Abstract
Prostate cancer is among the most prevalent life-threatening cancers diagnosed in the male population today. Various methods have been exploited in an attempt to treat this disease but these treatments, alongside preventative tactics, have been insufficient to control mortality rates and have usually resulted in detrimental adverse events. An opportunity to devise more-specific and potentially more-effective approaches for the eradication of prostate tumours can be found by targeting specific biological pathways. NUMB (protein numb homologue), a key regulator of cell fate, represents an attractive, actionable target in prostate cancer. NUMB participates in the observed deregulation of NOTCH (neurogenic locus notch homologue protein) signalling in prostate tumours, and the NUMB–NOTCH interaction regulates cell fate. NUMB has potential both as a target for control of prostate tumorigenesis and as a biomarker for identification of patients with prostate cancer who are likely to benefit from NOTCH inhibition.
Introduction
Many population forecasts predict a significant increase in the number of older people in the next 20 years.1 As men increase in age, their risk of developing prostate cancer rises exponentially—about six cases in 10 are diagnosed in men aged ≥65—so prostate cancer incidence is set to rise dramatically. In parallel, worldwide obesity has nearly doubled since 1980. In 2008, more than 1.4 billion adults ≥20 years of age were overweight, and ≥200 million men and nearly 300 million women were obese.2 Evidence suggests obesity is associated with an elevated incidence of aggressive prostate cancer, and increased risks of biochemical failure following radical prostatectomy and external-beam radiotherapy, cardiovascular complications and ‘metabolic syndrome’ following androgen-deprivation therapy and increased prostate-cancer-specific mortality.3 The design of strategies to improve the detection, diagnosis and treatment of prostate cancer, and survivorship of prostate cancer patients, is, therefore, essential.
Tumour growth is associated with tumour stem cell proliferation and tumour vascularization, so the eradication of cancer stem cells and complete vascular destruction are key to tumour control.4,5 Adaptation of the prostate cancer stem cell population to radiation therapy and chemotherapy is thought to be associated with loss of asymmetric cell divisions and an acceleration of differentiation, leading to progressive dominance of cells with a neuroendocrine phenotype.6,7 NUMB (protein numb homologue) is the human homologue of the protein numb that was initially discovered in Drosophila melanogaster as an adaptor protein responsible for recruiting proteins into different signalling pathways.8 NUMB is an evolutionarily conserved protein well-known for its multifaceted role in neurogenesis9,10 and cellular homeostasis within the peripheral and central nervous systems.8,11 The antagonistic influence of NUMB on the NOTCH pathway and the associated regulation of cell fate has drawn attention to the potential role of NUMB in tumorigenesis in a number of solid tumours, including those arising in the prostate.
The NOTCH (neurogenic locus notch homologue protein) pathway is an evolutionarily conserved signalling system that regulates cell proliferation, differentiation, cell-fate determination and self-renewal of stem cells and progenitor cells in both embryonic and adult organs.12,13 NOTCH inhibition is under increasing investigation as a novel anticancer strategy, and so the examination of the interaction between NUMB and NOTCH is warranted.
In 50% of human mammary carcinomas, the control of NOTCH signalling by NUMB is abrogated by ubiquitination and proteasomal degradation of NUMB.14 Although evidence for the role of NUMB in several types of cancer is accumulating,15,16 NUMB has not been extensively studied in relation to prostate cancer. In this Review we evaluate how NUMB is likely to participate in the observed deregulation of NOTCH signalling in prostate tumours, and we highlight the potential clinical implications of the NUMB–NOTCH interaction in prostate cancer.
The multiple regulatory functions of NUMB
The consistency of the biological functions of NUMB proteins within rats, chickens, birds, flies, humans and mice has highlighted their essential role in maintenance of cellular homeostasis within both the peripheral and central nervous systems.8,11 NUMB has diverse isoforms derived from alternative splicing of mRNA.17–19 Six human NUMB isoforms have been identified, with molecular weights between 50 and 75 kDa.18,20 In all these isoforms, the N-terminus carries a phosphotyrosine-binding (PTB) domain while the C-terminus contains Eps15 homology regions: aspartate-proline-phenylalanine tripeptide (DPF) and aspargine-proline-phenylalanine tripeptide (NPF) (Figure 1a). Studies in Drosophila have elucidated functional roles for these domains. The NPF domain can bind to the endocytic machinery components Eps15 and Eps15R.21 At the centre of the NUMB protein, a proline rich (PRR) domain exists in certain isoforms, containing Src homology binding sites involved in intracellular signal transduction.22 The PTB domain has a role in tyrosine-kinase-mediated signalling pathways and is also crucial for membrane localization.8,19,23 An endocytic function for NUMB proteins has been proposed on the basis of studies identifying a PTB domain that specifically binds to acidic phospholipids and that is rich in basic residues.8,23,24 The membrane localization of NUMB is mediated by G-protein-coupled receptor (GPCR)-activated protein hydrolysis and protein kinase C (PKC)-dependent phosphorylation interactions within the 218–366 amino acid regions of NUMB proteins.19,23
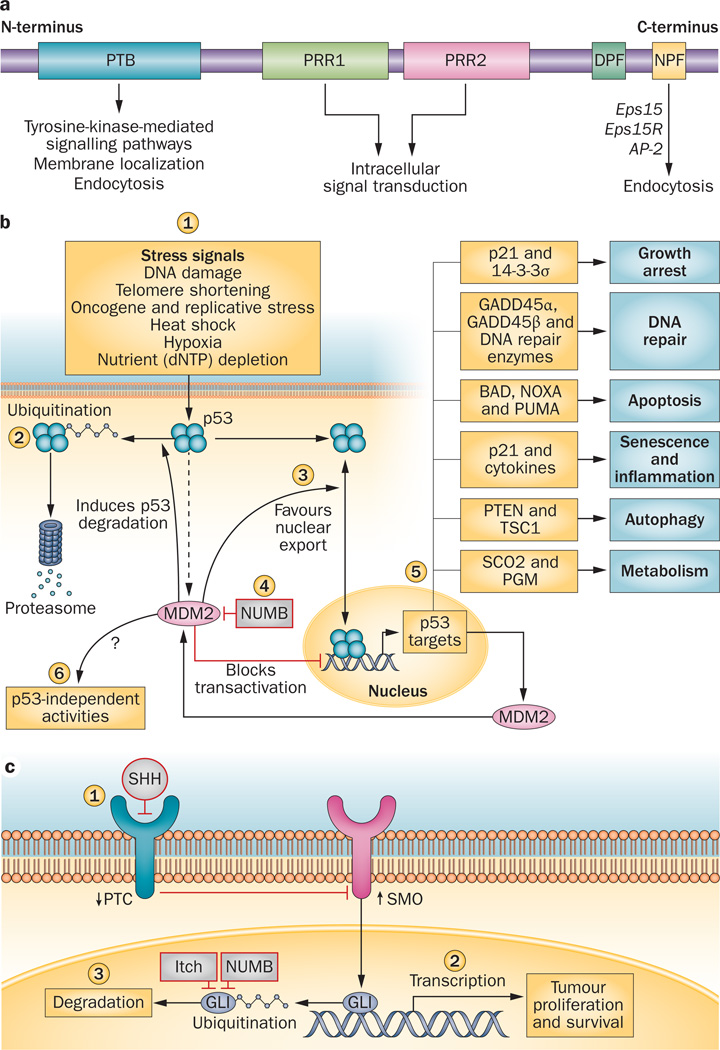
Figure 1.
