).
On the basis of these structural characteristics, the SV1 might be able to respond to GHRH and its antagonistic analogs and activate the signal transduction system of the tumor cells. The expression of the mRNA for SV1 has been detected, along with high-affinity binding sites for the radiolabeled GHRH antagonist JV-1-42, on a wide variety of human cancers, including gastroenteropancreatic, renal, lung, and prostatic tumor cell lines and surgical specimens of human prostate cancer (22-25). A comparison of the binding characteristics of GHRH, VIP, pituitary adenylate cyclase-activating polypeptide, and GHRH antagonists revealed that GHRH antagonists bind more powerfully to the membrane fraction of CAKI-I human renal cell adenocarcinoma than to the other peptides (17). High affinity of the antagonists to the tumoral receptor facilitates their selective uptake from the circulation onto the tumor tissue (17, 24). In addition, GHRH increased the proliferation of NIH 3T3 mouse fibroblasts transfected with the plasmid expressing the mRNA for SV1, and this effect was inhibited by the GHRH antagonist JV-1-38 in a dose-dependent manner (26).
All of these findings suggest that an isoform of the GHRH-R protein encoded by the mRNA for SV1 on the cell membrane could play a role in the proliferation of neoplastic cells. However, this receptor protein has not yet been isolated or identified by immunological methods. In this study, we describe the preparation and evaluation of a polyclonal antibody raised in rabbits against a peptide sequence specific for the N terminus of the putative SV1 protein. The specificity and sensitivity of this antibody on tumors was evaluated on RL and HT human lymphomas expressing binding sites for GHRH antagonists and the mRNA for the SV1.
Materials and Methods
Animals. All animals were housed under controlled conditions (24°C, 12-h light/12-h dark schedule). White female New Zealand rabbits (Charles River Laboratories) were allowed standard chow and water ad libitum. Six-week-old female athymic nude mice were purchased from the Frederick Cancer Research Facility of the National Cancer Institute (Frederick, MD), housed in a laminar-flow environment under pathogen-free conditions, and fed with autoclaved standard chow and water ad libitum. All experiments were performed in accord with institutional guidelines for the animal care.
Design and Synthesis of the SV1-Hapten and Other Peptides. To generate a sequence-specific antigen against the extracellular domain of the SV1 receptor, we synthesized a 27-aa peptide containing the N-terminal 1-25 amino acids of the putative SV1 receptor protein in which Cys-23 was replaced by Ala-23 to avoid an undesirable oxidation between peptide chains and Tyr and Cys residues were added at the C terminus in positions 26 and 27, respectively. Tyr-26 was incorporated to allow a subsequent iodination, and Cys-27 was inserted into the molecule to make it suitable for conjugation to maleimide-activated keyhole limpet hemocyanin (KLH) through its free SH-functional group. The sequence of the synthesized hapten was as follows: Met-Val-Pro-Gly-Thr-Pro-Ser-Pro-Leu-Leu-Gly-Arg-Gly-Lys-Glu-Leu-Trp-Leu-Glu-Ser-Leu-Ala-Ala-Leu-Pro-Tyr-Cys-NH2. The structure of this peptide can be abbreviated as [Ala-23]SV1 (1-25)-Tyr-Cys-NH2, based on the numbering of amino acid residues in SV1 according to ref. 16. Peptides corresponding to segments of SV1 and of the pituitary GHRH-R were also synthesized for crossreaction studies. One of these peptides, abbreviated as [Ala-28,41]GHRH-R (23-45)-Tyr-Cys-NH2, has the sequence His-Met-His-Pro-Glu-Ala-Asp-Phe-Ile-Thr-Gln-Leu-Arg-Glu-Asp-Glu-Ser-Ala-Ala-Leu-Gln-Ala-Ala-Tyr-Cys-NH2 and is an analog of segment 23-45 of the pituitary GHRH-R, with the numbering of amino acid residues in the pituitary GHRH-R being according to ref. 27. The other peptide can be abbreviated either as [Ala-48]SV1 (45-66)-Cys-NH2 or [Ala-112]GHRH-R (109-130)-Cys-NH2 and is an analog of segment 45-66 of SV1, which is identical to segment 109-130 of the pituitary GHRH-R. The sequence of this peptide is Pro-Val-Ala-Ala-Pro-Val-Pro-Leu-Glu-Leu-Leu-Ala-Glu-Glu-Glu-Ser-Tyr-Phe-Ser-Thr-Val-Lys-Cys-NH2. The peptides with amidated C termini were synthesized by standard solid-phase methods with Boc chemistry (28) by using manual solid-phase synthesis equipment on para-methylbenzhydryl-amine resin and purified by HPLC (29). All protected amino acids and reagents were purchased from Nova Biochem, and solvents were obtained from Fisher Scientific.
Preparation of the Immunogen. The synthetic SV1-hapten was conjugated to fatty acid-free BSA (Sigma) with glutaraldehyde (Sigma) as reported in ref. 30. The SV1-hapten-BSA complex was freshly prepared each time. For another series of immunizations, the hapten was conjugated to maleimide-activated KLH by using the Imject Maleimide-Activated mcKLH kit (Pierce) according to the manufacturer's instructions. Aliquots of the SV1-hapten-KLH conjugate were stored frozen at -70°C until used for immunization.
