Abstract
Background
Cryptosporidium and Giardia duodenalis are gastro-intestinal parasites that infect human and animals worldwide. Both parasites share a broad host range and are believed to be zoonosis. The aim of this study was to identify the species of Cryptosporidium and assemblages of G. duodenalis in lambs and to elucidate their role in zoonotic transmission.
Results
A total of 389 fecal samples were collected from lambs and screened by microscopy and nested PCR targeting the small-subunit ribosomal RNA for Cryptosporidium; and the small-subunit ribosomal RNA, triose phosphate isomerase, β-giardin, and glutamate dehydrogenase genes for G. duodenalis. The prevalence of Cryptosporidium and G. duodenalis was 2.1% (8/389) and 2.6% (10/389), respectively. The infection rate at the three study sites ranged from 1.3 to 3.1% for Cryptosporidium and 1.6 to 3.9% for G. duodenalis; but variation was not statistically significant (p > 0.05). The finding also showed that there is no sex and age group associated difference in the occurrence of Cryptosporidium and G. duodenalis infections in lambs. Sequence analysis revealed that lambs were mono-infection with C. ubiquitum and G. duodenalis assemblage E. The analysis also indicated the presence of genetic variation within isolates of assemblage E; with 4 of them are novel genotypes at the small-subunit ribosomal RNA, β-giardin, and glutamate dehydrogenase genes.
Conclusion
The findings of the current study showed that lambs are capable of harboring C. ubiquitum and G. duodenalis assemblage E. This finding suggests that lambs might be sources for potentially zoonotic Cryptosporidium species. This was first molecular study in lambs and contributes to a better understanding of the epidemiology of Cryptosporidium and G. duodenalis in central Ethiopia.
Keywords: Cryptosporidium, Giardia duodenalis, Zoonotic transmission, Lamb, Ethiopia
Background
Cryptosporidium and Giardia are protozoan parasites that cause diarrhea in humans and animals worldwide [1]. These parasites are transmitted via the fecal-oral route following contact with the infective stages of the parasites. Although the question of zoonotic transmission is clearer for Cryptosporidium than Giardia, both share a wide host range and are thought to be zoonosis [2]. Livestock have taken part in the disease cycle, and have been identified as the sources of numerous outbreaks of human cryptosporidiosis and giardiasis [3].
There is considerable genetic diversity within Cryptosporidium and Giardia duodenalis. Up till now, 26 Cryptosporidium species and more than 70 genotypes have been recognized [4]. G. duodenalis (syn. G. lamblia and G. intestinalis) is also considered a multispecies complex, with at least eight distinct genetic groups or assemblages (A–H) [5]. Assemblages A and B in humans and animals; assemblages C and D in dogs, assemblage E in cattle, sheep and pigs, assemblage F in cats, assemblage G in rats and assemblage H in seals have been described [1, 5].
Cryptosporidium and G. duodenalis infections in sheep are fairly common and have been reported globally. Genetic characterization of Cryptosporidium in sheep has demonstrated the occurrence of the zoonotic species C. parvum and C. ubiquitum; and the host-adapted species C. bovis, C. xiaoi and C. andersoni [6–11]. Three assemblages of G. duodenalis have been recognized in sheep, assemblage E, and the two zoonotic assemblages, assemblages A and B [6, 7, 12, 13]. These studies have provided the evidence that sheep may harbor species of Cryptosporidium and genotypes/assemblages of G. duodenalis which are potentially infectious to humans.
Reports from different parts of Ethiopia have showed different prevalence rate of cryptosporidiosis and giardiasis in humans [14–17]. Recent study conducted on giardiasis showed prevalence of 16.8% in children in Holetta, Sendafa and Chancho [18]. In addition, molecular studies conducted in humans identified C. parvum zoonotic subtype family IIa [19, 20] and G. duodenalis assemblage A and B [18, 21, 22] as the major cause of human cryptosporidiosis and giardiasis in Ethiopia. However, no molecular studies have conducted in sheep to assess the prevalence and genotypes that they harbor. Therefore, the aim of the present study was to identify the species of Cryptosporidium and assemblages of G. duodenalis infection in lambs using DNA sequence analysis to assess their role in zoonotic cycle of the diseases.
