Abstract Abstract
Wrasses (Labridae) are extremely diversified marine fishes, whose species exhibit complex interactions with the reef environment. They are widely distributed in the Indian, Pacific and Atlantic oceans. Their species have displayed a number of karyotypic divergent processes, including chromosomal regions with complex structural organization. Current cytogenetic information for this family is phylogenetically and geographically limited and mainly based on conventional cytogenetic techniques. Here, the distribution patterns of heterochromatin, GC-specific chromosome regions and Ag-NORs, and the organization of 18S and 5S rDNA sites of the Atlantic species Thalassoma noronhanum (Boulenger, 1890), Halichoeres poeyi (Steindachner, 1867), Halichoeres radiatus (Linnaeus, 1758), Halichoeres brasiliensis (Bloch, 1791) and Halichoeres penrosei Starks, 1913, belonging to the tribe Julidini were analyzed. All the species exhibited 2n=48 chromosomes with variation in the number of chromosome arms among genera. Thalassoma noronhanum has 2m+46a, while species of the genus Halichoeres Rüppell, 1835 share karyotypes with 48 acrocentric chromosomes. The Halichoeres species exhibit differences in the heterochromatin distribution patterns and in the number and distribution of 18S and 5S rDNA sites. The occurrence of 18S/5S rDNA syntenic arrangements in all the species indicates a functionally stable and adaptive genomic organization. The phylogenetic sharing of this rDNA organization highlights a marked and unusual chromosomal singularity inside the family Labridae.
Keywords: Chromosome evolution, Halichoeres, rDNA, syntenic genes, wrasses
Introduction
Wrasses (Labridae) are one of the most abundant and ecologically diversified fish groups in tropical reefs (Choat and Bellwood 1998). Their biodiversity is highlighted by nine tribes with 82 genera and 530 species (Westneat and Afaro 2005, Eschmeyer and Fong 2016), which exhibit extensive biological interactions in tropical reefs and temperate regions around the world (Choat and Bellwood 1998, Wainwright et al. 2004, Nelson 2006).
Cytogenetic analyses in Labridae have revealed particular trends in the karyotypic evolution of their clades (Sena and Molina 2007). In fact, pericentric inversions stand out as the major chromosomal rearrangements in the evolution of the tribes Hypsigenyini, Scarini and Julidini (Sena and Molina 2007, Molina et al. 2012). In turn, in the Novaculini, Cheilini, Pseudocheilini and Labrini tribes, both pericentric inversions and chromosome fusions have contributed for their karyotypic diversification (Ueno and Takai 2000).
In general, Labridae clades can be differentiated into four karyotypic patterns. The first one is characterized by conserved karyotypes, with 48 acrocentric chromosomes; the second by 48 chromosomes with an increase in the chromosome arms (NF); the third by a reduction in the number of chromosomes (<48 chromosomes) but with the same NF; and the fourth by reduced diploid number and NF (Alvarez et al. 1986, Sena and Molina 2007).
At the moment, Julidini is the clade with the largest amount of cytogenetic data in Labridae (Table 1). Nonetheless, they are based on conventional cytogenetic methods and very incipient yet, given its species’ diversity. This tribe falls mainly into the first and second karyotypic patterns, with conserved diploid values (2n=48), mostly acrocentric chromosomes, or with variations in the NF due to pericentric inversions. Different classes of repetitive DNAs are linked to chromosome rearrangements in many fish groups (Kidwell 2002, Cioffi and Bertollo 2012, Getlekha et al. 2016). Indeed, repetitive DNAs may clarify the occurrence of particular chromosome rearrangements and evolutionary relationships among different taxa (Shapiro and Sternberg 2005, Biémont and Vieira 2006, Artoni et al. 2015). However, the chromosome organization and the evolutionary dynamics of this important fraction of the genome are still poorly understood in Labridae fishes.
Table 1.
Variations in diploid values (2n) and number of chromosome arms (NF) among Labridae fishes (adapted from Sena and Molina 2007, Arai 2011).
| Tribe | N | 2n range/ Modal value | NF range/ Modal value | NF Average |
|---|---|---|---|---|
| Hypsigenyini | 7 | 48/48 | 56–86/78 | 76 |
| Pseudocheilini | 8 | 34–48/34 | 46–84/46 | 65 |
| Julidini | 32 | 48/48 | 48–86/48 | 52 |
| Labrini | 10 | 38–48/48 | 48–86/48 | 46 |
| Scarini | 5 | 46–48/48 | 66–88/66 | 74 |
| Cheilini | 8 | 32–48/48 | 38–84/60 | 66 |
| Labrichthyines | 1 | 48 | 48 | 48 |
| Novaculini | 8 | 22–48/48 | 40–56/48 | 47 |
| Pseudolabrini | 1 | 48 | 52 | 52 |
Among Julidini wrasses, Halichoeres Rüppell, 1835 is the most diversified and polyphyletic genus, comprising distinct components in the New World and Indo–Pacific Ocean (Barber and Bellwood 2005, Westneat and Alfaro 2005, Rocha et al. 2010). Thalassoma Swainson, 1839, phylogenetically close to Halichoeres, dates from 8–13 mya and contains 27 species, with a marked increase in diversification between 5–10 mya (Bernardi et al. 2004). Thalassoma noronhanum (Boulenger, 1890) is one of the smallest known species (Allen 1995), with a wide occurrence on the Brazilian coast and a number of oceanic islands in the Western Atlantic. Despite some cytogenetic data available for Halichoeres species (Sena and Molina 2007), there are no information for Thalassoma ones from the Atlantic (Arai 2011). In the present study, cytogenetic investigation on C-banding, Ag-NORs, base-specific fluorochrome staining and double-fluorescence in situ hybridization (FISH) with 18S rDNA and 5S rDNA probes, were realized in five Julidini species. The data were useful to clarify particular chromosomal processes and phylogenetic relationships of these marine fish species, besides evidencing an unusual co-localization of 18S and 5S rDNA clusters in all species.