Antiserum Production. Two rabbits (animal nos. 2,315 and 2,316) were immunized with the SV1-hapten-BSA and two other rabbits (animal nos. 2,317 and 2,318) were immunized with the SV1-hapten-KLH conjugate. The SV1-hapten-KLH conjugate and SV1-hapten-BSA conjugate were dissolved in 3 ml of distilled water and homogenized with 3 ml of complete Freund's adjuvant (Sigma) for the first immunization and with 1.5 ml each of complete and incomplete Freund's adjuvant (Sigma) for the booster immunizations. Three milliliters of this solution, equivalent to 150 μg of SV1-hapten per rabbit for the first immunization and 100 μg per rabbit for the boosters, was given to each rabbit. One milliliter of the suspension was injected s.c. into the interscapular region, 1 ml was injected i.p., and 0.5 ml was injected i.m. into each rear thigh. Booster injections were given 2 weeks later, then monthly, for 4 months. In the fifth month, only the SV1-hapten-KLH-immunized rabbits received booster injections. Blood was collected from the marginal vein of the ear 12 and 18 days after the third, fourth, fifth, and sixth booster. “Preimmune” serum was obtained from each rabbit before the initiation of the immunization. The serum was separated by centrifugation at 2,000 × g at 4°C for 20 min, then lyophilized and stored at 4°C until use. The serum was reconstructed in purified water (MilliQ Synthesis System, Millipore) at a concentration of 80 mg/ml and considered as undiluted antiserum.
RIA. The assay buffer consisted of 1% BSA, 0.025 M EDTA, 0.01 M sodium phosphate (pH 7.6), 0.14 M NaCl, and protease inhibitor mixture (Sigma) at a 1:800 final dilution. SV1-hapten aliquots of 37 μg, dissolved in 370 μl of 0.1 M acetic acid, were stored at -70°C and diluted with the assay buffer immediately before their use for the RIA. The SV1-hapten was iodinated on Tyr-26 by the lactoperoxidase method (31).
For testing the antisera, the following components were incubated overnight at 4°C: 500 μl of the assay buffer, 25,000 cpm of 125I-labeled SV1-hapten dissolved in 100 μl of assay buffer, and 100 μl of a batch of the antiserum for the SV1-hapten at final dilutions of 1:56,000 to 1:672,000. On the next day, 100 μl of normal rabbit serum at a 1:150 dilution and 100 μl of goat anti-rabbit Ig at a 1:15 dilution were added to the mixture. After 30 min of incubation, 500 μl of a 12.5% (vol/vol) solution of ice-cold polyethylene glycol 3350 (Sigma) was added to enhance the precipitation of the immunocomplexes. The contents of the tubes were mixed, and the immunocomplexes were separated by centrifugation at 3,000 × g for 30 min at 4°C. The supernatant was decanted, and the radioactivity in the sediment was counted by a Packard Cobra II Auto Gamma Counting System (PerkinElmer Life and Analytical Sciences). Binding of the SV1 hapten to the tested batch of the antiserum was calculated as the ratio of the radioactivity after the immunoreaction and the total radioactivity before the immunoreaction at the final dilutions of 1:56,000 to 1:672,000 of the antisera.
Immunoaffinity Purification of the Antisera. Initial screening of the antisera with RIA allowed the identification of the most sensitive batches. Batch no. 2317/7, which had a high sensitivity (see Tables 1 and 2), was purified by affinity chromatography with the synthetic SV1-hapten coupled to a SulfoLink gel column according to the instructions of the SulfoLink kit (Pierce). Fractions with the highest protein content, which also had the highest sensitivity in Western blots (see below) were pooled, concentrated by filtration with Ultrafree-4 concentrator (Fisher Scientific), and diluted with a phosphate buffer (pH 7.6) containing 0.14 M NaCl and 0.1% sodium azide. Purified antibodies were stored at -80°C until use.
Table 1. Binding of the antisera to radiolabeled synthetic hapten at a final dilution of 1:56,000.
| Binding values
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Rabbit no. | Batch 1 | Batch 2 | Batch 3 | Batch 4 | Batch 5 | Batch 6 | Batch 7 | Batch 8 |
| 2,315 | 24.1 | 26.0 | 25.3 | 14.3 | 19 | 18 | 19.7 | 24.2 |
| 2,316 | 25.4 | 20.4 | 32.4 | 31.1 | 35 | 35 | 26.2 | 34.2 |
| 2,317 | 68.5 | 67.0 | 56.6 | 64 | 75 | 69 | 71.2 | 63.1 |
| 2,318 | 64.0 | 68.3 | 53 | 55.6 | 58 | 52 | 53.5 | 50.1 |
Binding values are shown as the percentage of bound radioactivity to total radioactivity.