Methods
Study area and study animal
This cross-sectional study was conducted between January and June 2014 in three areas of Oromia Special Zone located within 40 km radius of the capital city, Addis Ababa. Based on the available climatological data, the mean annual rainfall of the Special Zone varies from 700 mm to 1400 mm in lowlands and highlands, respectively. The mean annual temperature of the Zone ranges between 20 and 25 °C in the lowlands and 10 to15 °C in the central highlands.
In Ethiopia, small ruminants are important components of the livestock subsector and kept as source of family income, meat, milk, food and wool by small holder farmers throughout the country. It is estimated that about 1,078,000 sheep are annually used for domestic meat consumption. In this study, lambs were included from three districts, Holetta, Sendafa and Chancho town.
Specimen collection
A total of 389 fecal samples were collected from lambs age younger than 5 months in separate and labeled stool containers. The specimens were taken either directly from the rectum of each lamb or from the ground immediately after defecation using sterile disposable gloves. During sample collection, the identification number, age and sex of each lamb were recorded. A part of the samples was used for microscopy and the remaining were preserved in 2.5% potassium dichromate in 1:1 ratio and stored at 4 °C before Deoxyribonucleic acid (DNA) extraction.
Microscopy
Fresh stool sample was tested using light microscope to detect trophozoites and cysts of G. duodenalis using the Lugol’s iodine staining at 10X and 40X magnifications. Thin smears were prepared from sediments of formol-ether concentrated stool samples. The smears were stained with modified Ziehl-Neelson staining method [23]. In brief, air-dried thin smears were fixed with absolute methanol for 5 min, air-dried and stained with carbol-fuchsin for 30 min. Smears were washed with tap water and decolorized with 1% acid-alcohol for 2 min. It was washed with tap water and counterstained with 1% methylene blue for another 2 min, rinsed again in tap water and air-dried. Finally, the stained smears were examined at 100X magnification to detect oocysts of Cryptosporidium.
DNA extraction
After washing the preserved fecal specimens with deionized water, the genomic DNA was extracted from each fecal sample using the E.Z.N.A.® Stool DNA kit (Omega Biotek Inc., Norcross, USA). Concisely, about 50–100 mg of fecal specimen was added in a 2 ml centrifuge tube containing 200 mg of glass beads and placed on ice. Following, 300 μl buffer SP1 and proteinase K were added, and incubated at 70 °C for 10 min. Subsequently, all the procedures outlined in product manual were performed according to the manufacturer’s protocol. Finally, DNA was eluted in 200 μl elution buffer and the extract was stored at −20 °C until used in PCR.
Nested PCR
Approximately 830 bp fragment of the small-subunit (ssu) ribosomal RNA gene of Cryptosporidium was amplified using nested PCR as described [24]. The amplification was done in 25 μl reaction volume containing 24 μl mixes and 1 μl DNA template. The PCR was performed using Applied Biosystems Thermal Cycler version 2.09. The reaction mixture was initially incubated at 94 °C for 5 min for denaturation and then a total 35 cycles of reaction were performed for final denaturation at 94 °C for 45 s, primer annealing at 55 °C for 45 s and strand extension at 72 °C for 1 min. The final extension was done at 72 °C for 10 min and cooled at 4 °C.
The nested PCR of G. duodenalis was conducted by amplification of the ssu rRNA, triose phosphate isomerase (tpi), β-giardin (bg), and glutamate dehydrogenase (gdh) genes. Previously described mixes and PCR conditions were used to amplify fragments of the ssu rRNA gene [25], the bg gene [26], the gdh gene [27] and the tpi gene [28], with some modifications. The primers used, their amplicon size, annealing temperature, and the main uses of the targeted genes are described [29].
The secondary PCR was run under the same reaction and cycling conditions as the primary PCR, except the amplicon of the primary PCR was used as a template. The secondary amplified products were separated by electrophoresis on 1% agarose gel and visualized under a trans-illuminator after staining with ethidium bromide.