Material and methods
Specimens and chromosomal preparation
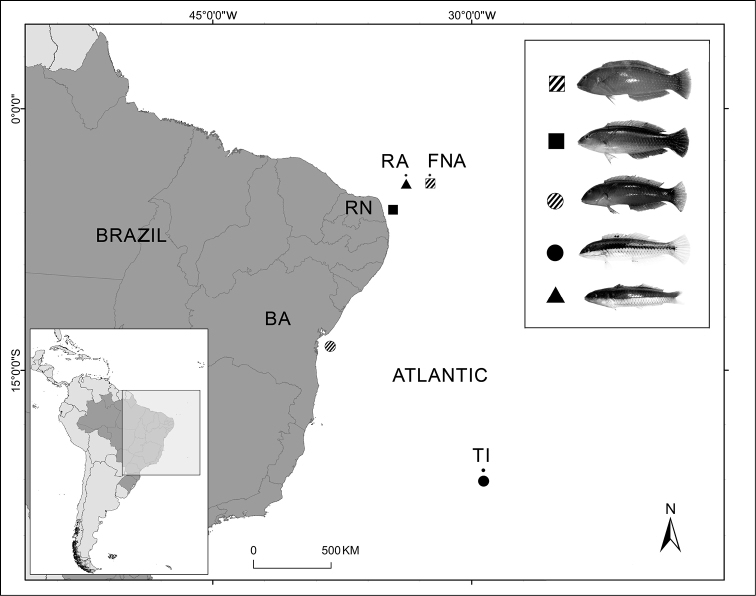
The specimens of Halichoeres poeyi (Steindachner, 1867) (N=13) and Halichoeres brasiliensis (Bloch, 1791) (N=6) were collected in the coast of Rio Grande do Norte (5°42'20"S, 35°11'38"W), Northeastern Brazil. Individuals of Halichoeres radiatus (Linnaeus, 1758) (N=16) were obtained from the Fernando de Noronha Archipelago (3°51'20"S, 32°25'32"W), Halichoeres penrosei Starks, 1913 (N=3) from the Trindade Island (20°30'13"S, 29°19'50"W) and Thalassoma noronhanum from the Rocas Atoll (N=22) (3°51'59"S, 33°48'20"W).
The specimens were submitted to intraperitoneal mitotic stimulation with fungal and bacterial antigen complexes (Molina et al. 2010). Mitotic chromosome preparations were obtained by in vitro methodology, using a cell suspension of kidney tissue fragments (Gold et al. 1990). The C-positive heterochromatin and (NORs) were visualized using the C-banding and Ag-NOR staining (Sumner 1972, Howell and Black 1980, respectively). Chromosomes were also stained with (MM) and (DAPI) fluorochromes, according to Schweizer (1976).
Obtaining probes for chromosomal hybridization
The 5S and 18S rDNA probes, containing approximately 200 pb and 1400 pb, respectively, were obtained by (PCR) from the nuclear DNA of Rachycentron canadum, using the primers A 5’-TAC GCC CGA TCT CGT CCG ATC-3’ and B 5’- CAG GCT GGT ATG GCC GTA AGC-3’ (Pendás et al. 1994), NS1 5’-GTA GTC ATA TGC TTG TCT C-3’ and NS8 5’-TCC GGT GCA TCA CCT ACG GA-3’ (White et al. 1990), respectively. The 18S rDNA and 5S rDNA probes were labeled with digoxigenin-11-dUTP (Roche, Mannheim, Germany) and biotin-14-dATP (InvitrogenTM), respectively, according to manufacturer’s specifications.
Chromosomal hybridization
(FISH) was performed according to Pinkel et al. (1986). Slides with metaphase chromosomes were first treated with RNAse (20 µg/ml in 2XSSC) at 37°C for 1 hour and with pepsin (0.005% in 10mM HCl), for 10 minutes, fixed with 1% formaldehyde for 10 minutes and dehydrated in alcohol baths (70%/85%/100%) for 5 minutes each. The chromosomes were then incubated in 70% formamide/2XSSC at 72°C, for 5 minutes and once again dehydrated in an alcohol series (70%/85%/100%). The hybridization was performed at 37°C for 16h, using a hybridization solution consisting of 50% formamide, 2XSSC, 10% dextran sulfate and the denatured probe (5 ng/µl), with a final volume of 30 µl. Post-hybridization washings were done in 15% formamide/0.2XSSC at 42°C, for 20 minutes, followed by washings in 0.1XSSC at 60°C for 15 minutes and in Tween 20 (0.5%/4XSSC) for 5 minutes, at 25°C. Next, the slides were incubated for 15 minutes in a blocking solution (5% NFDM /4xSSC) and washed with Tween 20 (0.5%/4XSSC) for 15 minutes. The hybridization signals were detected using anti-digoxigenin rhodamine (Roche, Mannheim, Germany) for the 18S rDNA probe and streptavidin-FITC (Vector Laboratories) for the 5S rDNA probe. The chromosomes were counterstained with Vectashield/DAPI (1.5 µg/ml) (Vector).
At least thirty metaphase spreads were analyzed to confirm the diploid chromosome numbers, karyotype structure and FISH results. The best metaphases were photographed using an OlympusTM BX50 epifluorescence microscope, coupled to an Olympus DP73 digital capture system. The chromosomes were classified as (sm) and (a), according to the arm ratio (Levan et al. 1964), and arranged in decreasing order of size in the karyotypes.
Results
All species showed a high number of acrocentric chromosomes. Thalassoma noronhanum has 2n=48, with 2sm+46a (NF=50) (Fig. 2a). The species Halichoeres radiatus, Halichoeres poeyi, Halichoeres brasiliensis and Halichoeres penrosei have symmetric karyotypes, with 2n=48 acrocentric chromosomes (NF=48) (Fig. 2b-e). The heterochromatin occupies the centromeric and pericentromeric regions of all chromosomes, and also the telomeric regions of a few (Fig. 2a–e).
Figure 2.