Table 2. Binding values of the antisera generated against the SV-hapten-KLH complex at different dilutions.
| Binding values at dilution
|
||||
|---|---|---|---|---|
| Antiserum | 1:224,00 | 1:336,000 | 1:448,000 | 1:672,000 |
| JH-2317/3 | 50.1 | 39.7 | n.t. | n.t. |
| JH-2317/4 | 47.3 | 41.4 | n.t. | n.t. |
| JH-2317/5 | 67.0 | 61.0 | 51.4 | n.t. |
| JH-2317/6 | 55.5 | 50.3 | 40.9 | n.t. |
| JH-2317/7 | 62.8 | 51.4 | 47.8 | 36.7 |
| JH-2317/8 | 66.3 | 56.8 | 49.3 | 42.1 |
| JH-2318/3 | 43.1 | 29.1 | n.t. | n.t. |
| JH-2318/4 | 40.4 | 37.3 | n.t. | n.t. |
| JH-2318/5 | 39.2 | 33.1 | 30.8 | n.t. |
| JH-2318/6 | 34.4 | 28.9 | 25.2 | n.t. |
| JH-2318/7 | 32.8 | 28.3 | 24.2 | 17.2 |
| JH-2318/8 | 36.5 | 33.0 | 26.5 | n.t. |
Binding values are shown as percentage of bound activity to total activity. n.t., not tested.
Receptor Binding Studies. Radioiodinated derivative of GHRH antagonist JV-1-42 was prepared by the chloramine-T method as described in ref. 32. Preparation of the membrane fractions from the tumor samples was carried out as reported (17, 25). Binding sites for GHRH were determined by in vitro ligand competition assays based on the binding of radiolabeled JV-1-42 to tumor membrane fractions (17, 25). The characteristics of the specific ligand binding were evaluated with ligand-pc curve-fitting software and Scatchard analysis.
Cell Culture and Initiation of Experimental Tumor Models. RL and HT human non-Hodgkin's lymphoma cell lines were obtained from the American Type Culture Collection. MDA-MB-231 estrogen-independent breast cancer cell line was also provided by the American Type Culture Collection. All of the media and supplements were purchased from GIBCO/BRL. The culture medium for HT and RL cells was RPMI medium 1640 containing 1 mM pyruvate. Iscove's modified Eagle's medium was used for MDA-MB-231 cells. Each medium was supplemented with 10% FBS, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B. The cultures were kept in humidified atmosphere containing 5% CO2/95% air at 37°C. The cells were passaged twice a week and monitored routinely for the presence of mycoplasma by using a test kit from Boehringer Mannheim. The xenografts were initiated by injecting 1 × 107 RL or HT lymphoma cells into both flanks of two male nude mice or 1 × 107 MDA-MB-231 cells into the right flank of a female nude mouse.
Western Blot. One milligram of the tumor tissue was homogenized in a lysis buffer containing 50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 2 mM EGTA, 2 mM MgCl2, 1% Triton X-100 (vol/vol), 10% glycerol (vol/vol), 10 mM DTT, 1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 5 μg/ml pepstatin A, 50 mM NaF, 10 mM NaP2O7, and 3 mM H2O2 (Sigma). The whole-cell lysate was centrifuged at 12,000 × g for 20 min at 4°C, and the supernatant was kept for Western blot analysis. The preparation of membrane fractions was also performed as described in ref. 25. Protein concentrations were measured by using a protein assay kit from Bio-Rad (25). Five- to 50-μg aliquots of protein fractions were suspended within Laemmli buffer (Bio-Rad). The samples were heated for 5 min at 95°C and resolved by SDS/PAGE by using 4-20% linear gradient Tris·HCl Criterion precasted gels (Bio-Rad). Proteins were transferred to nitrocellulose membranes (Bio-Rad), which were soaked in a buffer containing 39 mM Gly, 48 mM Tris-Base (pH 8.3), 0.037% (vol/vol) SDS, and 20% methanol (Sigma) before the transfer. Excess protein-binding sites were saturated with 5% nonfat dried milk suspended in a buffer containing 10 mM Tris·HCl (pH 7.5), 50 mM NaCl, and 0.1% Tween 20. Optimal dilution of the SV1 antiserum was determined in preliminary studies. The blotted membranes were incubated for 1 h at room temperature with the crude or purified rabbit SV1 antisera diluted 1:1,000 to 1:4,000, the anti-actin goat serum diluted 1:1,000 (Santa Cruz Biotechnology), or with the 2317/7 batch of the SV1 antiserum overnight incubated with 100 pg/ml SV1-hapten. All of the antibodies were dissolved in the 10 mM Tris·HCl (pH 7.5)/50 mM NaCl/0.1% Tween 20/5% nonfat dried milk containing buffer. Negative controls included omission of the proteins and of the primary or secondary Ig from the Western blots. The signal for the immunoreactive proteins was developed with peroxidase-conjugated anti-rabbit or anti-goat IgG antibodies (Santa Cruz Biotechnology) and visualized by a chemiluminescent detection system on Hyperfilm ECL. (Amersham Pharmacia Bioscience). The developed bands were captured with a Kodak DC290 digital camera, and the pictures were analyzed with kodak 1d image analysis software. Densitometric analyses of developed films were done three times, and the results are presented as average ± standard deviation.