The multiple functions of NUMB. a | Structure of the NUMB protein. All six isoforms of the human NUMB protein have a similar structure. At the N-terminus, a phosphotyrosine-binding (PTB) domain facilitates tyrosine kinase signalling, endocytosis and membrane localization. Eps15 homology regions DPF and NPF are located at the C-terminus. The NPF domain can bind to the endocytic machinery components Eps15 and Eps15R. At the centre of the NUMB protein, proline rich (PRR) domains exist in certain isoforms, containing Src homology binding sites involved in intracellular signal transduction. b | Control of p53 activity by NUMB. p53 activity is induced by many stress-related signals (1). A negative regulatory feedback loop controls cellular levels of p53: p53-dependent transcription of MDM2 promotes p53 degradation (2). MDM2 also inhibits p53 activity by blocking p53 transcriptional activity and favouring p53 nuclear export (3). By inhibiting MDM2 activity, NUMB amplifies p53 activity (4). Transcription of target genes promotes p53 tumour-suppressor properties (5). NUMB-MDM2 interaction might also alter p53-independent MDM2 activities (6). c | Regulation of the Hedgehog/GLI pathway by NUMB. Sonic hegehog (SHH) inhibits the repression of Smoothened (SMO) by Patched (PTC) (1). Activation of SMO transmits the signal intracellularly and triggers the nuclear action of the GLI proteins. GLI proteins regulate target genes that participate in tumour proliferation and survival (2). NUMB interacts with Itch to promote ubiquitination and degradation of GLI (3). In prostate cancer cells, GLI proteins bind to the androgen receptor and affect androgen signalling. Abbreviations: BAD, BcL-2-associated agonist of cell death; GADD45, growth arrest and DNA-damage-inducible; Itch, E3 ubiquitin–protein ligase Itchy homologue; MDM2, E3 ubiquitin–protein ligase Mdm2; NOXA, PMA-induced protein 1; PGM, phosphoglycerate mutase; PTEN, phosphatase and tensin homolog; PUMA, Bcl-2-binding component 3; SCO2, synthesis of cytochrome c oxidase 2; TSC1, tuberous sclerosis 1. Permission for part b obtained from Nature Publishing Group © Chène, P. Nat. Rev. Cancer 3, 102–109 (2003) and Dotto, G. P. Nat. Rev. Cancer 9, 587–595 (2009). Permission for part c obtained from Nature Publishing Group © Ruiz i Altaba, A. et al. Nat. Rev. Cancer 2, 361–372 (2002).
The C-terminus of NUMB directly interacts with the endocytic machinery. Adaptor protein 2 (AP-2) is an integral component of clathrin-coated pits25 and functions in endocytic trafficking of transmembrane receptors for internalization and relocalization.21 During asymmetric cell division in neurogenesis, NUMB colocalizes with AP-2 through binding to α-adaptin. Mutation of α-adaptin results in NUMB loss-of-function effects such as loss of neurons and glia and gain of hairs and sockets at external sensory organs.26–30 NUMB can also interact with adaptor protein 1 (AP-1), which is involved in protein sorting and recycling.31
NUMB has a prominent role in the control of cell fate within the Hedgehog and p53 signalling pathways (Figure 1b, 1c).23 In breast cancer, inactivity of the tumour suppressor protein p53 through loss of NUMB results in activation of NOTCH and reduction of tumour suppression.26 NUMB inhibits E3 ubiquitin–protein ligase Mdm2, blocking degradation of p53 transcription factors.23,26,27 Direct methylation of the PTB binding domain of NUMB by lysine methyltransferase Set8 proteins results in p53 ubiquitination and degradation.28 By maintaining high levels of p53 within the cell, an aggressive tumour phenotype can be suppressed, whereas a p53-dependentphenotype is strengthened.27 With a p53-dependent phenotype, genes associated with apoptotic responses to cellular stresses can be expressed and can impede tumorigenesis.26,29 Although p53 has been primarily studied in breast cancer, disruption of normal p53 pathways has been reported in most, if not all, cancers,26 including prostate cancer.30
Inhibition by NUMB of the Hedgehog signalling pathway could also contribute to regulation of prostate tumorigenesis. Hedgehog signalling is the primary regulator of embryonic and developmental tissue patterning and cell proliferation.32 In prostate cancer, activation of the pathway is required for regeneration of the prostate epithelium, and continuous activation induces tumorigenic properties in prostate progenitor cells.33 Several Hedgehog pathway inhibitory drugs are under clinical development, with one (vismodegib) recently approved by the FDA for the treatment of basal cell carcinoma.34–36 Activation of Hedgehog signalling is triggered by protein–protein interactions between Hedgehog ligands (SHH, IHH and DHH) and Patched (PTC) transmembrane receptors that are regulated by Hedgehog ligand gradient formation.32,37 This binding results in signalling by the GPCR Smoothened protein that activates downstream GLI family transcription factors (GLI1–3).37 NUMB promotes ubiquitination and degradation of GLI1 through recruitment of Itch, the E3 ubiquitin–protein ligase Itchy homologue (Figure 1c).37,39
The importance of GLI proteins in prostate cancer has been reported:40 high expression of hedgehog ligands and GLI2 correlate with metastasis potential and therapeutic resistance. Inhibition of the Smoothened protein reduced cell viability and spheroid formation, and induced apoptosis, in prostate cancer stem cells in NOD/SCID IL2Rγ null mice.38 GLI proteins also interact with the androgen receptor and might contribute to androgen independence.40,41 Normal (PNT-2) and tumorigenic (DU145 and PC3) androgen-independent cells displayed higher GLI activity than androgen-dependent LNCaP prostate cancer cells.42 GLI1 was shown to act as a corepressor of the androgen receptor and to block transactivation mediated by the androgen receptor.43 In LnCaP cells, androgen R1881 strongly suppressed the expression of Hedgehog ligands and GLI2 expression.44 LnCaP cells overexpressing GLI1 were viable in the presence of the androgen receptor inhibitor bicalutamide and the transcriptome of these cells was significantly closer to DU145 and PC3 cells than to untransfected controls.42 Further examination is warranted of the relationship between NUMB and GLI expression levels within the context of androgen independence.