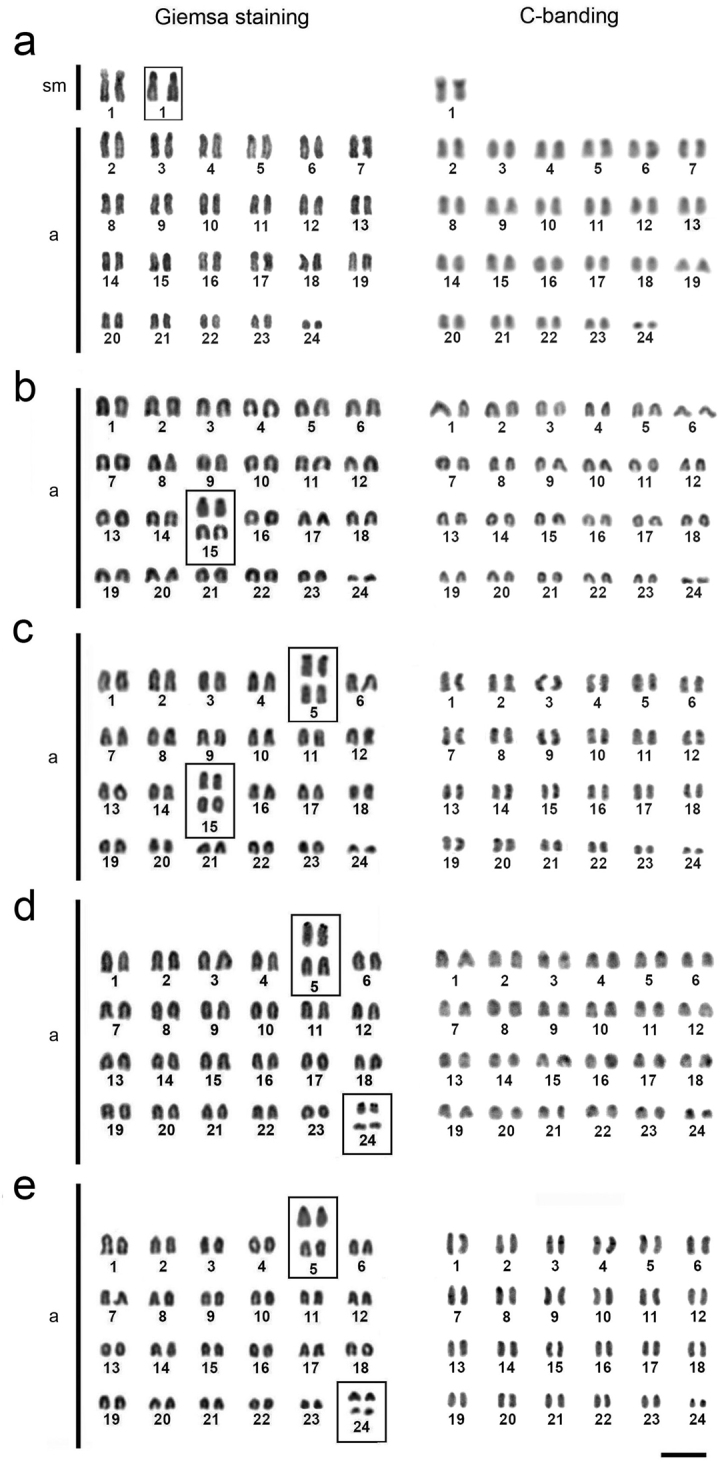
Karyotypes of Thalassoma noronhanum (a), Halichoeres penrosei (b), Halichoeres poeyi (c), Halichoeres radiatus (d), and Halichoeres brasiliensis (e). The chromosomal pairs bearing Ag-NORs are boxed, the silver staining in the upper row. Bar: 5 μm.
Figure 1.
Collection points of the Labridae species analyzed. (FNA); (RA); (RN); (BA); and (TI). Halichoeres radiatus (FNA), Halichoeres brasiliensis (RN), Halichoeres poeyi (BA), Halichoeres penrosei (TI) and Thalassoma noronhanum (RA).
The Ag-NORs are positioned on the short arms of the single submetacentric pair of Thalassoma noronhanum (Fig. 2a). In Halichoeres species, these sites are located in two chromosome pairs, except in Halichoeres penrosei, where they are located on the short arms of pair 15 (Fig. 2b, highlighted). In Halichoeres poeyi, the Ag-NORs occupy the short arms of pairs 5 and 15 (Fig. 2c) and in Halichoeres radiatus and Halichoeres brasiliensis the short arms of pairs 5 and 24 (Fig. 2d, e).
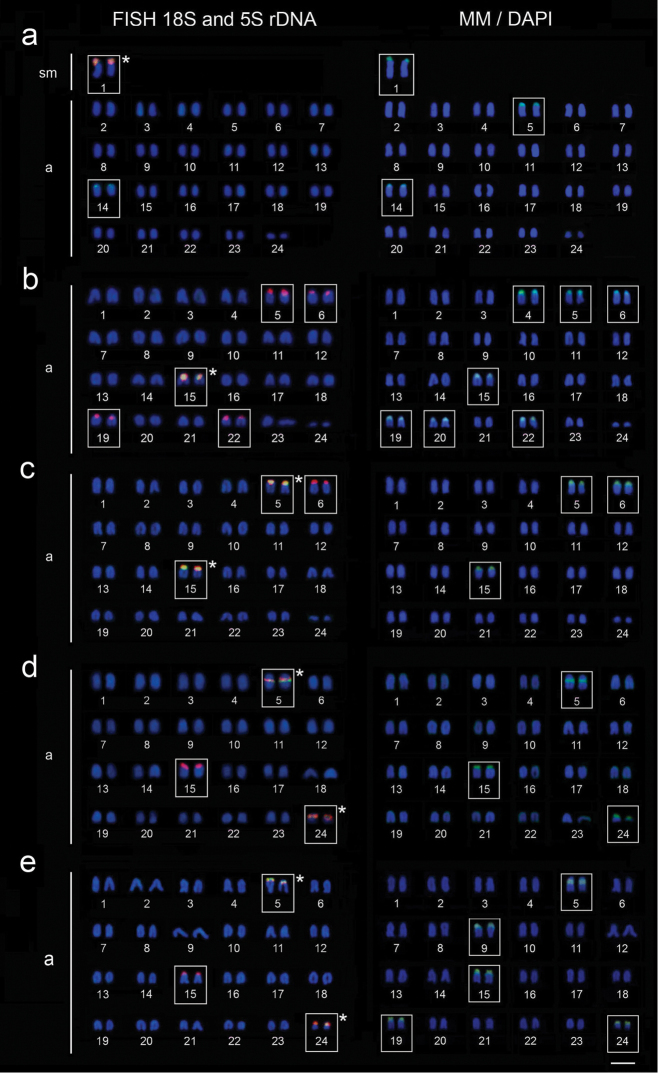
The mapping of 18S rDNA sequences showed single sites in Thalassoma noronhanum, coincident with the Ag-NORs (Fig. 3a). On the contrary, all Halichoeres species have multiple 18S rDNA sites. They occur in the terminal position of the short arms of pairs 5, 6, 15, 19 and 22 in Halichoeres penrosei (Fig. 3b), of pairs 5, 6 and 15 in Halichoeres poeyi (Fig. 3c), and of pairs 5, 15 and 24 of Halichoeres brasiliensis (Fig. 3e). However, in Halichoeres radiatus they are found in interstitial position on pair 5 and in terminal position on pairs 15 and 24 (Fig. 3d).