RNA Extractions and RT-PCR. Total RNA was extracted with TRIReagent (Sigma) according to the manufacturer's protocol. Of the total RNA, 4-600 μg was further purified with the MicroPoly(A)-Pure kit (Ambion, Austin, TX). One microgram of the poly(A) RNA was reverse-transcribed into cDNA with the oligo(dT16) primers and the reagents of the PCR Core kit (Applied Biosystems) according to the instructions of the manufacturer. cDNA for the SV1 was amplified by gene-specific primers according to the nested PCR protocol described in ref. 16, except for transferring 1 μl of the first PCR product to the second PCR without dilution. The primers and the PCR protocol for human GAPDH have also been described in ref. 11. Negative controls included a reverse transcription and a PCR omitting the RNA and the cDNA, respectively. PCR products were separated on 1.5% agarose gel followed by ethidium bromide staining and visualized under UV light. Documentation and analysis of data were performed with the instrument and software specified above.
Results
Antisera. The polypeptide hapten consisting of an analog of the first 25 aa of SV1, in which Cys-23 was replaced by Ala and Tyr-26 and Cys-27 were added to its C-terminal sequence, was linked to either BSA or KLH to produce an effective antigen. The immunization process in four rabbits produced 32 batches of antiserum that were then evaluated for binding to the radiolabeled hapten.
Evaluation of the Antisera by RIA. Tyr-26 in the peptide sequence of the hapten was radiolabeled to determine the binding of the individual batches of the antisera to this SV1-hapten and the labeled peptide was purified. All batches of the antisera showed a significant binding to the radiolabeled hapten at a high 1:56,000 final dilution of the antiserum. The binding of the antisera raised against KLH- or BSA-conjugated hapten was different (Table 1). Antisera of the rabbits immunized with the BSA-conjugated hapten showed binding values of 35% or less. However, the binding values exceeded 50% with sera generated against the KLH-SV1-hapten. To select the batches with the best binding, we tested at higher dilutions the batches of serum obtained from the third to the eighth bleeding from rabbits 2,317 and 2,318. Batches 5, 7, and 8 obtained from rabbit 2,317 proved to be the best, producing 51.4%, 47.8%, and 49.3% binding, respectively, at a final dilution of 1:448,000 (Table 2). The standard curve of the RIA showed the following characteristics: The minimal detectable dose was found to be 2 fmol (5.78 pg) per tube for SV1-hapten, based on the dose estimated by a 95% confidence limit for the counts per minute of the zero standard. Nonspecific binding was 1.1%. The coefficient of correlation was 0.9905. Interassay variation was <15%, and intraassay variation was <10%. The crossreaction was estimated as the relative amounts of the test peptide and the unlabeled compound, which induce 50% reduction in binding of the labeled compound (31). By using the 2317/7 batch specific for SV1 in the initial dilution of 1:70,000, we did not find a significant crossreaction with [Ala-28,41]GHRH-R (23-45)-Tyr-Cys-NH2 peptide (0.015%), which is an analog of segment 23-45 of the pituitary GHRH-R. Similarly, no significant crossreaction (0.016%) was found with peptide [Ala-48]SV1 (45-66)-Cys-NH2, corresponding to [Ala-112]GHRH-R (109-130)-Cys-NH2. The crossreactivity was also tested with various natural peptides. No detectable crossreactivity was found with luteinizing hormone-releasing hormone, somatostatin, [Tyr-4]bombesin, and GHRH(1-29)NH2. Antiserum-2317/7 appeared to be specific for SV1-hapten.
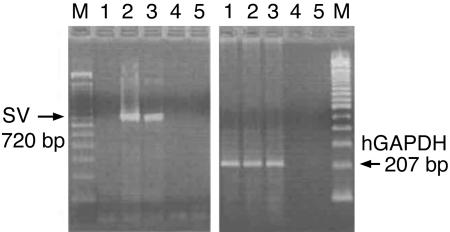
Receptor Assays on the Tumor Tissues. RT-PCR analysis revealed the presence of a single 720-bp band corresponding to the mRNA for the SV1 of the GHRH-R in RL and HT human non-Hodgkin's lymphomas (Fig. 1). The other three SVs could not be amplified. In accordance with our earlier findings, no PCR products were detected in the estrogen-independent human breast carcinoma cell line MDA-MB-231 for SV1 (15). In addition, no PCR products were amplified in the controls when the RNA in reverse transcription or the cDNA in the PCR were omitted. Similarly, we could not amplify PCR products from the negative controls. Binding studies also confirmed high-affinity binding sites for GHRH analog JV-1-42 in the membrane preparations of RL and HT lymphomas, but no JV-1-42 binding could be detected in MDA-MB-231 tumors (Table 3).
Fig. 1.
RT-PCR analysis of human cancer lines for the expression of mRNA for the SV1 of the GHRH-R. Lanes: M, 100-bp molecular mass marker; 1, MDA-MB-231 breast cancer; 2 and 3, RL and HT human non-Hodgkin's lymphomas; 4, RT negative control; 5, PCR negative control.