NUMB proteins are regulated by alternative splicing, and transcriptional and post-transcriptional modifications. Musashi1 (Msi1) is a neural RNA-binding protein that represses NUMB translation,45 preserving the immature neural stem cell state,46 and increasing NOTCH signalling.46 Low-level expression of Msi1 has been reported in prostate cancer cells.47 Several RING finger proteins are involved in ubiquitin-dependent degradation of NUMB, including LNX,48 Siah-149 and Mdm2.49 Specific microRNAs such as MiR-146a are involved in transcriptional repression of NUMB, which inhibits NOTCH signalling in muscle cells by delaying myogenic differentiation.50 In prostate cancer, MiR-146a is associated with increased cancer risk,51 and loss of MiR-146a function is associated with suppression of tumour growth in castration-resistant disease.52,53 In mammals, phosphorylation of NUMB by protein kinase C (PKC) or regulation of membrane binding by GPCR-mediated lipid hydrolysis, lead to relocation of NUMB from the cortical membrane to the cytosol,19 rendering it unable to complete its function. This loss of function prevents NUMB-mediated NOTCH inhibition and could explain the increase in oncogenic expression of NOTCH in the prostate.54
NOTCH is a key pathway in prostate cancer
The NOTCH pathway is increasingly recognized as a major deregulated pathway in many solid malignancies including renal and prostate cancers.55,56 The pivotal function of NOTCH in tumorigenesis stems from the control of cell fate decisions, stem cell maintenance, cellular proliferation and apoptosis.13 Additionally, NOTCH is involved in cellular differentiation in multicellular organisms, angiogenesis, epithelial–mesenchymal transitions and cell adhesion,55 and is responsive to hypoxia.57
The human NOTCH pathway is composed of ligands (Jagged-1, Jagged-2 and Delta-like proteins 1, 3 and 4) and receptors (Notch 1–4).58 Activation, regulation and degradation of the Notch signal require endosomal trafficking of both ligands and receptors.59,60 Activation of the NOTCH pathway begins with binding of type 1 ligands from the DSL families (Delta, Jagged) to Notch transmembrane receptors,61–64 triggering a series of proteolytic cleavages within the plasma membrane and intracellular compartment, and initiating signal transduction. ADAM family proteins TACE (ADAM 17) and Kuzbanian (ADAM 10) mediate regulation of Notch extracellular domain shedding.65,66 The final cleavage occurs by a γ–secretase complex releasing the Notch intracellular domain (NICD).62,63,65,67–70 The NICD functions as a transcription factor within the nucleus, interacting with recombining binding protein suppressor of hairless (RBPJ) transcription factor and regulating target genes including those of several helix–loop–helix transcription factors such as Hes-1 and Hey-1.62,63,71,72 During NICD–RBPJ interaction, the transcriptional activator protein Mastermind is also recruited to initiate transcription (Figure 2). The NOTCH pathway contributes to prostate tumorigenesis,73,74 has been proposed to influence the outcome of anticancer hormonal75,76 and cytotoxic (docetaxel)77 treatments and might be more active in patients with prostate cancer who have a high BMI.78 The NOTCH pathway is essential to regulation of blood vessel structure,79,80 which is altered within tumours.81,82
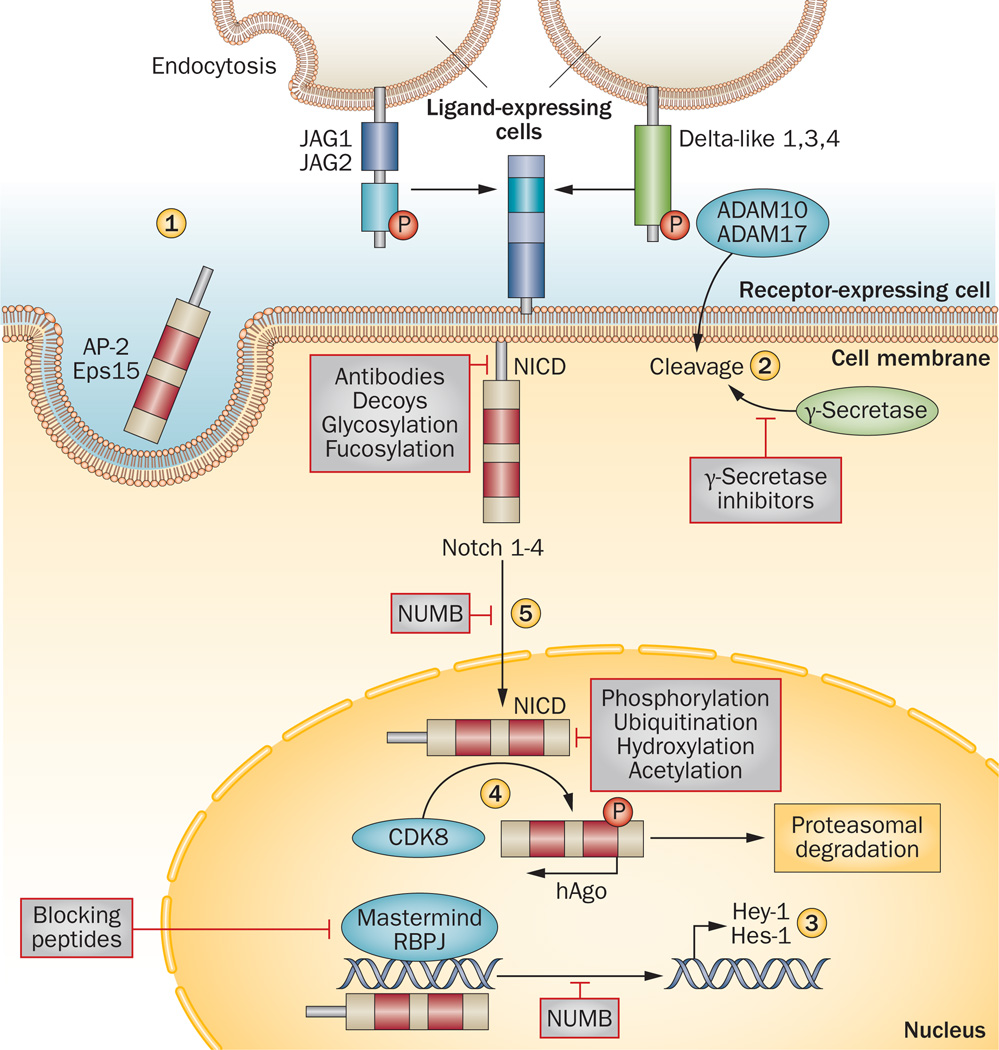
Figure 2.
Inhibition of the NOTCH pathway by NUMB. NUMB interacts with AP-2 and Eps15 to disrupt endocytosis of Notch ligands and receptors (1). The binding of ligands (Delta, Jagged [JAG]) to Notch transmembrane receptors 1–4 triggers a series of proteolytic cleavages by ADAM family proteins (ADAM 10, ADAM 17) and a γ–secretase complex that lead to the release of the Notch intracellular domain (NICD) (2). The NICD is translocated into the nucleus, interacts with the recombining binding protein suppressor of hairless (RBPJ) transcription factor and the transcriptional activator protein Mastermind to regulate target genes such as Hes-1 and Hey-1. NUMB might disturb this transcriptional machinery and inhibit Notch-dependent gene expression (3). Phosphorylation of NICD by CDK8 leads to hAgo-dependent proteosomal degradation of NICD (4). NUMB prevents translocation of NCID into the nucleus (5). Strategies under development for the therapeutic inhibition of the NOTCH pathway are indicated in boxes.
Inhibition of NOTCH signalling
Possible strategies for NOTCH inhibition include receptor activation inhibition, modification of receptor–ligand interactions, inhibition by antibodies, disruption of γ-secretase cleavage of Notch, alteration of NICD post-translational modifications, inhibition of protein–protein interactions within the nucleus and disruption of assembly of the coactivator NICD–RBPJ complex (Figure 2).
Receptor activation inhibition
Ligand-induced ADAM-type metalloprotease cleavage of Notch transmembrane receptors at the extracellular negative regulatory region (NRR) is essential to initiation of the regulated intramembrane proteolytic (RIP) cascade that is required for release of NICD by γ-secretase cleavage.83 If the ligand-induced Notch receptor conformational change is blocked, the NOTCH pathway remains inactivated. Inhibition of receptor cleavage by ADAM proteases65 or γ-secretase84 also abolishes NOTCH signalling.