DNA sequence analysis
The PCR products purified using Montage PCR filters (Millipore, Bedford, MA) were sequenced in both directions using forward and reverse primers with an ABI BigDye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) on an ABI 3100 automated sequencer. The sequence and chromatograms obtained from each strand were analyzed using ClustalX software. Consensus sequences were then compared with sequences in GenBank database using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast/) to identify the species of Cryptosporidium and assemblages of G. duodenalis.
Statistical analysis
Data were entered into the compute by EpiData version 3.1 and analysed using STATA software. Chi square test was used to verify possible association between infections with the parasites and different factors. Values were considered to be statistically significant when the p value was less than 0.05.
Results
From the total of 389 lambs, 103, 158 and 128 were sampled from Holetta, Sendafa and Chancho areas, respectively. Of these, 201 were female and 188 were male lambs with female to male ratio of 1:0.9 (Table 1). The mean age of the lambs were 2.1 (ranged: 0.1 to 5 months). The study lambs were apparently healthy and no symptom of disease was observed during sample collection.
Table 1.
Prevalence of Cryptosporidium species and G. duodenalis in lambs by study site, sex and age group in Oromia Special Zone, central Ethiopia (January–June, 2014)
| Parameters | Samples examined (N = 389) | Parasites studied | |||||
|---|---|---|---|---|---|---|---|
| Cryptosporidium | G. duodenalis | ||||||
| No. of samples positives n (%) | χ2 | p value | No. of samples positives n (%) | χ2 | p value | ||
| Study Site | |||||||
| Holetta | 103 | 2 (1.9) | 4 (3.9) | ||||
| Sendafa | 158 | 2 (1.3) | 5.285 | 0.071 | 4 (2.5) | 1.229 | 0.542 |
| Chancho | 128 | 4 (3.1) | 2 (1.6) | ||||
| Sex | |||||||
| Male | 188 | 3 (1.6) | 0.383 | 0.536 | 2 (1.1) | 3.298 | 0.069 |
| Female | 201 | 5 (2.5) | 8 (4.0) | ||||
| Age group | |||||||
| <5 weeks | 90 | 3 (3.3) | 0 (0.0) | ||||
| 5–8 weeks | 163 | 3 (1.8) | 0.998 | 0.607 | 5 (3.1) | 3.199 | 0.202 |
| >8 weeks | 136 | 2 (1.5) | 5 (3.7) | ||||
Key: χ2 and p values compare the prevalence between sex, age and breed groups in calves
Prevalence of Cryptosporidium and G. duodenalis infections
The overall prevalence of Cryptospodidium and G. duodenalis infection in lambs based on microscopy and nested PCR was 2.1% (8/389) and 2.6% (10/389), respectively (Fig. 1). The infection rate at the three study sites ranged from 1.3 to 3.1% for Cryptosporidium and 1.6 to 3.9% for G. duodenalis. The variation was statistically not significant (p > 0.05) between the study sites (Table 1). The finding also showed that there is no sex and age group associated difference in the occurrence of Cryptosporidium and G. duodenalis infections in lambs.
Fig. 1.
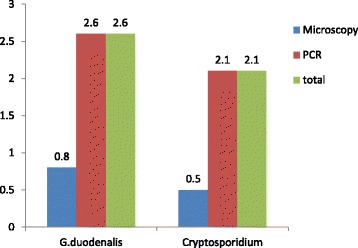
Prevalence of Cryptosporidium and G. duodenalis infection in lambs (n = 389) by microscopy and PCR
Molecular characterization
Eight isolates of Cryptosporidium identified from lambs by PCR amplification of ssu rRNA gene were successfully sequenced. Sequence analysis showed that all the isolates belonged to C. ubiquitum. Comparison with ssu rRNA sequences available in the GenBank shown 100% sequence identity with sequences of isolates previously recognized from sheep, lemurs, wildlife and storm water (accession numbers: EU827398, AF442484, EF641018 and AY737592).