Table 3. Binding characteristics of the radiolabeled GHRH antagonist JV-1-42 to RL and HT human lymphomas and MDA-MB-231 human breast cancer xenografts grown in nude mice.
| Tumor | Kd,* nM | Bmax,† fmol/mg membrane protein |
|---|---|---|
| MDA-MB-231 | N.D. | N.D. |
| RL | 4.8 ± 0.7 | 406.1 ± 24.8 |
| HT | 6.6 ± 0.1 | 365.2 ± 18.1 |
N.D., not detected.
Binding affinity.
Binding capacity.
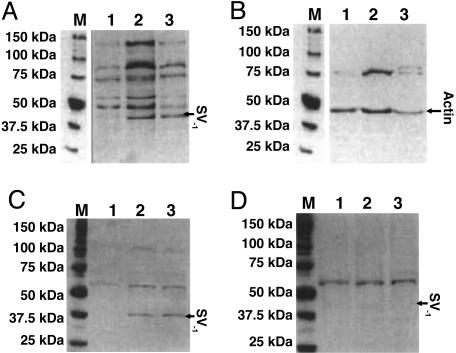
Western Blot Analysis. The specificity of batch 2317/7, which was one of the best according to the results of the RIA tests, was further characterized by immunoblotting on RL and HT lymphomas, shown to be positive for the SV1 receptor with the receptor assays and with MDA-MB-231 as a negative control. The optimal dilution for the primary SV1 antiserum was found to be 1:4,000 in the preliminary experiments, and the antiserum was probed first on the whole-cell lysate of the tumors. Western blot analysis showed a band in the RL and HT lymphoma samples with a molecular mass of ≈40 kDa, which was not detected in the MDA-MB-231 tumors (Fig. 2A). Other bands were also present in all of the samples, at ≈51, 58, 75, 98, and 124 kDa. Only the 75-kDa signal appeared on the blots incubated with the preimmune serum (data not shown). We detected a 43-kDa band in all of the tested samples with the anti-actin serum. A band of 75 kDa also appeared after incubation with the anti-actin serum (Fig. 2B). In addition, the antisera with good RIA binding from the batches harvested after early boosters (2315/2, 2316/2, 2317/2, and 2318/2) were compared with antisera from the later batches (2315/8, 2316/6, 2317/7, and 2318/7) on the positive and negative control samples. In these tests, we detected only a weak 40-kDa band with batches 2317/2 and 2315/8 (data not shown), but the strongest signal was achieved with the 2317/7 batch. From the bands obtained with batch 2317/7, only the 75-kDa band was present in the tests on the other batches of the antisera in all samples.
Fig. 2.
Western blot analysis of MDA-MB-231 breast carcinoma and RL and HT human non-Hodgkin's lymphomas. (A and B) Immunoreactive signals from whole-cell lysate were detected with the nonpurified SV1 antiserum (A) or with anti-actin Igs (B). (C and D) Blots with crude membrane extracts from the tumors were incubated with the purified SV1 antiserum (C) or with the purified SV1 antiserum saturated with 100 pg/ml of the SV1-hapten (D). Lanes: M, Precision Plus molecular mass marker; 1, MDA-MB-231 breast cancer; 2 and 3, RL and HT human non-Hodgkin's lymphomas.
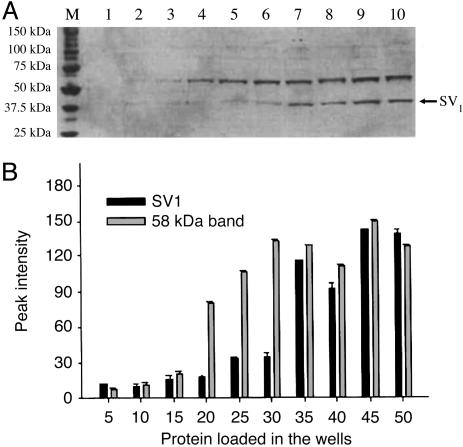
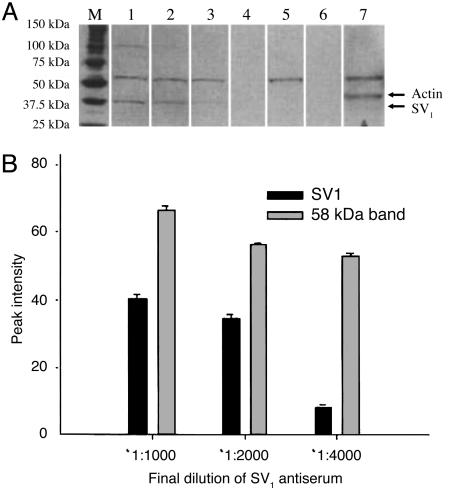
The purified SV1-antiserum also recognized the 40-kDa band from the extract of membrane proteins of RL and HT non-Hodgkin's lymphomas. The intensity of other immunoreactive signals decreased significantly, except for the band at 58 kDa, if the blots were probed with the purified SV1-antiserum (Fig. 2C). Overnight preincubation of the purified antiserum with 100 pg/ml of the synthetic SV1-hapten abolished only the 40-kDa band (Fig. 2D). The intensity of the 40-kDa band corresponding to the SV1 and of the 58-kDa band was gradually enhanced as the amount of membrane protein loaded into the gel was increased up to 30 and 35 μg, respectively. A greater concentration of membrane proteins resulted in a plateau (Fig. 3). A linear decrease in the immunoreactive signal of the 40-kDa band was observed within the dilution range of 1:1,000 to 1:4,000 of the purified SV1-antiserum with 40 μg of membrane proteins (Fig. 4). The intensity of 58-kDa signal decreased slightly at higher dilutions (Fig. 4). Furthermore, the 58-kDa band was also present if the SV1-antiserum was omitted from Western blots or blots were incubated with anti-actin Ig (Fig. 4A).