Figure 3.
Double-FISH with18S rDNA (red) and 5S rDNA (green) probes and MM/DAPI fluorochromes staining in the chromosomes of Thalassoma noronhanum (a), Halichoeres penrosei (b), Halichoeres poeyi (c), Halichoeres radiatus (d) and Halichoeres brasiliensis (e). Asterisks indicate the chromosome pairs with 18S/5S rDNA arrays. Bar: 5 μm.
The 5S rDNA sites occur in an 18S/5S rDNA array in pair 1 and exclusively on the short arms of pair 14 in Thalassoma noronhanum (Fig. 3a). On the other hand, in all Halichoeres species, the 5S rDNA sites are co-located with the 18S rDNA ones. They occur in the terminal position of the short arms of pair 15 in Halichoeres penrosei (Fig. 3b), of pairs 5 and 15 in Halichoeres poeyi (Fig. 3c), of pairs 5 and 24 in Halichoeres brasiliensis (Fig. 3e). In Halichoeres radiatus they are found in interstitial position in pair 5 and terminal position in pair 24 (Fig. 3d). The Ag-NOR marks were located exclusively on the 18S/5S rDNA arrays.
The sequential staining with MM/DAPI fluorochromes showed a larger number of GC-rich regions than rDNA sites in Thalassoma noronhanum, Halichoeres penrosei and Halichoeres brasiliensis. However, in all the species, the 18S and 5S rDNA sites and 18S/5S rDNA arrays were coincident with GC-rich regions (Fig. 3a–e).
Discussion
The rates of chromosome diversification can vary significantly among marine fish families (Molina et al. 2014) and, in some cases, they are linked to the evolutionary dynamics of the rDNA sequences. Indeed, groups with marked karyotype conservatism (Molina 2007) usually exhibit low diversification in the frequency and organization of ribosomal sites (Motta-Neto et al. 2011, Calado et al. 2013), while those with moderate or higher rates of chromosomal diversification (Molina et al. 2014) may display marked variations in the rDNA regions (Lima-Filho et al. 2014a, b).
In contrast with several Perciformes groups, Labridae show considerable variation in the diploid values (2n=22 to 48), as well as in the number of chromosome arms (NF=38 to 92) (Sena and Molina 2007, Arai 2011). The evolutionary rates of chromosomes differ significantly among clades (Table 1), reflecting their different histories linked to a deep association with coral reefs (Wainwright et al. 2004).
The cytogenetic patterns of the five analyzed wrasses suggest a greater karyotype conservatism in Julidini than in other Labridae clades. Indeed, Halichoeres and Thalassoma species exhibit karyotypes with 2n=48 chromosomes, mostly or entirely formed by acrocentric chromosomes, small amount of heterochromatin and one or two pairs bearing Ag-NORs (Sena and Molina 2007, present paper), a characteristic recognized as basal for Perciformes (Brum and Galetti 1997, Galetti et al. 2000).
The chromosomal divergences in Julidini are mainly due to a small number of pericentric inversions (Table 1). In Thalassoma noronhanum, the presence of an exclusive pair of biarmed chromosomes demonstrates a variant condition with respect to six other species previously described in this genus, all of them with 2n=48a. On the other hand, in Halichoeres species the presence of few biarmed chromosomes (1 to 3 pairs) is relatively more frequent (Sena and Molina 2007, Arai 2011), albeit not identified in the Atlantic species here investigated. However, despite the similarities in the karyotype structure of Thalassoma noronhanum, Halichoeres penrosei, Halichoeres poeyi, Halichoeres radiatus and Halichoeres brasiliensis, a dynamic evolutionary condition concerning the rDNA regions occurs among these species, which contribute to understanding the karyotypic evolution in Julidini. In fact, the chromosome mapping of rDNA sequences showed a significant variation in frequency, distribution and organization, especially in the Halichoeres species.
Chromosomes with homogeneous and small amounts of repetitive DNAs have been found in fish species with little karyotype diversification (Molina 2007, Motta-Neto et al. 2011). On the other hand, heterogeneous and large amounts of repetitive DNAs are related in several families with notable levels of chromosomal rearrangements and differentiation (Moreira-Filho and Bertollo 1991, Souza et al. 2001, Favarato et al. 2016). Among the repetitive DNAs, rDNA has a major role in karyotype diversification. In fact, species from various fish families exhibit 18S and 5S rDNAs sequences involved in chromosome fusion points (Molina and Galetti 2002, Ziemniczak 2011, Jacobina et al. 2013, Getlekha et al. 2016), indicating their probable involvement in the chromosomal reorganization. In this sense, the presence of an 18S rDNA site in the interstitial position on pair 5 in Halichoeres radiatus, in contrast to its terminal position in the homeologous chromosomes of the remaining species, puts in evidence a cryptic paracentric inversion in that chromosome pair.