The multi-locus analysis of G. duodenalis obtained from lamb specimens are shown in Table 2. Among the 10 positive specimens, 5 were successfully amplified and sequenced at tpi locus only. The remaining positive specimens were identified by the genes in common. Sequence analysis of the isolates at the ssu rRNA, tpi, bg and gdh, genes revealed the presence of G. duodenalis assemblage E.
Table 2.
Assemblages of G. duodenalis as determined by sequence analysis of ssu rRNA, tpi bg and gdh genes in lambs
| Study site | Isolate code | Lambs | Assemblages by thte four genes | ||||
|---|---|---|---|---|---|---|---|
| Sex | Age | ssu rRNA | tpi | bg | gdh | ||
| Holetta | HL-45 | F | 2 | - | E | - | |
| HL-51 | M | 3.5 | E* | - | - | E* | |
| HL-87 | F | 4 | E* | - | - | E* | |
| HL-91 | F | 3 | E* | - | E | - | |
| Sendafa | SL-12 | F | 3 | - | E | - | - |
| SL-86 | M | 2 | - | E | E* | - | |
| Chancho | CL-152 | F | 1.5 | - | E | - | - |
| CL-169 | F | 3.5 | - | E | - | - | |
| DL-07 | M | 2 | - | E | - | - | |
| AL-02 | F | 3 | - | E | E* | - | |
Key: Asterisks (*) indicate novel genotypes; hyphens (-) indicate PCR-negative results
BLAST analysis of sequences obtained from the three isolates at the ssu rRNA gene showed 100% similarity to each other, but had 99% similarity to sequence registered in the GenBank (accession number AY655701). They had one additional nucleotide (G) at position 271, and named as ET-L1.
The analysis of the 7 isolates at the tpi gene revealed the presence of three distinct genotypes, named as E1, E2 and E3, for a convenient description (Table 3). These isolates showed 100% sequence identity to assemblage E identified from cattle, sheep and goat kids with GenBank accession numbers: KJ363351, JF792419 and EU189333, respectively. Isolates E2 and E3 had 2–3 SNPs compared to the reference (JF792419).
Table 3.
Genetic variants within sub-genotypes of G. duodenalis assemblage E at the tpi, bg and gdh gene in lambs
| Genetic variants | GenBank accession no | No. of isolates | Nucleotide at positions | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 36 | 50 | 57 | 78 | 131 | 144 | 200 | 254 | 512 | |||
| tpi gene | |||||||||||
| E (Ref.) | JF792419 | G | A | C | C | C | A | A | A | G | |
| E1 | KT922261 | 4 | * | * | * | * | * | * | * | * | * |
| E2 | KT922262 | 2 | A | * | T | * | * | G | * | * | * |
| E3 | KT922260 | 1 | * | * | T | T | * | * | * | * | * |
| bg gene | |||||||||||
| E (Ref.) | EU189361 | G | C | G | A | G | T | A | A | G | |
| E4 | KT922250 | 1 | * | * | * | * | * | * | * | * | * |
| ET-L2 | KT922251 | 2 | * | * | * | * | * | * | * | G | * |
| gdh gene | |||||||||||
| E (Ref.) | KR048474 | G | C | G | T | G | G | G | C | T | |
| ET-L3 | KT922256 | 1 | * | * | * | * | * | * | A | * | * |
| ET-L4 | KU196418 | 1 | * | T | * | * | A | * | * | * | C |
Key: Asterisks (*) represent nucleotide identity
Isolate with accession number KT922250 identified at the bg gene showed 100% similarity with assemblage E (accession numbers: EU189361, DQ116625, KP334150, and KC960639). The other isolates were novel (named as ET-L2); and showed 99% similarity to isolate with accession number EU189361. It had substitution of one nucleotide (A-to-G) at nucleotide position 254 (Table 3).
Analysis of the two lamb isolates identified at the gdh gene revealed the presence of two genotypes (Table 3). When these sequences were compared by BLAST analysis of sequences in the GenBank database, they revealed 99% nucleotide sequence uniqueness to the assemblage E registered in the database, and named as ET-L3 and ET-L4. Multiple alignments of these sequences with the reference sequence (KR048474) showed substitution at 50, 131, 200 and 512.