Modification of receptor–ligand interaction
Blocking ligand ubiquitination and transendocytosis, which serve to activate ligands,85 and decreasing the availability of O-Fucose, which is required for normal NOTCH signalling,86 renders Notch transmembrane receptors unable to interact with ligands and to initiate signal transduction. Creating a soluble NOTCH decoy peptide composed of epidermal growth factor (EGF)-like repeats enables the ligand-binding region of Notch transmembrane receptors to be mimicked, preventing NOTCH pathway signalling by competitive inhibition.87 These approaches have not yet been examined in the setting of prostate cancer. Decoys can potentially abrogate NOTCH signalling through disruption of receptor–ligand interactions, with effects that depend on pharmacokinetics, biodistribution and decoy abundance. Decoys are soluble structures created to imitate the biological structure of extracellular protein receptors and their ligands. Decoys of DSL ligands88 and Jagged89 have produced promising results in establishing NOTCH inhibition, but the levels of decoys required to consistently impede receptor–ligand interactions have not yet been determined.
Antibody inhibition of NOTCH
Two classes of blocking monoclonal antibodies (mAbs) have been developed to inhibit NOTCH. The first class inhibits Notch receptors by maintaining Notch in an inactive conformation on binding to the NRR domain.90,91 The second class impedes ligand binding though competitive inhibition at the Notch EGF-repeat region.90 Other mAbs have also been shown to inhibit NOTCH signalling by targeting ligands, such as DLL4.92 These blocking antibodies have inhibited NOTCH signalling in endothelial cells.92
γ-Secretase inhibitors
Activation of NOTCH signalling is highly dependent upon γ-secretase initiating the final cleavage to release NICD,93 and so γ-secretase inhibitors are being investigated as potential anticancer agents. These compounds have shown promising results in various carcinomas,56 despite their off-target effects.
Post-transcriptional modification of NICD
Post-transcriptional modifications of NICD could be targets for therapeutic regulation of NOTCH activity. Although NICD is a short-lived protein with a half-life of approximately 4 hours,94,95 it can be modified by phosphorylation, ubiquitination, hydroxylation, glycosylation and acetylation.
Phosphorylation
Phosphorylation of NICD was first discovered in Drosophila,67 and phosphorylated NICD isoforms have also been reported in mammalian cells.95–98 Phosphorylation can either activate or deactivate NICD, depending on the number of cleavages the Notch transmembrane proteins experience, and the specific kinase protein that is involved. Several phosphorylated forms of NICD can exist within the nucleus. Glycogen synthase kinase-3β (GSKβ or Shaggy) is a serine/threonine kinase that increases the half-life of NICD but inhibits induction of nuclear genes, such as Hes-1.99 However, when granulocyte colony stimulating factor (G-CSF) phosphorylates NICD at the Ser2,078 residue, NICD is inactivated.100 Hyperphosphorylation has been identified within the nucleus and is stringently associated with NICD interaction with RBPJ proteins,98 NICD nuclear accumulation96,97,101 and NICD transformations.101
Ubiquitination
Despite the activation of NOTCH signalling by monoubiquitination,102 several cytoplasmic proteins promote NOTCH downregulation by inducing endocytosis followed by lysosomal degradation.103 Itch has an evolutionarily conserved role as a negative regulator of NOTCH.104 However, Deltex, a homologue of Itch, is a positive regulator of NOTCH signalling and binds to NICD. Therefore, the suppressor of Deltex, E3 ubiquitin–protein ligase Su(dx), is a negative regulator of NOTCH signalling.66,105–108 Within Su(dx) molecules, WW domains select target proteins for ubiquitination, and WW modules can also be found in the developmentally down-regulated NEDD-4 (E3 ubiquitin–protein ligase NEDD4) family proteins that are expressed in neural precursor cells.105 The loss of NEDD-4 function activates NOTCH and Deltex mutant phenotypes while NEDD-4 hyper-activation promotes degradation of Notch and Deltex.109 Other ubiquitin–protein ligases, such as c-Cbl (casitas B-lineage lymphoma), target receptors for endosomal and lysosomal sorting by facilitating receptor translocation from the cell surface to the degradation machinery.110,111 In comparison, Mam-1 (Mastermind-like protein 1) recruits CDK8 kinase for phosphorylation of NICD that is followed by E3 ubiquitin–protein ligase hAgo-dependent degradation of Notch.112,113 Furthermore, β-arrestin/Kurtz114 can also promote Notch inactivation by inducing proteasomal degradation, but has yet to be studied in the setting of prostate cancer.
Hydroxylation
Under hypoxic conditions, cells will adapt by initiating a cellular response involving the stabilization of HIF-1α (hypoxia-inducible factor 1α), activating transcription, increasing angiogenesis, increasing erythropoiesis and utilizing glycolytic metabolism.115,116 During this response, regulation of NOTCH signalling occurs when FIH-1 (factor inhibiting HIF-1) negatively regulates NICD by hydroxylating the Asn1,945 and Asn2,012 residues that are associated with Notch-mediated transcriptional activation, resulting in severe impairment of NICD function.116,117 NICD function can be restored by Mam-1.116 Preclinical studies involving mice and targeting FIH-1 have not yet resulted in significant overexpression of NOTCH phenotypes, suggesting a complex relationship exists between FIH-1 and NOTCH.118
Glycosylation
Glycosylation is essential for normal functioning of NOTCH transmembrane glycoproteins.119 O-fucosylation by O-FucT-1 (GDP-fucose protein O-fucosyltransferase 1) at NOTCH EGF domains facilitates ligand–receptor binding,119 and elongation of O-linked Fucose residues at Notch EGF-like sequences by both mFng (β-1,3-N-acetylglucosaminyltransferase manic fringe) and lFng (β-1,3-N-acetylglucosaminyltransferase lunatic fringe) impedes receptor–ligand binding.120 Specifically, mFng and lFng prevent Notch activation by Jagged ligands but leave Delta–Notch interactions unaffected.120,121 Fringe proteins can positively and negatively regulate the ability of ligands to activate the Notch receptor,122 so Fringe is able to inhibit activation of Notch by Serrate and Jagged ligands but stimulates Notch activation by Delta ligands.121 The diverse roles of Fringe in Notch activation and inactivation need to be further examined in the setting of prostate tumorigenesis.
Acetylation
Despite the ability of Mam-1 to recruit CDK8 for NICD phosphorylation, resulting in proteasomal degradation, the presence of Mam-1 in the nucleus is also highly correlated with decreased ubiquitination of Notch and increased Mam-1-dependent acetylation of Notch by p300 HAT (histone acetyltransferase p300).123 Acetylation by p300 HAT stabilizes NICD, whereas phosphorylation by CDK8 induces NICD degradation. Protein acetylation is reversible, and is regulated by histone deacetylases (HDACs), NAD-dependent protein deacetylases and histone acetyltransferases.124 Preclinical studies have reported that HDAC inhibitors that stabilize protein hyperacetylation have resulted in a significant decrease in both in vivo and in vitro growth of T-cell leukemia in NOTCH3 transgenic mice.124 By comparison, hSIRT1 in association with NICD, by functioning as an NICD deacetylase, promotes NICD destabilization and alters NICD protein turnover in endothelial cells lacking active SRT1 protein.125 Inhibition of NICD1 has been seen with both p300 and Tip60 HATs,125,126 but NICD3 inhibition is apparently p300 HAT-dependent.124 Prostate cancer therapy by acetylation-based regulation of NICD activity will depend on the relative involvement of specific NICD isoforms in prostate tumorigenesis.