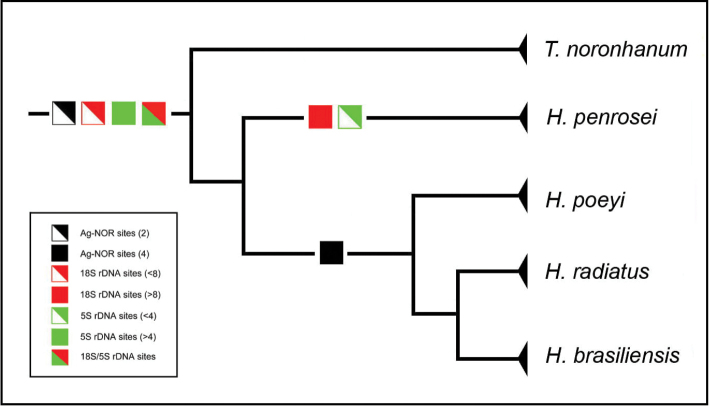
From the phylogenetic view, a single Ag-NOR/18S rDNA site in Thalassoma noronhanum likely represents an ancestral condition for Julidini species. Halichoeres penrosei, the most basal species analyzed in this genus (possibly belonging to the genus Thalassoma, according to Rocha et al. 2010), shows intermediate features, with a single Ag-NOR and multiple 18S rDNA sites. This indicates that multiple rDNA regions is an ancestral condition and that the rDNA dynamics is an ancient trait in Thalassoma and Halichoeres genera (Fig. 4). In fact, the multiple Ag-NORs present in Halichoeres poeyi, Halichoeres radiatus and Halichoeres brasiliensis and the large number of rDNA sites present in Halichoeres suggest that the dispersal process of these sequences precedes their diversification. The wide variation in distribution and organizational patterns of these sequences in Julidini are compatible with birth-and-death processes (Rooney and Ward 2005) acting in a stochastic evolutionary model.
Figure 4.
Evolutionary patterns of ribosomal sites in Thalassoma noronhanum, Halichoeres radiatus, Halichoeres poeyi, Halichoeres brasiliensis and Halichoeres penrosei, from the phylogenetic perspective (evolutionary relationships adapted from Rocha et al. 2010).
In turn, the 5S rDNA sequences can present a conserved chromosomal distribution, even among phylogenetically non-related taxa (Perina et al. 2011). In most eukaryotes, these sequences are organized in tandem repetitions, in which the (NTS) present high interspecific variations, due to insertions/deletions, minisatellites and pseudogenes (Nelson and Honda 1985, Leah et al. 1990, Alves-Costa et al. 2008). However, although evolutionarily conserved, stochastic events may promote a great dispersal of the 5S rDNA sequences in a large number of chromosomes in some Perciformes species (Affonso and Galetti 2005, Lima-Filho et al. 2014b).
The location of 45S and 5S rDNA sites in different chromosomes is the most common condition in vertebrates (Lucchini et al. 1993, Suzuki et al. 1996, Gornung 2013), indicating independent evolution of these loci. The syntenic arrangement of these rDNA classes, as found in Halichoeres and Thalassoma, is not a common feature, although it has already been reported in some main fish orders, such as Perciformes (Ghigliotti et al. 2008, Merlo et al. 2013), Characiformes (Vicari et al. 2006, Bellafronte et al. 2009, Cioffi et al. 2009), Siluriformes (Mariotto et al. 2011, Ziemniczak et al. 2012), Anguilliformes (Deiana et al. 2006), Salmoniformes (Pendás et al. 1994), Nototheniformes (Ghigliotti et al. 2007) and Tetraodontiformes (Martinez et al. 2010). In fishes, 45S/5S rDNA arrays are phylogenetically stochastic and limited to few species of a clade (Almeida-Toledo et al. 2002), and preferentially explained by random events in the course of the evolutionary trajectory of the genome (Calado et al. 2014). Thus, the phylogenetic spread of these arrangements in the Julidini clade indicates a noteworthy evolutionary stability. In fact, although the non-syntenic organization of these rDNA classes might be interpreted as a functional advantage (Martins and Galetti 2001), the persistent 18S/5S rDNA arrays in Thalassoma and Halichoeres indicates that they are feasible and, in this case, suggesting a probable adaptive condition for this multigene organization. In addition, syntenic rDNA genes may exhibit adjacent or interspersed arrangements (Artoni et al. 2015). In Julidini, hybridization signals are apparently superimposed, suggesting the occurrence of the latter kind of organization. Further fiber-FISH analyses will allow better understanding of the organization of these arrangements.
Final remarks
The uncommon pattern of 18S and 5S rDNA synteny presented by Julidini species indicates a shared ancestral condition, in contrast to stochastic and taxonomically restricted occurrences found in other fish groups (Drouin and Sá 1995, Calado et al. 2013). In addition to phylogenetic sharing patterns, these arrangements suggest a possible adaptive organization, given that they are all active ribosomal sites (Ag-NOR positive) in this species. The differentiated 18S/5S rDNA regions in Halichoeres species are particularly useful in identifying phylogenetic homeologies (pairs 5 and 15), but also sufficiently divergent to represent effective cytotaxonomic markers for this genus. Although a conserved karyotypic pattern is maintained in some Labridae species, the present data reveal a significant dynamism of the ribosomal sequences, in accordance to the moderate/high rate of chromosomal diversification in this family.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The experimental work fulfills all ethical guidelines regarding the handling of specimens. The collection and handling of specimens followed protocols approved by the Ethics Committee on the Use of Animals of the Federal University of Rio Grande do Norte (Process 044/2015). All authors consent to participate in the publication and are in agreement with the article content.
Acknowledgements
The authors thank CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) for financial assistance (Process 442664/2015-0), IBAMA (Instituto Brasileiro do Meio Ambiente e Recursos Naturais) for the license to collect specimens (Process 19135/1), the UFRN (Universidade Federal do Rio Grande do Norte) for providing the means to conduct the study, and José Garcia Júnior for the taxonomic identification of the species.