Phylogenetic analysis
Phylogenetic analysis was conducted using sequences obtained in this study at the bg and gdh genes and those available in GenBank database to clarify the genetic relationship among the genotypes. The novel genotype at the bg gene, ET-L2 (KT922251), had formed separate cluster from the reference sequences; whereas the known genotype with accession number EU189361 was clustered with the reference sequences (Fig. 2a). Analysis of one of the two new genotypes obtained at gdh gene, ET-L3 (KT922256), was clustered with reference sequences of assemblage E with accession numbers: EF507645, KP334147, KC960647, KF843925, AY178740 and KP635110. On the other hand, the second new genotype ET-L4 (KU196418) was clustered with sequence obtained from pig (accession number AY178741) (Fig. 2b).
Fig. 2.
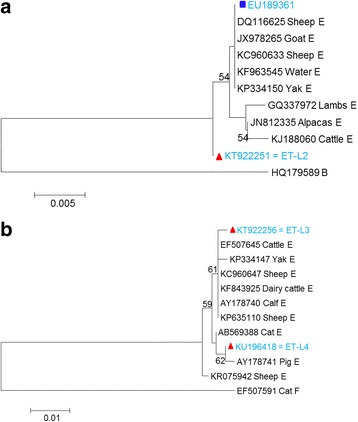
Phylogenetic tree based on nucleotide sequences of the bg gene (a) and gdh gene (b) of G. duodenalis. Trees were constructed using the neighbor-joining method based on genetic distance calculated by the Kimura 2-parameter model, employed in MEGA version 5.2. Bootstrap values >50% from 1,000 replicates is shown on nodes. Isolates showed known and novel sequences obtained from this study are marked by rectangle (blue) and triangles (red), respectively
Discussion
Cryptosporidium species and G. duodenalis infections are relatively common and have been reported worldwide in sheep [6, 8, 13, 30, 31]. Although some reports suggest that sheep are probably not an important reservoirs for zoonotic Cryptosporidium species and G. duodenalis [6, 8], molecular characterization of the two parasites indicated that they can harbor species or genotypes/assemblages which are potentially infectious to humans [7, 8, 11–13, 30, 31].
The present study is the first to report the occurrence and genetic characterization of Cryptosporidium and G. duodenalis in sheep in Ethiopia. The overall infection rates were 2.1 and 2.6% for Cryptospodidium species and G. duodenalis, respectively, which was much lower than those reported from other parts of the world [6, 8, 12, 30, 31]. Even though Cryptosporidium is thought to be more prevalent in sheep than G. duodenalis [11], the present study suggests that this may not always be the case, as the prevalence of these protozoan infections was not significantly different. This situation was also supported by report from Greece, where the prevalence of Cryptosporidium infection was much lower both in lambs and goat kids than the anticipated as compared to G. duodenalis [31].
Results of the present molecular analyses reveal that C. ubiquitum is the only species identified in lambs. Likewise, studies conducted in various parts of the world have identified C. ubiquitum as the common and the most prevalent species in sheep observed in different magnitude [6–8, 10–12, 30]. However, several studies have also reported species of Cryptosporidium other than C. ubiquitum [8, 10, 11, 30]. The zoonotic species, C. parvum, was not detected in lambs in the present study which is similar with the previous report [6]. This finding suggests that lambs may not be an important reservoir for C. parvum in Ethiopia.
C. ubiquitum has been reported in a wide variety of hosts including humans. The first report of C. ubiquitum in humans was found in fecal samples from patients with clinical symptoms of cryptosporidiosis [32]. Another study conducted to assess the importance of dairy cattle as a source of human Cryptosporidium infections in Ontario, Canada has also been reported C. ubiquitum in humans [33]. Subsequently, sporadic cases of this species affecting humans have been described [34, 35]. Therefore, C. ubiquitum should be considered a potential emerging zoonotic pathogen, and lambs can be considered as an important reservoir for this species in Ethiopia.