Nuclear protein–protein interactions
Following nuclear translocation, NICD can interact with the Notch transcriptional activation complex (NTC), which is composed of DNA-bound transcription factor RBPJ and Mastermind-like family proteins, to begin targeting specific genes.127 Disruption of this NTC complex by blocking peptides can negatively regulate NOTCH signalling.128
Natural compounds
Several natural compounds have been investigated for the ability to inhibit NOTCH signalling, but their effectiveness is not yet confirmed. Genistein, a product of soybeans, can inhibit NOTCH signalling by inducing cellular apoptosis and significantly reducing cell viability in prostate cancer cells.129 Sulforaphane, a compound identified in broccoli, increases chemotherapeutic drug effects in both prostate cancer cells and pancreatic cancer stem cells.130 Other substances, such as resveratrol, a phytoalexin compound found in grapes and red wine,131 and curcumin, a food flavouring compound found in Curcuma longa,132 have all caused downregulation of NOTCH signalling pathways but have yet to be tested in prostate cancer.
The NUMB–NOTCH interaction
An inverse relationship between NUMB and NOTCH has been described: cells inheriting NUMB are usually deficient in NOTCH activity while cells inheriting NOTCH lack active NUMB.133–135 Although the precise mechanisms for Notch transmembrane receptor inhibition by NUMB remain unknown, several models have been proposed (Figure 2).
The endocytosis model
NOTCH signalling requires endosomal trafficking for activation, regulation and degradation of the signal.59,60 NUMB interacts with the AP-2 adaptor complex and the endocytic proteins α-adaptin and Eps15, in clathrin-coated pits and in endosomes.133 This ability of NUMB to localize in endocytic organelles, cotraffic with internalizing receptors and physically interact with endocytic machinery has raised the possibility of NOTCH inhibition by NUMB through endocytosis.
The postinternalization model
The NUMB–NOTCH interaction was proposed as a key regulator of asymmetric cell division in Drosophila peripheral neurogenesis, through regulation of Sanpodo.21 Sanpodo, a membrane-associated protein that interacts with both NUMB and Notch, is responsible for Notch transmembrane protein trafficking and endocytosis.31,136 A model was proposed suggesting that NUMB could inhibit AP-1-mediated recycling of Notch-Sanpodo complexes in pllb cells postinternalization.21,136 In Drosophila neural stem cells (NSCs), Numb/Notch signalling plays a key role in the balance between self-renewal and differentiation. While this model has been proposed in Drosophila, the transition between symmetric and asymmetric cell division is critical to the regulation of the cancer stem cell population.137 In colorectal cancer, NUMB was reported as a key factor in this switch.138 Examination of this NUMB–NOTCH interaction could have implications for (prostate) cancer stem cell biology.
The downstream inhibition model
Following NICD internalization, NUMB could inhibit NOTCH signal transduction by preventing NICD from initiating gene transcription. In the NOTCH signalling pathway, NICD interacts with RBPJ to form a complex in the nucleus, involving Mastermind transcriptional activation proteins, histone deacetylase transformations into histone acetyl transferases and chromatin modifications.58,63,66,139 Ubiquitin ligases also contribute to function of the complex.140,141 The RBPJ–NICD complex binds to DNA within the nucleus, activating gene transcription.142 With cotransfection of NOTCH and RBPJ, RBPJ localizes to the cytoplasm, and exposure to Delta ligands leads to RBPJ relocalization to the nucleus.143,144 Preliminary data collected by Frise et al.143 show that, with coexpression of NUMB and Notch, binding of Delta to Notch results in suppression of RBPJ translocation into the nucleus. NUMB can inhibit NOTCH signalling by binding directly to the NICD domain, preventing access of NICD to the nucleus.143–145 NUMB is also able to directly inhibit NOTCH by recruiting Itch for polyubiquitination and degradation of Notch proteins.146
Clinical implications
The clinical potential of inhibition of the NOTCH pathway is under increasing investigation. However, the intricate relationship between NOTCH and NUMB has been poorly reported. Determination of NUMB expression in patients with prostate cancer could be central to the identification of patients most likely to benefit from NOTCH inhibition. For instance, loss of NUMB has been associated with resistance to cisplatin in malignant pleural mesothelioma,147 and to docetaxel in breast cancer cells.148 Treatment with the γ-secretase inhibitor PF-03084014 resensitized cells to docetaxel.148 The two NUMB isoforms hNUMB5 and hNUMB6 are encoded by mRNAs that lack exon 10 and are expressed in cells known for polarity and migratory behavior, such as human amniotic fluid cells, glioblastoma and metastatic tumour cells. These isoforms seem to be less antagonistic to NOTCH signalling than other NUMB isoforms and could have a more prominent role in carcinogenesis.20 In patients diagnosed with non-small cell lung cancer, tumour-associated increases in NUMB exon 9 inclusion correlate with reduced levels of NUMB protein expression and activation of the NOTCH signalling pathway.149 Loss of NUMB is associated with tumour aggressiveness and BRCA1 status in patients with breast cancer,150 and poor prognosis in patients with salivary gland carcinomas.151
Conclusions
Identifying a molecular target with a significant role in cancer cell proliferation and survival is essential for the development of a more specific, more effective and potentially less toxic approach to treating cancer. As both NOTCH and NUMB have evolutionarily conserved roles in cell-fate determination and tumour angiogenesis, their biological pathway is currently being examined as a potential target for anticancer therapeutics. NUMB is able to control various oncogenic signalling pathways, including the p53, Hedgehog and NOTCH pathways. Although the precise mechanism of NOTCH inhibition by NUMB is not yet known, the complexity of the isoforms and functions of NUMB enable it to encompass a variety of functions within different signalling pathways. It might be possible to selectively target the involvement of NUMB in prostate cancer, and to identify patients most likely to benefit from NOTCH inhibition. However, NUMB is poorly characterized in the setting of prostate cancer despite being firmly established as a tumour suppressor in various other carcinomas.152 The role of NUMB in prostate cancer, and specifically its role in the regulation of NOTCH signalling, should be examined as an issue of the utmost importance.
Key points.
-
▪
NUMB is a complex protein with multiple cellular functions
-
▪
NUMB negatively regulates the NOTCH signalling pathway
-
▪
The NUMB–NOTCH interaction regulates cell fate in prostate tumours
-
▪
Targeting NUMB has potential to control prostate tumorigenesis
-
▪
NUMB profiling could assist the identification of patients with prostate cancer who are likely to benefit from NOTCH-inhibition strategies
Review criteria.
The PubMed database was searched using combinations of the search terms “NOTCH”, “NUMB”, “prostate neoplasm” and “inhibition”. Peer-reviewed English-language papers were considered for inclusion in the manuscript. The reference lists of identified publications were searched for additional articles.