Citation
Amorim KDJ, Cioffi MB, Bertollo LAC, Soares RX, Souza AS, Costa GWWF, Molina WF (2016) Co-located 18S/5S rDNA arrays: an ancient and unusual chromosomal trait in Julidini species (Labridae, Perciformes). Comparative Cytogenetics 10(4): 555–570. doi: 10.3897/CompCytogen.v10i4.10227
References
- Affonso PRAM, Galetti PM., Jr (2005) Chromosomal diversification of reef fishes from genus Centropyge (Perciformes, Pomacanthidae). Genetica 123(3): 227–233. doi: 10.1007/s10709-004-3214-x [DOI] [PubMed] [Google Scholar]
- Allen GR. (1995) Thalassoma robertsoni, a new species of wrasse (Labridae) from Clipperton Island, tropical eastern Pacific Ocean. Revue française d’aquariologie 22: 3–4. doi: 10.2305/IUCN.UK.2015.RLTS.T183703A85698513 [Google Scholar]
- Almeida-Toledo LF, Ozouf-Costaz C, Foresti F, Bonillo C, Porto-Foresti F, Daniel-Silva MF. (2002) Conservation of the 5S-bearing chromosome pair and co-localization with major rDNA clusters in five species of Astyanax (Pisces, Characidae). Cytogenetic and Genome Research 97(3–4): 229–33. doi: 10.1159/000066609 [DOI] [PubMed] [Google Scholar]
- Alvarez MC, Garcia E, Thode G. (1986) Contribution to the karyotypes of Ctenolabrus rupestris and Symphodus ocellatus. Caryologia 39(3–4): 353–357. doi: 10.1080/00087114.1986.10797797 [Google Scholar]
- Alves-Costa FA, Martins C, Matos FDC, Foresti F, Oliveira C, Wasko AP. (2008) 5S rDNA characterization in twelve Sciaenidae fish species (Teleostei, Perciformes): Depicting gene diversity and molecular markers. Genetics and Molecular Biology 31(1): 303–307. doi: 10.1590/S1415-47572008000200025 [Google Scholar]
- Arai R. (2011) Fish Karyotypes: A Check List. Springer, 340 pp. doi: 10.1007/978-4-431-53877-6 [Google Scholar]
- Artoni RF, Castro JP, Jacobina UP, Lima-Filho PA, Costa GWWF, Molina WF. (2015) Inferring diversity and evolution in fish by means of integrative molecular cytogenetics. The Scientific World Journal 2015(3): 365787. doi: 10.1155/2015/365787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber PH, Bellwood DR. (2005) Biodiversity hotspots: Evolutionary origins of biodiversity in wrasses (Halichoeres: Labridae) in the Indo-Pacific and new world tropics. Molecular Phylogenetics and Evolution 35(1): 235–53. doi: 10.1016/j.ympev.2004.10.004 [DOI] [PubMed] [Google Scholar]
- Bellafronte E, Vicari MR, Artoni RF, Margarido VP, Moreira-Filho O. (2009) Differentiated ZZ/ZW sex chromosomes in Apareiodon ibitiensis (Teleostei, Parodontidae): Cytotaxonomy and biogeography. Journal of Fish Biology 75(9): 2313–2325. doi: 10.1111/j.1095-8649.2009.02488.x [DOI] [PubMed] [Google Scholar]
- Bernardi G, Bucciarelli G, Costagliola D, Robertson DR, Heiser JB. (2004) Evolution of coral reef fish Thalassoma spp. (Labridae). 1. Molecular phylogeny and biogeography. Marine Biology 144: 369–75. doi: 10.1007/s00227-003-1199-0 [Google Scholar]
- Biémont C, Vieira C. (2006) Genetics: junk DNA as an evolutionary force. Nature 443(7111): 521–524. doi: 10.1038/443521a [DOI] [PubMed] [Google Scholar]
- Brum MJI, Galetti PM., Jr (1997) Teleostei ground plan karyotype. Journal of Comparative Biology 2: 91–102. [Google Scholar]
- Calado LL, Bertollo LAC, Cioffi MB, Costa GWWF, Jacobina UP, Molina WF. (2014) Evolutionary dynamics of rDNA genes on chromosomes of the Eucinostomus fishes: cytotaxonomic and karyoevolutive implications. Genetics and Molecular Research 13(4): 9951–9959. doi: 10.4238/2014.November.27.24 [DOI] [PubMed] [Google Scholar]
- Calado LL, Bertollo LAC, Costa GWWF, Molina WF. (2013) Cytogenetic studies of Atlantic mojarras (Perciformes - Gerreidae): Chromosomal mapping of 5S and 18S ribosomal genes using double FISH. Aquaculture Research 44(5): 829–835. doi: 10.1111/j.1365-2109.2012.03089.x [Google Scholar]
- Choat H, Bellwood D. (1998) Wrasses & Parrotfishes. In: Eschmeyer W, Paxton J. (Eds) Encyclopedia of fishes - second edition Academic Press, San Diego, CA, 209–213. [Google Scholar]
- Cioffi MB, Bertollo LAC. (2012) Chromosomal distribution and evolution of repetitive DNAs in fish. In: Garrido-Ramos MA. (Eds) Repetitive DNA. Genome Dyn. Basel, Karger, vol 7: 197–221. doi: 10.1159/000337950 [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Martins C, Centofante L, Jacobina U, Bertollo LAC. (2009) Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: mapping of three classes of repetitive DNAs. Cytogenetic and Genome Research 125(2) 132–141. doi: 10.1159/000227838 [DOI] [PubMed] [Google Scholar]
- Deiana AM, Coluccia E, Cannas R, Pesci P, Fonnesu A, Salvadori S. (2006) Colocalization of the ribosomal gene families in Conger conger (Anguilliformes, Congridae). Italian Journal of Zoology 73(1): 1–5. doi: 10.1080/11250000500502038 [Google Scholar]
- Drouin G, Sá MM. (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Molecular Biology and Evolution 12(3): 481–493. [DOI] [PubMed] [Google Scholar]
- Eschmeyer WN, Fong JD. (2016) Catalog of Fishes electronic version (4 January 2016), California Academy of Sciences, San Francisco: http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp#Labridae [Google Scholar]