Sequence analysis of 10 G. duodenalis isolates revealed mono-infection with G. duodenalis assemblage E at the four loci, which is similar with the previous report [31]. This assemblage is usually found in hoofed animals including cattle and sheep [6–8, 10–12, 30], although the zoonotic genotypes A and B have also been observed in sheep in other studies [6, 8, 12, 13]. This observation indicates that lambs may not be an important reservoir for zoonotic assemblages of G. duodenalis in Ethiopia. However, further investigations are needed if this observation holds true in other parts of the country preferably involving larger sample size for a better understanding of epidemiology of cryptosporidiosis and giardiasis in lambs.
Molecular and phylogenetic analysis indicated that there is genetic variation within genotype E. Earlier allozyme data based on 23 allozyme loci shown that genotype E comprised three clusters of isolates; a pig cluster, a sheep cluster and a cattle cluster [36]. In the present study, all the sheep-derived isolates were showed clear clustering of isolates into host groups except one separate cluster in assemblage E. This is based on phylogenetic analysis of sequence data from two loci, bg and gdh.
Conclusion
The present study demonstrates that lambs are capable of harboring C. ubiquitum and G. duodenalis assemblage E. These finding indicate that lambs may be important sources of potentially zoonotic C. ubiquitum but are not sources of zoonotic assemblages of G. duodenalis in the study localities. This is the first report of Cryptosporidium and G. duodenalis infection in lambs, and the obtained result would serve as a baseline data for further investigation for better understanding of the epidemiology of cryptosporidiosis and giardiasis in Ethiopia.
Acknowledgements
We would like to express our gratitude to colleagues of International Joint Research Laboratory for Zoonotic Diseases, Henan Agricultural University (China) for their technical assistance during the molecular work. Our special thanks go to Dr. Tesfaye Habtemariam for language edition.
Funding
This study was supported by the Office of Graduate Studies and the Office of the Vice President for Research and Technology Transfer, Thematic Research Fund, Addis Ababa University; and the State Key Program of National Natural Science Foundation of China (No. 31330079), and Innovation Scientists and Technicians Troop Construction Projects of Henan Province (No. 134200510012). The contribution of Arba Minch University was also remarkable. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The dataset supporting the conclusions of this article is available in the GenBank database repository under the accession numbers: KT922236 for Cryptosporidium; and KT922250, KT922251, KT922256, KT922260, KT922261, KT922262, KT922264 and KU196418 for G. duodenalis.
Authors’ contributions
TW, HA, BE and GT conceived and designed the experiments; TW, MRK and JL performed the experiments; TW, JL, LZ and GT analyzed the data; MRK, BE, LX and GT contributed reagents/materials/analysis tools; TW, HA, BE, LZ and GT wrote the manuscript. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not Applicable.
Ethics approval and consent to participate
Ethical clearance was obtained from the Institutional Review Board of ALIPB, Addis Ababa University and the National Health Research Ethics Review Committee, Federal Ministry of Science and Technology of Ethiopia. Support letters were obtained from concerned agricultural offices and administrative authorities at community level. Before collection of the fecal specimens, the objective of the study was explained to the owners of the animals and informed consent was obtained.
Contributor Information
Teklu Wegayehu, Email: tekluweg2007@yahoo.com.
Md Robiul Karim, Email: vet_robiul@yahoo.com.
Junqiang Li, Email: lijunqiangcool@126.com.
Haileeyesus Adamu, Email: haile27@gmail.com.
Berhanu Erko, Email: berhanu.erko@aau.edu.et.
Longxian Zhang, Email: zhanglx8999@gmail.com.
Getachew Tilahun, Email: getachew.tilahun@aau.edu.et.