Acknowledgments
We would like to acknowledge support from the Irish Cancer Society (grant code PCA12MAR). Victoria Anastasia Belle is a Mount Sinai International Exchange Program minority student participant. Her work was supported in part by grant MD001452 from the National Center on Minority Health and Health Disparities of the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
V.A.B. and L.M. researched the data for the article, discussed the content and wrote the article. All authors contributed to review and editing of the manuscript before submission.
References
- 1.Jemal A, et al. Cancer statistics, 2007. CA Cancer. J. Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organisation; 2014. Obesity and Overweight, Fact Sheet No. 311. [online], http://www.who.int/mediacentre/factsheets/fs311/en/ [Google Scholar]
- 3.Allott EH, Masko EM, Freedland SJ. Obesity and prostate cancer: weighing the evidence. Eur. Urol. 2013;63:800–809. doi: 10.1016/j.eururo.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann M, Krause M, Thames H, Trott K, Zips D. Cancer stem cells and radiotherapy. Int. J. Radiat. Biol. 2009;85:391–402. doi: 10.1080/09553000902836404. [DOI] [PubMed] [Google Scholar]
- 5.Kioi M, et al. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J. Clin. Invest. 2010;120:694–705. doi: 10.1172/JCI40283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danza G, et al. Notch signaling modulates hypoxia-induced neuroendocrine differentiation of human prostate cancer cells. Mol. Cancer. Res. 2012;10:230–238. doi: 10.1158/1541-7786.MCR-11-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu SD, Liu FY, Wang QR. Notch inhibitor: a promising carcinoma radiosensitizer. Asian. Pac. J. Cancer Prev. 2012;13:5345–5351. doi: 10.7314/apjcp.2012.13.11.5345. [DOI] [PubMed] [Google Scholar]
- 8.Dho SE, French MB, Woods SA, McGlade CJ. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 1999;274:33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 9.Kelsom C, Lu W. Uncovering the link between malfunctions in Drosophila neuroblast asymmetric cell division and tumorigenesis. Cell Biosci. 2012;2:38. doi: 10.1186/2045-3701-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 11.Pece S, Confalonieri S, R Romano P, Di Fiore PP. NUMB-ing down cancer by more than just a NOTCH. Biochim. Biophys. Acta. 2011;1815:26–43. doi: 10.1016/j.bbcan.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 13.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 14.Pece S, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Marcotullio L, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat. Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 16.Greer RL, Staley BK, Liou A, Hebrok M. Numb regulates acinar cell dedifferentiation and survival during pancreatic damage and acinar to ductal metaplasia. Gastroenterology. 2013;145:1088–1097. doi: 10.1053/j.gastro.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salcini AE, et al. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdi JM, et al. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 1996;6:1134–1145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- 19.Dho SE, Trejo J, Siderovski DP, McGlade CJ. Dynamic regulation of mammalian numb by G protein-coupled receptors and protein kinase C activation: Structural determinants of numb association with the cortical membrane. Mol. Biol. Cell. 2006;17:4142–4155. doi: 10.1091/mbc.E06-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karaczyn A, et al. Two novel human NUMB isoforms provide a potential link between development and cancer. Neural Dev. 2010;5:31. doi: 10.1186/1749-8104-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichardt I, Knoblich JA. Cell biology: Notch recycling is numbed. Curr. Biol. 2013;23:R270–R272. doi: 10.1016/j.cub.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y, Abagyan R. How and why phosphotyrosine-containing peptides bind to the SH2 and PTB domains. Fold. Des. 1998;3:513–522. doi: 10.1016/S1359-0278(98)00067-4. [DOI] [PubMed] [Google Scholar]
- 23.Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Exp. Cell Res. 2010;316:900–906. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Ravichandran KS, et al. Evidence for a requirement for both phospholipid and phosphotyrosine binding via the Shc phosphotyrosine-binding domain in vivo. Mol. Cell. Biol. 1997;17:5540–5549. doi: 10.1128/mcb.17.9.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giebel B, Wodarz A. Notch signaling: numb makes the difference. Curr. Biol. 2012;22:R133–R135. doi: 10.1016/j.cub.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Carter S, Vousden KH. A role for Numb in p53 stabilization. Genome Biol. 2008;9:221. doi: 10.1186/gb-2008-9-5-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colaluca IN, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 28.Dhami GK, et al. Dynamic methylation of Numb by Set8 regulates its binding to p53 and apoptosis. Mol. Cell. 2013;50:565–576. doi: 10.1016/j.molcel.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 29.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 30.Cappello F, et al. The value of immunohistochemical research on PCNA, p53 and heat shock proteins in prostate cancer management: a review. Eur. J. Histochem. 2006;50:25–34. [PubMed] [Google Scholar]
- 31.Couturier L, Mazouni K, Schweisguth F. Numb localizes at endosomes and controls the endosomal sorting of notch after asymmetric division in Drosophila. Curr. Biol. 2013;23:588–593. doi: 10.1016/j.cub.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev. Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karhadkar SS, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 34.Axelson M, et al. U. S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin. Cancer Res. 2013;19:2289–2293. doi: 10.1158/1078-0432.CCR-12-1956. [DOI] [PubMed] [Google Scholar]
- 35.Drenkhahn SK, Jackson GA, Slusarz A, Starkey NJ, Lubahn DB. Inhibition of hedgehog/Gli signaling by botanicals: a review of compounds with potential hedgehog pathway inhibitory activities. Curr. Cancer Drug Targets. 2013;13:580–595. doi: 10.2174/15680096113139990003. [DOI] [PubMed] [Google Scholar]
- 36.Sandhiya S, Melvin G, Kumar SS, Dkhar SA. The dawn of hedgehog inhibitors: Vismodegib. J. Pharmacol. Pharmacother. 2013;4:4–7. doi: 10.4103/0976-500X.107628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Marcotullio L, et al. Multiple ubiquitin-dependent processing pathways regulate hedgehog/gli signaling: implications for cell development and tumorigenesis. Cell Cycle. 2007;6:390–393. doi: 10.4161/cc.6.4.3809. [DOI] [PubMed] [Google Scholar]
- 38.Nanta R, et al. NVP LDE-225 (Erismodegib) inhibits epithelial-mesenchymal transition and human prostate cancer stem cell growth in NOD/SCID IL2Rgamma null mice by regulating Bmi-1 and microRNA-128. Oncogenesis. 2013;2:e42. doi: 10.1038/oncsis.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Marcotullio L, et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene. 2011;30:65–76. doi: 10.1038/onc.2010.394. [DOI] [PubMed] [Google Scholar]
- 40.Chen M, Carkner R, Buttyan R. The hedgehog/Gli signaling paradigm in prostate cancer. Expert Rev. Endocrinol. Metab. 2011;6:453–467. doi: 10.1586/EEM.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirab N, et al. Androgens regulate Hedgehog signalling and proliferation in androgen-dependent prostate cells. Int. J. Cancer. 2012;131:1297–1306. doi: 10.1002/ijc.27384. [DOI] [PubMed] [Google Scholar]
- 42.Nadendla SK, et al. GLI1 confers profound phenotypic changes upon LNCaP prostate cancer cells that include the acquisition of a hormone independent state. PLoS ONE. 2011;6:e20271. doi: 10.1371/journal.pone.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen G, et al. GLI1, a crucial mediator of sonic hedgehog signaling in prostate cancer, functions as a negative modulator for androgen receptor. Biochem. Biophys. Res. Commun. 2011;404:809–815. doi: 10.1016/j.bbrc.2010.12.065. [DOI] [PubMed] [Google Scholar]