Fig. 3.
Western blot (A) and densitometric analysis (B) of increasing amounts of RL membrane protein detected with the purified SV1 antiserum. Lanes: M, Precision Plus molecular mass marker; 1-10, 5-50 μg of total protein, respectively, loaded on the gel raised by 5 μg per lane.
Fig. 4.
Western blot (A) and densitometric analysis (B) of membrane proteins from RL tumors detected with different dilutions of purified SV1 antiserum. Lanes: M, Precision Plus molecular mass marker; 1, SV1 antiserum at a 1:1,000 dilution; 2, SV1 antiserum at a 1:2,000 dilution; 3, SV1 antiserum at a 1:4,000 dilution; 4, protein omitted from Western blot; 5, SV1 antiserum omitted from Western blot; 6, secondary anti-rabbit antibody omitted from Western blot; 7, crude membrane extract from RL tumor probed with anti-actin serum.
Discussion
GHRH antagonists effectively inhibit the growth of a broad variety of human cancer cell lines xenografted into nude mice and could provide a new approach to cancer treatment (1-9, 17, 23). The inhibitory effects of GHRH antagonists on tumor growth are exerted in part indirectly through the suppression of the secretion of growth hormone and the resulting reduction in levels of hepatic insulin-like growth factor I. However, in vitro and in vivo findings indicate that the principal action of GHRH antagonists is direct and based on the suppression of the autocrine and/or paracrine production of insulin-like growth factors I and II in tumors and/or to the blockade of the stimulatory action of tumoral GHRH. The direct actions of GHRH antagonists are mediated by specific tumoral receptors, which are different from the pituitary form of receptors (2, 16, 17, 26). mRNAs for four of these SVs of the GHRH-R were found in many cancers, SV1 being more prevalent than the other forms (16, 17). The presence of a specific binding site for GHRH antagonists on human tumors is supported by studies with a radiolabeled GHRH antagonist, which binds with high affinity to various cancers (17). Thus, it was proposed that a receptor protein encoded by SV1 is the binding protein for GHRH antagonists on tumors.
This paper reports the development of polyclonal antibodies to GHRH-R isoforms and the immunological detection of the SV1 receptor protein. Previous antigens for the detection of the full-length GHRH-R were designed to generate antibodies that recognize the C-terminal intracellular domain of the receptor. Bands with molecular masses of 50, 52, and 57 kDa were detected by these antibodies on Western blots by using human pituitary membrane preparations (33, 34). The 52-kDa signal corresponds to the glycosylated form of the GHRH-R, as it had been demonstrated with an antiserum directed against the N terminus of the full-length pituitary GHRH-R (35). The antisera generated against the C terminus of the GHRH-R should detect the SVs described by us, but the 50- and 57-kDa molecular masses do not correlate with the predicted 40-kDa molecular mass of the SV1 (ExPASy Molecular Biology Server, Swiss Institute of Bioinformatics; http://us.expasy.org/tools/pi_tool.html) (16). Furthermore, the antiserum raised against the N terminus of the full-length pituitary GHRH-R (35) should not recognize the SV1 isoform, because its target sequence is not present in the SV1. One of the aims of this study was to demonstrate the expression of the SV1 receptor protein. We thus designed and synthesized a peptide sequence suitable for the generation of a polyclonal antiserum in rabbits for the immunological detection of the SV1 protein. Because only the first 25 aa at the N terminus of the SV1 protein differ from the full-length GHRH-R, our antisera were directed against this sequence. This 25-aa sequence is also present in protein isoforms of the GHRH-R encoded by SV2 and SV4. No significant similarity of the SV1-hapten to any other amino acid sequence of human proteins deposited in databanks was found by blast search (National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/blast).