References
- 1.Caccio SM, Ryan UM. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Xiao L, Fayer R. Molecular characterization of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol. 2008;38:1239–1255. doi: 10.1016/j.ijpara.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks-an update 2004–2010. Water Res. 2011;45:6603–6614. doi: 10.1016/j.watres.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Ma J, Cai J, Ma J, Feng Y, Xiao L. Occurrence and molecular characterization of Cryptosporidium spp. in yaks (Bos grunniens) in China. Vet Parasitol. 2014;202:113–118. doi: 10.1016/j.vetpar.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan UM, Bath C, Robertson I, Read C, Elliot A, McInnes L, et al. Sheep may not be an important zoonotic reservoir for Cryptosporidium and Giardia parasites. Appl Environ Microbiol. 2005;71:4992–4997. doi: 10.1128/AEM.71.9.4992-4997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geurden T, Thomas P, Casaert S, Vercruysse J, Claerebout E. Prevalence and molecular characterization of Cryptosporidium and Giardia in lambs and goat kids in Belgium. Vet Parasitol. 2008;155:142–145. doi: 10.1016/j.vetpar.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Yang R, Jacobson C, Gordon C, Ryan U. Prevalence and molecular characterization of Cryptosporidium and Giardia species in pre-weaned sheep in Australia. Vet Parasitol. 2009;161:19–24. doi: 10.1016/j.vetpar.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Fayer R, Santin M. Cryptosporidium xiaoi n.sp. (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries) Vet Parasitol. 2009;164:192–200. doi: 10.1016/j.vetpar.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Feng Y, Cui B, Jian F, Ning C, Wang R, et al. Cervine genotype is the major Cryptosporidium genotype in sheep in China. Parasitol Res. 2010;106:341–347. doi: 10.1007/s00436-009-1664-x. [DOI] [PubMed] [Google Scholar]
- 11.Robertson LJ, Gjerde BK, Furuseth HE. The zoonotic potential of Giardia and Cryptosporidium in Norwegian sheep: a longitudinal investigation of 6 flocks of lambs. Vet Parasitol. 2010;171:140–145. doi: 10.1016/j.vetpar.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Santin M, Trout JM, Fayer R. Prevalence and molecular characterization of Cryptosporidium and Giardia species and genotypes in sheep in Maryland. Vet Parasitol. 2007;146:17–24. doi: 10.1016/j.vetpar.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Zhang X, Wang R, Liu A, Shen Y, Ling H, et al. Genetic characterizations of giardia duodenalis in sheep and goats in Heilongjiang province, china and possibility of zoonotic transmission. PLoS Negl Trop Dis. 2012;6(9):e1826. doi: 10.1371/journal.pntd.0001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adamu H, Endeshaw T, Teka T, Kifle A, Petros B. The prevalence of intestinal parasites in paediatric diarrhoeal and non-diarrhoeal patients in Addis Ababa hospitals, with special emphasis on opportunistic parasitic infections and with insight into the demographic and socio-economic factors. Ethiop J Health Dev. 2006;20:39–46. doi: 10.4314/ejhd.v20i1.10010. [DOI] [Google Scholar]
- 15.Ayalew D, Boelee E, Endeshaw T, Petros B. Cryptosporidium and Giardia infection and drinking water source among Children in Lege Dani, Ethiopia. Trop Med Inter Health. 2008;13(4):472–475. doi: 10.1111/j.1365-3156.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- 16.Tigabu E, Petros B, Endeshaw T. Prevalence of Giardiasis and Cryptosporidiosis among children in relation to water sources in Selected Village of Pawi Special District in Benishangul-Gumuz Region, Northwestern Ethiopia. Ethiop J Health Dev. 2010;24(3):205–213. [Google Scholar]