- 44.Chen M, et al. Androgenic regulation of hedgehog signaling pathway components in prostate cancer cells. Cell Cycle. 2009;8:149–157. doi: 10.4161/cc.8.1.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawahara H, et al. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imai T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toda M, et al. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 48.Nie J, et al. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002;21:93–102. doi: 10.1093/emboj/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Susini L, et al. Siah-1 binds and regulates the function of Numb. Proc. Natl Acad. Sci. USA. 2001;98:15067–15072. doi: 10.1073/pnas.261571998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuang W, et al. Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochem. Biophys. Res. Commun. 2009;378:259–263. doi: 10.1016/j.bbrc.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 51.Xu B, et al. A functional polymorphism in Pre miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate. 2010;70:467–472. doi: 10.1002/pros.21080. [DOI] [PubMed] [Google Scholar]
- 52.Xu B, et al. MiR-146a suppresses tumor growth and progression by targeting EGFR pathway and in a p ERK dependent manner in castration-resistant prostate cancer. Prostate. 2012;72:1171–1178. doi: 10.1002/pros.22466. [DOI] [PubMed] [Google Scholar]
- 53.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Groth C, Fortini ME. Therapeutic approaches to modulating Notch signaling: current challenges and future prospects. Semin. Cell Dev. Biol. 2012;23:465–472. doi: 10.1016/j.semcdb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 56.Espinoza I, Miele L. Notch inhibitors for cancer treatment. Pharmacol. Ther. 2013;139:95–110. doi: 10.1016/j.pharmthera.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marignol L, Rivera-Figueroa K, Lynch T, Hollywood D. Hypoxia, notch signalling, and prostate cancer. Nat. Rev. Urol. 2013;10:405–413. doi: 10.1038/nrurol.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- 59.Barth JM, Kohler K. How to take autophagy and endocytosis up a notch. Biomed. Res. Int. 2014;2014:960803. doi: 10.1155/2014/960803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto S, Charng WL, Bellen HJ. Endocytosis and intracellular trafficking of Notch and its ligands. Curr. Top. Dev. Biol. 2010;92:165–200. doi: 10.1016/S0070-2153(10)92005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 62.McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 2009;284:26427–26438. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brou C. Intracellular trafficking of Notch receptors and ligands. Exp. Cell Res. 2009;315:1549–1555. doi: 10.1016/j.yexcr.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 64.Schweisguth F. Regulation of notch signaling activity. Curr. Biol. 2004;14:R129–R138. [PubMed] [Google Scholar]
- 65.Brou C, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 66.Wilkin MB, Baron M. Endocytic regulation of Notch activation and down-regulation (review) Mol. Membr. Biol. 2005;22:279–289. doi: 10.1080/09687860500129778. [DOI] [PubMed] [Google Scholar]
- 67.Kidd S, Lieber T, Young MW. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 69.Lieber T, Kidd S, Young MW. Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma secretase like proteolytic activation of Notch1. Mol. Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 71.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 72.Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- 73.Leong KG, Gao WQ. The Notch pathway in prostate development and cancer. Differentiation. 2008;76:699–716. doi: 10.1111/j.1432-0436.2008.00288.x. [DOI] [PubMed] [Google Scholar]
- 74.Wang XD, et al. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev. Biol. 2006;290:66–80. doi: 10.1016/j.ydbio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Villaronga MA, Bevan CL, Belandia B. Notch signaling: a potential therapeutic target in prostate cancer. Curr. Cancer Drug Targets. 2008;8:566–580. doi: 10.2174/156800908786241096. [DOI] [PubMed] [Google Scholar]
- 76.Kong D, et al. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR DIM treatment. Am. J. Transl. Res. 2012;4:14–23. [PMC free article] [PubMed] [Google Scholar]
- 77.Domingo-Domenech J, et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell. 2012;22:373–388. doi: 10.1016/j.ccr.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sharad S, et al. Prostate cancer gene expression signature of patients with high body mass index. Prostate Cancer Prostatic Dis. 2011;14:22–29. doi: 10.1038/pcan.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson LM, Gibbons GH. Notch: a mastermind of vascular morphogenesis. J. Clin. Invest. 2007;117:299–302. doi: 10.1172/JCI31288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gridley T. Notch signaling in vascular development and physiology. Development. 2007;134:2709–2718. doi: 10.1242/dev.004184. [DOI] [PubMed] [Google Scholar]
- 81.Pallares J, et al. Study of microvessel density and the expression of the angiogenic factors VEGF, bFGF and the receptors Flt-1 and FLK-1 in benign, premalignant and malignant prostate tissues. Histol. Histopathol. 2006;21:857–865. doi: 10.14670/HH-21.857. [DOI] [PubMed] [Google Scholar]
- 82.Bostwick DG, Iczkowski KA. Microvessel density in prostate cancer: prognostic and therapeutic utility. Semin. Urol. Oncol. 1998;16:118–123. [PubMed] [Google Scholar]
- 83.Gordon WR, et al. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 84.Kopan R, Ilagan MX. Gamma-secretase: proteasome of the membrane? Nat. Rev. Mol. Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 85.Pitsouli C, Delidakis C. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development. 2005;132:4041–4050. doi: 10.1242/dev.01979. [DOI] [PubMed] [Google Scholar]
- 86.Okajima T, Irvine KD. Regulation of notch signaling by o linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 87.Nickoloff BJ, et al. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF kappaB and PPARgamma. Cell Death Differ. 2002;9:842–855. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- 88.Varnum-Finney B, et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J. Cell Sci. 2000;113:4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 89.Small D, et al. Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src-dependent chord-like phenotype. J. Biol. Chem. 2001;276:32022–32030. doi: 10.1074/jbc.M100933200. [DOI] [PubMed] [Google Scholar]
- 90.Aste-Amézaga M, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS ONE. 2010;5:e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li K, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J. Biol. Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- 92.Ridgway J, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 93.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu. Rev. Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 95.Tremblay I, Paré E, Arsenault D, Douziech M, Boucher MJ. The MEK/ERK pathway promotes NOTCH signalling in pancreatic cancer cells. PLoS ONE. 2013;8:e85502. doi: 10.1371/journal.pone.0085502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat. Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- 97.Shimizu K, et al. Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2. Mol. Cell. Biol. 2000;20:6913–6922. doi: 10.1128/mcb.20.18.6913-6922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Foltz DR, Nye JS. Hyperphosphorylation and association with RBP of the intracellular domain of Notch1. Biochem. Biophys. Res. Commun. 2001;286:484–492. doi: 10.1006/bbrc.2001.5421. [DOI] [PubMed] [Google Scholar]
- 99.Foltz DR, Santiago MC, Berechid BE, Nye JS. Glycogen synthase kinase-3beta modulates notch signaling and stability. Curr. Biol. 2002;12:1006–1011. doi: 10.1016/s0960-9822(02)00888-6. [DOI] [PubMed] [Google Scholar]
- 100.Inglés-Esteve J, Espinosa L, Milner LA, Caelles C, Bigas A. Phosphorylation of Ser2078 modulates the Notch2 function in 32D cell differentiation. J. Biol. Chem. 2001;276:44873–44880. doi: 10.1074/jbc.M104703200. [DOI] [PubMed] [Google Scholar]