- Favarato RM, Silva M, Oliveira RR, Artoni RF, Feldberg E, Matoso DA. (2016) Cytogenetic diversity and the evolutionary dynamics of rDNA genes and telomeric sequences in the Ancistrus genus (Loricariidae: Ancistrini). Zebrafish 13(2): 103–111. doi: 10.1089/zeb.2015.1140 [DOI] [PubMed] [Google Scholar]
- Galetti PM, Jr, Aguilar CT, Molina WF. (2000) An overview of marine fish cytogenetics. Hydrobiologia 420(1): 55–62. doi: 10.1023/A:1003977418900 [Google Scholar]
- Ghigliotti L, Mazzei F, Ozouf-Costaz C, Bonillo C, Williams R, Cheng CHC, Pisano E. (2007) The two giant sister species of the Southern Ocean, Dissostichus eleginoides and Dissostichus mawsoni, differ in karyotype and chromosomal pattern of ribosomal RNA genes. Polar Biology 30(5): 625–634. doi: 10.1007/s00300-006-0222-6 [Google Scholar]
- Ghigliotti L, Mazzei F, Ozouf-costaz C, Christiansen JS, Fevolden SE, Pisano E. (2008) First cytogenetic characterization of the sub-arctic marine fish. Genetics and Molecular Biology 31: 180–187. doi: 10.1590/S1415-47572008000200003 [Google Scholar]
- Getlekha N, Molina WF, Cioffi MB, Yano CF, Maneechot N, Bertollo LAC, Supiwong W, Tanomtong A. (2016) Repetitive DNAs highlight the role of chromosomal fusions in the karyotype evolution of Dascyllus species (Pomacentridae, Perciformes). Genetica 144(2): 203–211. doi: 10.1007/s10709-016-9890-5 [DOI] [PubMed] [Google Scholar]
- Gold JR, Li YC, Shipley NS, Powers PK. (1990) Improved methods for working with fish chromosomes with a review of metaphase chromosome banding. Journal of Fish Biology 37(4): 563–575. doi: 10.1111/j.1095-8649.1990.tb05889.x [Google Scholar]
- Gornung E. (2013) Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: A review of research. Cytogenetic and Genome Research 141(2-3): 90–102. doi: 10.1159/000354832 [DOI] [PubMed] [Google Scholar]
- Howell WM, Black DA. (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia 36(8): 1014–1015. doi: 10.1007/BF01953855 [DOI] [PubMed] [Google Scholar]
- Jacobina UP, Vicari MR, Martinez PA, Cioffi MB, Bertollo LAC, Molina WF. (2013) Atlantic moonfishes: independent pathways of karyotypic and morphological differentiation. Helgoland Marine Research 67(3) 499–506. doi: 10.1007/s10152-012-0338-8 [Google Scholar]
- Kidwell MG. (2002) Transposable elements and the evolution of genome size in eukaryotes. Genetica 115(1): 49–63. doi: 10.1023/A:1016072014259 [DOI] [PubMed] [Google Scholar]
- Leah R, Frederiksen S, Engberg J, Sorensen PD. (1990) Nucleotide sequence of a mouse 5S rRNA variant gene. Nucleic Acids Research 18(24): 7441. doi: 10.1093/nar/18.24.7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg A. (1964) Nomenclature for centromeric position at chromosomes. Hereditas 52(2): 201–220. doi: 10.1111/j.1601-5223.1964.tb01953.x [Google Scholar]
- Lima-Filho PA, Amorim KD J, Cioffi MB, Bertollo L AC, Molina WF. (2014a) Chromosomal mapping of repetitive DNAs in Gobionellus oceanicus and G. stomatus (Gobiidae; Perciformes): A shared XX/XY system and an unusual distribution of 5S rDNA sites on the Y chromosome. Cytogenetic and Genome Research 144(4): 333–40. doi: 10.1159/000373909 [DOI] [PubMed] [Google Scholar]
- Lima-Filho PA, Bertollo LAC, Cioffi MB, Costa GWWF, Molina WF. (2014b) Karyotype divergence and spreading of 5S rDNA sequences between genomes of two species: Darter and emerald gobies (Ctenogobius, Gobiidae). Cytogenetic and Genome Research 142(3): 197–203. doi: 10.1159/000360492 [DOI] [PubMed] [Google Scholar]
- Lucchini SD, Nardi I, Barsacchi G, Batistoni R, Andronico F. (1993) Molecular cytogenetics of the ribosomal (18S + 28S and 5S) DNA loci in primitive and advanced urodele amphibians. Genome 36(4): 762–773. doi: 10.1139/g93-101 [DOI] [PubMed] [Google Scholar]
- Mariotto S, Centofante L, Vicari MR, Artoni RF, Moreira-Filho O. (2011) Chromosomal diversification in ribosomal DNA sites in Ancistrus kner, 1854 (Loricariidae, Ancistrini) from three hydrographic basins of Mato Grosso, Brazil. Comparative Cytogenetics 5(4): 289–300. doi: 10.3897/CompCytogen.v5i4.1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez PA, Araujo WC, Molina WF. (2010) Derived cytogenetic traits, multiple NORs and B chromosomes in the compact karyotype of Canthigaster figueiredoi (Tetraodontiformes). Marine Genomics 3(2): 85–89. doi: 10.1016/j.margen.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Martins C, Galetti PM., Jr (2001) Two 5S rDNA arrays in Neotropical fish species: Is it a general rule for fishes? Genetica 111(1): 439–446. doi: 10.1023/A:1013799516717 [DOI] [PubMed] [Google Scholar]
- Merlo MA, Cross I, Manchado M, Cárdenas S, Rebordinos L. (2013) The 5S rDNA high dynamism in Diplodus sargus is a transposon-mediated mechanism. Comparison with other multigene families and Sparidae species. Journal of Molecular Evolution 76(3): 83–97. doi: 10.1007/s00239-013-9541-8 [DOI] [PubMed] [Google Scholar]
- Molina WF. (2007) Chromosomal changes and stasis in marine fish groups. In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG. (Eds) Fish Cytogenetics. Science Publishers, Enfield, 69–110.