- 17.Wegayehu T, Adamu H, Petros B. Prevalence of Giardia duodenalis and Cryptosporidium species infections among children and cattle in North Shewa Zone, Ethiopia. BMC Infect Dis. 2013;13:419. doi: 10.1186/1471-2334-13-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegayehu T, Karim MR, Li J, Adamu H, Erko B, Zhang L, Tilahun G. Multilocus genotyping of Giardia duodenalis isolates from children in Oromia Special Zone, central Ethiopia. BMC Microbiol. 2016;16:89. doi: 10.1186/s12866-016-0706-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamu H, Petros P, Hailu A, Petry F. Molecular characterization of Cryptosporidium isolates from humans in Ethiopia. Acta Trop. 2010;115:77–83. doi: 10.1016/j.actatropica.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, et al. Distribution and Clinical Manifestations of Cryptosporidium Species and Subtypes in HIV/AIDS Patients in Ethiopia. PLoS Negl Trop Dis. 2014;8(4):e2831. doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flecha MJ, Benavides CM, Tissiano G, Tesfamariam A, Cuadros J, de Lucio A, Bailo B, Cano L, Fuentes I, Carmena D. Detection and molecular characterization of Giardia duodenalis, Cryptosporidium spp. and Entamoeba spp. among patients with gastrointestinal symptoms in Gambo Hospital, Oromia Region, southern Ethiopia. Trop Med Int Health. 2015;20:1213–1222. doi: 10.1111/tmi.12535. [DOI] [PubMed] [Google Scholar]
- 22.Gelanew T, Lalle M, Hailu A, Pozio E, Caccio SM. Molecular characterization of human isolates of Giardia duodenalis from Ethiopia. Acta Trop. 2007;120:92–99. doi: 10.1016/j.actatropica.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Henriksen SA, Pohlenz JF. Staining of Cryptosporidia by a modified Ziehl- Neelsen technique. Acta Vet Scand. 1981;22:594–596. doi: 10.1186/BF03548684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl Environ Microbiol. 2001;67:1097–1101. doi: 10.1128/AEM.67.3.1097-1101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Appelbee AJ, Frederick LM, Heitman TL, Olson ME. Prevalence and genotyping of Giardia duodenalis from beef calves in Alberta, Canada. Vet Parasitol. 2003;112:289–294. doi: 10.1016/S0304-4017(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 26.Caccio SM, de Giacomo M, Pozio E. Sequence analysis of the b-giardin gene and development of a PCR-RFLP assay to genotype Giardia duodenalis cysts from human faecal samples. Int J Parasitol. 2002;32:1023–1030. doi: 10.1016/S0020-7519(02)00068-1. [DOI] [PubMed] [Google Scholar]
- 27.Read CM, Monis PT, Thompson RC. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Zhao G, Chen G, Jian F, Zhang S, Feng C, et al. Multilocus Genotyping of Giardia duodenalis in Dairy Cattle in Henan, China. PLoS One. 2014;9(6):e100453. doi: 10.1371/journal.pone.0100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paz e Silva FM, Lopes RS, Bresciani KDS, Amarante AFT, Araujo JP., Jr High occurrence of Cryptosporidium ubiquitum and Giardia duodenalis genotype E in sheep from Brazil. Acta Parasitol. 2014;59(1):193–196. doi: 10.2478/s11686-014-0223-5. [DOI] [PubMed] [Google Scholar]
- 31.Tzanidakis N, Sotiraki S, Claerebout E, Ehsan A, Voutzourakis N, Kotsiopoulos D, et al. Occurrence and molecular characterization of Giardia duodenalis and Cryptosporidium spp. in sheep and goats reared under dairy husbandry systems in Greece. Parasite. 2014;21:45–52. doi: 10.1051/parasite/2014048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong CS, Eisler DL, Alikhani A, Fung VW, Tomblin J, Bowie WR, et al. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg Infect Dis. 2002;8:263–268. doi: 10.3201/eid0803.010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trotz-Williams LA, Martin DS, Gatei W, Cama V, Peregrine AS, Martin SW, et al. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol Res. 2006;99:346–352. doi: 10.1007/s00436-006-0157-4. [DOI] [PubMed] [Google Scholar]
- 34.Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, McEvoy JM. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbiol. 2006;44:4303–4308. doi: 10.1128/JCM.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhea in England between 1985 and 2000. J Med Microbiol. 2006;55:703–707. doi: 10.1099/jmm.0.46251-0. [DOI] [PubMed] [Google Scholar]
- 36.Monis PT, Andrews RH, Mayrhofer G, Ey PL. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect Genet Evol. 2003;3:29–38. doi: 10.1016/S1567-1348(02)00149-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is available in the GenBank database repository under the accession numbers: KT922236 for Cryptosporidium; and KT922250, KT922251, KT922256, KT922260, KT922261, KT922262, KT922264 and KU196418 for G. duodenalis.