- 101.Ronchini C, Capobianco AJ. Notch(ic)-ER chimeras display hormone-dependent transformation, nuclear accumulation, phosphorylation and CBF1 activation. Oncogene. 2000;19:3914–3924. doi: 10.1038/sj.onc.1203719. [DOI] [PubMed] [Google Scholar]
- 102.Gupta-Rossi N, et al. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J. Cell Biol. 2004;166:73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nichols JT, Miyamoto A, Weinmaster G. Notch signaling—constantly on the move. Traffic. 2007;8:959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- 104.Qiu L, et al. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 105.Cornell M, et al. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics. 1999;152:567–576. doi: 10.1093/genetics/152.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fostier M, Evans DA, Artavanis-Tsakonas S, Baron M. Genetic characterization of the Drosophila melanogaster Suppressor of deltex gene: A regulator of notch signaling. Genetics. 1998;150:1477–1485. doi: 10.1093/genetics/150.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matsuno K, et al. Human deltex is a conserved regulator of Notch signalling. Nat. Genet. 1998;19:74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- 108.Yong T, Sun A, Henry MD, Meyers S, Davis JN. Down regulation of CSL activity inhibits cell proliferation in prostate and breast cancer cells. J. Cell. Biochem. 2011;112:2340–2351. doi: 10.1002/jcb.23157. [DOI] [PubMed] [Google Scholar]
- 109.Sakata T, et al. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr. Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 110.Jehn BM, Dittert I, Beyer S, von der Mark K, Bielke W. c Cbl binding and ubiquitin-dependent lysosomal degradation of membrane associated Notch1. J. Biol. Chem. 2002;277:8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- 111.Levkowitz G, et al. c Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oberg C, et al. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 113.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 114.Mukherjee A, et al. Regulation of Notch signalling by non-visual beta-arrestin. Nat. Cell Biol. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- 115.Gustafsson MV, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 116.Zheng X, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc. Natl Acad. Sci. USA. 2008;105:3368–3373. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Coleman ML, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J. Biol. Chem. 2007;282:24027–24038. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- 118.Zhang N, et al. The asparaginyl hydroxylase factor inhibiting HIF-1alpha is an essential regulator of metabolism. Cell Metab. 2010;11:364–378. doi: 10.1016/j.cmet.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O fucosyltransferase 1 and fringe. J. Biol. Chem. 2003;278:42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- 120.Shimizu K, et al. Manic fringe and lunatic fringe modify different sites of the Notch2 extracellular region, resulting in different signaling modulation. J. Biol. Chem. 2001;276:25753–25758. doi: 10.1074/jbc.M103473200. [DOI] [PubMed] [Google Scholar]
- 121.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 122.Moloney DJ, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 123.Popko-Scibor AE, Lindberg MJ, Hansson ML, Holmlund T, Wallberg AE. Ubiquitination of Notch1 is regulated by MAML1 mediated p300 acetylation of Notch1. Biochem. Biophys. Res. Commun. 2011;416:300–306. doi: 10.1016/j.bbrc.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 124.Palermo R, et al. Acetylation controls Notch3 stability and function in T cell leukemia. Oncogene. 2012;31:3807–3817. doi: 10.1038/onc.2011.533. [DOI] [PubMed] [Google Scholar]
- 125.Guarani V, et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473:234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim MY, et al. Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol. Cell. Biol. 2007;27:6506–6519. doi: 10.1128/MCB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc. Natl Acad. Sci. USA. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jung J, et al. Regulation of Notch1 signaling by Delta-like ligand 1 intracellular domain through physical interaction. Mol. Cells. 2011;32:161–165. doi: 10.1007/s10059-011-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Z, et al. Down-regulation of Notch-1 is associated with Akt and FoxM1 in inducing cell growth inhibition and apoptosis in prostate cancer cells. J. Cell. Biochem. 2011;112:78–88. doi: 10.1002/jcb.22770. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130.Kallifatidis G, et al. Sulforaphane increases drug-mediated cytotoxicity toward cancer stem-like cells of pancreas and prostate. Mol. Ther. 2011;19:188–195. doi: 10.1038/mt.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cecchinato V, et al. Resveratrol-induced apoptosis in human T cell acute lymphoblastic leukaemia MOLT-4 cells. Biochem. Pharmacol. 2007;74:1568–1574. doi: 10.1016/j.bcp.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 132.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006;106:2503–2513. doi: 10.1002/cncr.21904. [DOI] [PubMed] [Google Scholar]
- 133.Santolini E, et al. Numb is an endocytic protein. J. Cell Biol. 2000;151:1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 135.Rebay I, Fehon RG, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 136.Couturier L, Mazouni K, Schweisguth F. Inhibition of Notch recycling by Numb: relevance and mechanism(s) Cell Cycle. 2013;12:1647–1648. doi: 10.4161/cc.24983. [DOI] [PubMed] [Google Scholar]
- 137.Caussinus E, Hirth F. Asymmetric stem cell division in development and cancer. Prog. Mol. Subcell. Biol. 2007;45:205–225. doi: 10.1007/978-3-540-69161-7_9. [DOI] [PubMed] [Google Scholar]
- 138.Lerner RG, Petritsch C. A microRNA-operated switch of asymmetric to symmetric cancer stem cell divisions. Nat. Cell Biol. 2014;16:212–214. doi: 10.1038/ncb2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lai EC. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3:840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baron M, et al. Multiple levels of Notch signal regulation (review) Mol. Membr. Biol. 2002;19:27–38. doi: 10.1080/09687680110112929. [DOI] [PubMed] [Google Scholar]
- 141.Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jarriault S, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 143.Frise E, Knoblich JA, Younger-Shepherd S, Jan LY, Jan YN. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl Acad. Sci. USA. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gho M, Lecourtois M, Géraud G, Posakony JW, Schweisguth F. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development. 1996;122:1673–1682. doi: 10.1242/dev.122.6.1673. [DOI] [PubMed] [Google Scholar]
- 145.Berdnik D, Török T, González-Gaitán M, Knoblich JA. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- 146.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 147.Kang Y, et al. Overexpression of Numb suppresses tumor cell growth and enhances sensitivity to cisplatin in epithelioid malignant pleural mesothelioma. Oncol. Rep. 2013;30:313–319. doi: 10.3892/or.2013.2429. [DOI] [PubMed] [Google Scholar]
- 148.Zhang CC, et al. Synergistic effect of the gamma-secretase inhibitor PF-03084014 and docetaxel in breast cancer models. Stem Cells Transl. Med. 2013;2:233–242. doi: 10.5966/sctm.2012-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Misquitta-Ali CM, et al. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol. Cell. Biol. 2011;31:138–150. doi: 10.1128/MCB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rennstam K, et al. Numb protein expression correlates with a basal-like phenotype and cancer stem cell markers in primary breast cancer. Breast Cancer Res. Treat. 2010;122:315–324. doi: 10.1007/s10549-009-0568-x. [DOI] [PubMed] [Google Scholar]
- 151.Maiorano E, et al. Prognostic implications of NUMB immunoreactivity in salivary gland carcinomas. Int. J. Immunopathol. Pharmacol. 2007;20:779–789. doi: 10.1177/039463200702000414. [DOI] [PubMed] [Google Scholar]
- 152.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]