- Molina WF, Alves DE, Araújo WC, Martinez PA, Silva MF, Costa GW. (2010) Performance of human immunostimulating agents in the improvement of fish cytogenetic preparations. Genetics and Molecular Research 9(3): 1807–1814. doi: 10.4238/vol9-3gmr840 [DOI] [PubMed] [Google Scholar]
- Molina WF, Galetti PM., Jr (2002) Robertsonian rearrangements in the reef fish Chromis (Perciformes, Pomacentridae) involving chromosomes bearing 5S rRNA genes. Genetics and Molecular Biology 25(4): 373–77. doi: 10.1590/S1415-47572002000400004 [Google Scholar]
- Molina WF, Martinez PA, Bertollo LAC, Bidau CJ. (2014) Evidence for meiotic drive as an explanation for karyotype changes in fishes. Marine Genomics 15: 29–34. doi: 10.1016/j.margen.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Molina WF, Motta-Neto CC, Sena DCS, Cioffi MB, Bertollo LAC. (2012) Karyoevolutionary aspects of Atlantic hogfishes (Labridae-Bodianinae), with evidence of an atypical decondensed argentophilic heterochromatin. Marine Genomics 6: 25–31. doi: 10.1016/j.margen.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Moreira-Filho O, Bertollo L. (1991) Astyanax scabripinnis (Pisces, Characidae): A species complex. Genetics and Molecular Biology 14: 331–357. [Google Scholar]
- Motta-Neto CC, Cioffi MB, Bertollo LAC, Molina WF. (2011) Extensive chromosomal homologies and evidence of karyotypic stasis in Atlantic grunts of the genus Haemulon (Perciformes). Journal of Experimental Marine Biology and Ecology 401: 75–79. doi: 10.1016/j.jembe.2011.02.044 [Google Scholar]
- Nelson DW, Honda BM. (1985) Genes coding for 5S ribosomal RNA of the nematode Caenorhabditis elegans. Gene 38(1–3): 245–51. doi: 10.1016/0378-1119(85)90224-0 [DOI] [PubMed] [Google Scholar]
- Nelson SJ. (2006) Fishes of the World. John Wiley and Sons Inc, 601 pp. [Google Scholar]
- Pendás AM, Moran P, Freije JP, Garcia-Vazquez E. (1994) Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenetics and Cell Genetics 67(1): 31–36. doi: 10.1159/000133792 [DOI] [PubMed] [Google Scholar]
- Perina A, Seoane D, González-Tizón AM, Rodríguez-Fariña F, Martínez-Lage A. (2011) Molecular organization and phylogenetic analysis of 5S rDNA in crustaceans of the genus Pollicipes reveal birth-and-death evolution and strong purifying selection. BMC Evolutionary Biology 11: 304. doi: 10.1186/1471-2148-11-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW. (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proceedings of the National Academy of Sciences of the United States of America 83(9): 2934–38. doi: 10.1073/pnas.83.9.2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha LA, Pinheiro HT, Gasparini JL. (2010) Description of Halichoeres rubrovirens, a new species of wrasse (Labridae: Perciformes) from the Trindade and Martin Vaz Island group, southeastern Brazil, with a preliminary mtDNA molecular phylogeny of new world Halichoeres. Zootaxa 2422: 22–30. doi: 10.11646/%25x [Google Scholar]
- Rooney AP, Ward TJ. (2005) Evolution of a large ribosomal RNA multigene family in filamentous fungi: birth and death of a concerted evolution paradigm. Proceedings of the National Academy of Sciences of the United States of America 102(14): 5084–89. doi: 10.1073/pnas.0409689102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA, Sternberg RV. (2005) Why repetitive DNA is essential to genome function. Biological Reviews of the Cambridge Philosophical Society 80(2): 227–50. doi: 10.1017/S1464793104006657 [DOI] [PubMed] [Google Scholar]
- Schweizer D. (1976) Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 58(4): 307–24. doi: 10.1007/BF00292840 [DOI] [PubMed] [Google Scholar]
- Sena DCS, Molina WF. (2007) Chromosomal rearrangements associated with pelagic larval duration in Labridae (Perciformes). Experimental Marine Biology and Ecology 353(2): 203–10. doi: 10.1016/j.jembe.2007.08.020 [Google Scholar]
- Souza IL, Galián J, La Rúa PD, Bertollo LAC, Moreira-Filho O. (2001) Non-random distribution of the GC-rich heterochromatin and nucleolar rDNA sites on Astyanax scabripinnis chromosomes. Cytologia 66(2001): 85–91. doi: 10.1508/cytologia.66.85 [Google Scholar]
- Sumner AT. (1972) A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 75(1): 304–6. doi: 10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Moriwaki K, Sakurai S. (1996) Rat rDNA sapacer sequences and chromosomal assignment of the genes to the extreme terminal region of chromosome 19. Cytogenetics and Cell Genetics 72: 1–4. doi: 10.1159/000134149 [DOI] [PubMed] [Google Scholar]
- Ueno K, Takai A. (2000) Chromosome evolution involving Robertsonian rearrangements in Xyrichthys fish (Labridae, Perciformes). Cytobios 103(402): 7–15. [PubMed] [Google Scholar]
- Vicari MR, Moreira-Filho O, Artoni RF, Bertollo LAC. (2006) ZZ/ZW sex chromosome system in an undescribed species of the genus Apareiodon (Characiformes, Parodontidae). Cytogenetic and Genome Research 114: 163–68. doi: 10.1159/000093333 [DOI] [PubMed] [Google Scholar]
- Wainwright PC, Bellwood DR, Westneat MW, Grubich JR, Hoey AS. (2004) A functional morphospace for the skull of labrid fishes: Patterns of diversity in a complex biomechanical system. Biological Journal of the Linnean Society 82(1): 1–25. doi: 10.1111/j.1095-8312.2004.00313.x [Google Scholar]
- Westneat MW, Alfaro ME. (2005) Phylogenetic relationships and evolutionary history of the reef fish family Labridae. Molecular Phylogenetics and Evolution 36(2): 370–90. doi: 10.1016/j.ympev.2005.02.001 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing offungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols: A Guide to Methods and Applications. Academic Press Inc, 315–322. [Google Scholar]
- Ziemniczak K, Barros A, Rosa K, Nogaroto V. (2012) Comparative cytogenetics of Loricariidae (Actinopterygii: Siluriformes): emphasis in Neoplecostominae and Hypoptopomatinae. Italian Journal of Zoology 79(4): 492–501. doi: 10.1080/11250003.2012.676677 [Google Scholar]
- Ziemniczak K. (2011) Cytogenetic study in species of Loricariidae (Pisces, Siluriformes) of the sources of the Ribeira and Tibagi rivers, Ponta Grossa - PR. MSc dissertation, Universidade Federal do Paraná, Ponta Grossa, Brazil, 84 pp [In portuguese] [Google Scholar]