The tests on the serum from each bleeding of the immunized rabbits confirmed the binding of the polyclonal antibodies to the hapten. Although every batch had an acceptable binding even at a high final dilution, the BSA-conjugated SV1 fragment seemed to be a weaker immunogen in these tests than that with the KLH carrier. The antisera with the best binding in the RIA were further tested in Western blot studies. Because the RL and HT human non-Hodgkin's lymphomas express SV1 and display high-affinity receptors for GHRH antagonists, we selected these cell lines for testing the capacity of the antisera to detect the protein corresponding to SV1. We chose as negative controls MDA-MB-231 mammary carcinoma tumors known to be negative for GHRH binding sites and the mRNA for the SV1. Western blot analysis of both lymphomas revealed the expression of a protein with a molecular mass of ≈40 kDa. This band corresponds closely to the predicted 40-kDa molecular mass of the SV1 protein (16). Further analysis of the predicted protein sequence suggested that the SV1 receptor protein is not a subject of significant posttranslational modifications, such as the cleavage of a signal peptide (SignalIP 3.0 Server, Center for Biological Sequence Analysis at Technical University of Denmark, Www.Cbs.Dtu.Dk/Services/Signalp) or glycosylation (NetNGly 1.0 Server, Center for Biological Sequence Analysis at Technical University of Denmark, www.cbs.dtu.dk/services/NetNGlyc, and NetOGlyc 3.1 Server, Center for Biological Sequence Analysis at Technical University of Denmark, www.cbs.dtu.dk/services/NetOGlyc), which explains the close similarity between the predicted and the detected molecular masses. We could not find SV2 or SV4 of GHRH-R in the two lymphoma models investigated by RT-PCR analysis. Similarly, Western blots could not detect the corresponding bands at the predicted 15 or 7 kDa (ExPASy Molecular Biology Server, Swiss Institute of Bioinformatics; http://us.expasy.org/tools/pi_tool.html). The bands at higher molecular mass cannot be considered as specific signals, because their intensity greatly decreased if the purified antiserum was probed on the crude membrane fractions of tumors. The intensity of the signal for the 40-kDa band was saturable in a range from 35 to 50 μg of protein loaded on the gel and decreased with the dilution of the SV1-antiserum, which may indicate a specific binding of the SV1 antiserum to this band. Although the intensity of the band with a molecular mass of 58 kDa became stronger with increasing protein concentration, this band was still present when the antiserum was saturated with an excess of the antigen. Immunoblotting with the actin antibody led to the visualization of the 58- and 75-kDa bands. This finding could be explained by a nonspecific interaction of one of the Western blot reagents with the 58-kDa band. The signal of the 75-kDa band is also nonspecific, because it appeared on the blots probed with the preimmune serum and the anti-actin serum.
Our results demonstrate that, in RL and HT non-Hodgkin's lymphomas grown in nude mice, the mRNAs for SV1 and binding sites for radiolabeled antagonist JV-1-42 were associated with the expression of a protein that specifically reacted with the antibody directed against the isoform of the GHRH-R. This finding indicates the presence of the protein encoded by SV1 in these tumors. This receptor protein can be a putative receptor for GHRH analogs and consequently should be considered a potential target for the therapy of cancer. Further studies on experimental human tumor models and surgically excised tumor specimens are required to define human malignancies possessing the receptor protein encoded by SV1.
Acknowledgments
We thank Elena Glotser for excellent technical help. This work was supported by the Medical Research Service of the Department of Veterans Affairs (A.V.S.).
Abbreviations: GHRH, growth hormone-releasing hormone; GHRH-R, GHRH receptor; SV, splice variant; VIP, vasoactive intestinal peptide; KLH, keyhole limpet hemocyanin.
References
- 1.Schally, A. V. & Varga, J. L. (1999) Trends Endocrinol. Metab. 10, 383-391. [DOI] [PubMed] [Google Scholar]
- 2.Schally, A. V., Comaru-Schally, A. M., Nagy, A., Kovacs, M., Szepeshazi, K., Plonowski, A., Varga, J. L. & Halmos, G. (2001) Front. Neuroendocrinol. 22, 248-291. [DOI] [PubMed] [Google Scholar]
- 3.Schally, A. V., Szepeshazi, K., Nagy, A., Comaru-Schally, A. M. & Halmos, G. (2004) Cell. Mol. Life Sci. 61, 1042-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schally, A. V. & Comaru-Schally, A. M. (2003) in Cancer Medicine, eds. Kufe, D. W., Pollock, R. E., Weichselbaum, R. R., Bast, R. C., Jr., Gansler, T. S., Holland, J. F. & Frei, E., III (Dekker, Hamilton, Ontario) 6th Ed., pp. 911-926.
- 5.Szepeshazi, K., Schally, A. V., Groot, K., Armatis, P., Herbert, F. & Halmos, G. (2000) Eur. J. Cancer 36, 128-136. [DOI] [PubMed] [Google Scholar]
- 6.Szepeshazi, K., Schally, A. V., Armatis, P., Groot, K., Herbert, F., Feil, A., Varga, J. L. & Halmos, G. (2001) Endocrinology 142, 4371-4378. [DOI] [PubMed] [Google Scholar]
- 7.Jungwirth, A., Schally, A. V., Pinski, J., Groot, K., Armatis, P. & Halmos, G. (1997) Proc. Natl. Acad. Sci. USA 94, 5810-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinski, J., Schally, A. V., Jungwirth A., Groot, K., Armatis, P. & Vadillo-Buenfill, M. (1999) Int. J. Oncol. 9, 1099-1105. [DOI] [PubMed] [Google Scholar]
- 9.Pinski, J., Schally, A. V., Halmos, G., Szepeshazi, K., Zarandi, M. & Armatis, P. (1995) J. Natl. Cancer Inst. 87, 1787-1794. [DOI] [PubMed] [Google Scholar]
- 10.Lamharzi, N., Schally, A. V., Koppan, M. & Groot, K. (1998) Proc. Natl. Acad. Sci. USA 95, 8864-8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahán, Z., Varga, J. L., Schally, A. V., Rekasi, Z., Armatis, P., Chatzistamou, I., Czompoly, T. & Halmos, G. (2000) Breast Cancer Res. Treat. 60, 71-79. [DOI] [PubMed] [Google Scholar]
- 12.Kiaris, H., Schally, A. V., Varga, J. L., Groot, K. & Armatis, P. (1999) Proc. Natl. Acad. Sci. USA 96, 14894-14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rekasi, Z., Varga, J. L., Schally, A. V., Halmos, G., Armatis, P., Groot, K. & Czompoly, T. (2000) Endocrinology 141, 2120-2128. [DOI] [PubMed] [Google Scholar]
- 14.Chopin, K. L. & Herington, A. C. (2001) Prostate 49, 116-121. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Fernandez, O. M., Schally, A. V., Varga, J. L., Groot, K. & Busto, R. (2003) Breast Cancer Res. Treat. 77, 15-26. [DOI] [PubMed] [Google Scholar]
- 16.Rekasi, Z., Czompoly, T., Schally, A. V. & Halmos, G. (2000) Proc. Natl. Acad. Sci. USA 97, 10561-10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halmos, G., Schally, A. V., Varga, J. L., Plonowski, A., Rekasi, Z. & Czompoly, T. (2000) Proc. Natl. Acad. Sci. USA 97, 10555-10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeAlmeida, V. I. & Mayo, K. E. (1998) Mol. Endocrinol. 12, 750-765. [DOI] [PubMed] [Google Scholar]
- 19.Tang, J., Lagace, G., Castagne, J. & Collu, R. (1995) J. Clin. Endocrinol. Metab. 80, 2381-2387. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto, K., Koga, M., Motomura, T., Kasayama, S., Kouhara, H., Ohnishi, T., Arita, N., Hayakawa, T., Sato, B. & Kishimoto, T. (1995) J. Clin. Endocrinol. Metab. 80, 2933-2938. [DOI] [PubMed] [Google Scholar]
- 21.Motomura, T., Hashimoto, K., Koga, M., Arita, N., Hayakawa, T., Kishimoto, T. & Kasayama, S. (1998) Metabolism 47, 804-808. [DOI] [PubMed] [Google Scholar]
- 22.Busto, R., Varga, J. L., Garcia-Fernandez, O. M., Armatis, P. & Szepeshazi, K. (2002) Proc. Natl. Acad. Sci. USA 99, 11866-11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanashiro, C. A., Schally, A. V., Groot, K., Armatis, P., Bernardino, A. L. & Varga, J. L. (2003) Proc. Natl. Acad. Sci. USA 100, 15836-15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plonowski, A., Schally, A. V., Busto, R., Krupa, M., Varga, J. L. & Halmos, G. (2002) Peptides 23, 1127-1133. [DOI] [PubMed] [Google Scholar]
- 25.Halmos, G., Schally, A. V., Czompoly, T., Krupa, M., Varga, J. L. & Rekasi, Z. (2002) J. Clin. Endocrinol. Metab. 87, 4707-4714. [DOI] [PubMed] [Google Scholar]
- 26.Kiaris, H., Busto, R., Artavanis-Tsakonas, S. & Varga, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaylinn, B. D., Harrison, J. K., Zysk, J. R., Lyons, C. E., Jr., Lynch, K. R. & Thorner, M. D. (1993) Mol. Endocrinol. 7, 77-84. [DOI] [PubMed] [Google Scholar]
- 28.Stewart, J. M. & Young, J. D. (1984) Solid Phase Peptide Synthesis (Pierce, Rockford, IL), 2nd Ed.
- 29.Zarandi, M., Csernus, V. J., Bokser, L., Bajusz, S., Groot, K. & Schally, A. V. (1990) Int. J. Peptide Protein Res. 36, 499-503. [DOI] [PubMed] [Google Scholar]
- 30.Mason-Garcia, M., Vaccarella, M., Horvath, J., Redding, T. W., Groot, K., Orsolini, P. & Schally, A. V. (1988) Proc. Natl. Acad. Sci. USA 85, 5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groot, K., Horvath, J., Cai, R. Z. & Schally, A. V. (1995) Int. J. Peptide Protein Res. 45, 561-566. [DOI] [PubMed] [Google Scholar]
- 32.Csernus, V. J., Szende, B., Groot, K., Redding, T. W. & Schally, A. V. (1990) Arzneim.-Forsch. 40, 111-118. [PubMed] [Google Scholar]
- 33.Fujinaka, Y., Yokogoshi, Y., Zhang, C., Okura, T., Kitagawa, K. & Saito, S. (1993) FEBS Lett. 394, 1-4. [DOI] [PubMed] [Google Scholar]
- 34.Boulanger, L., Andersen, P. H. & Gaudreau, P. (1999) Neuroendocrinology 70, 117-127. [DOI] [PubMed] [Google Scholar]
- 35.Gaylinn, B. D., DeAlmeida, V. I., Lyons, C. E., Jr., Wu, K. C., Mayo, K. E. & Thorner, M. O. (1999) Endocrinology 140, 5066-5074. [DOI] [PubMed] [Google Scholar]