Abstract Abstract
Corydoras Lacepède, 1803 is the most specious genus of Corydoradinae subfamily and many of its species are still unknown in relation to molecular cytogenetic markers. However, the diploid number and karyotypic formula were recorded for many species of this group. In current study, we provided the first cytogenetic information of Corydoras carlae Nijssen & Isbrücker, 1983, an endemic fish species from Iguassu River basin, Paraná State, Brazil. The individuals were collected in Florido River, a tributary of Iguassu River and analysed with respect to diploid number, heterochromatin distribution pattern, Ag-NORs and mapping of 5S and 18S ribosomal genes. The karyotype of this species comprises 46 chromosomes arranged in 22m+22sm+2st. The heterochromatin is distributed in centromeric and pericentromeric positions in most of the chromosomes, and also associated with NORs. The Ag-NORs were detected in the terminal position on the long arm of the metacentric pair 6. The double-FISH technique showed that 5S rDNA and 18S rDNA were co-localized in the terminal portion on the long arm of the metacentric pair 6. This condition of co-localization of ribosomal genes in Corydoras carlae seems to represent a marker for this species.
Keywords: Ag-NORs, cytogenetic markers, ribosomal DNA, heterochromatin, karyotype
Introduction
In higher eukaryotes, rDNA is organized into two distinct gene classes: major class (45S rDNA), which contains the genes that code for the 18S, 5.8S and 28S rRNAs, and the minor class (5S rDNA), which contains the genes that code for 5S rRNA. Fish species have been analyzed for 5S and 18S rDNA location in chromosomes using (FISH). The major rDNA sequences detected by FISH always coincided with (Ag-NORs) location, although in species with multiple Ag-NORs the number of markings was usually smaller than the regions detected by the DNA probes.
The most common condition in the karyotype of different fish groups is the positioning of ribosomal genes in different chromosome pairs (Galetti Jr. and Martins 2004). However, syntenic localization of the major rDNA clusters and the 5S sites were observed for the first time in the genus Corydoras Lacepède, 1803 (present study) and Callichthys callichthys (Linnaeus, 1758) (Konerat et al. 2014), the other integrant of the family Callichthyidae. In Loricariidae, Kronichthys lacerta (Nichols, 1919), Isbrueckerichthys duseni (Miranda Ribeiro, 1907), Parotocinclus maculicauda (Steindachner, 1877) and Trichomycterus sp. (Ziemniczak et al. 2012) also presented syntenic localization of ribosomal genes. Thus, the mapping of ribosomal genes has added important information about the chromosomal diversification in Corydoras, as in other groups of Siluriformes.
Callichthyidae is a family of the order Siluriformes widely distributed in Neotropical region, which has 215 valid species, divided in two subfamilies, Callichthyinae with 17 valid species and Corydoradinae with 198 valid species (Eschmeyer and Fong 2016). Corydoras is the most specious and cytogenetically studied genus of Corydoradinae, demonstrating different diploid numbers, which may vary from 2n = 40 chromosomes in Corydoras nattereri Steindachner, 1876 (Oliveira et al. 1990, 1993) to 2n = 134 chromosomes in Corydoras aeneus (Gill, 1858) (Turner et al. 1992).
Considering aspects related to number and morphology of chromosomes, as well as analysis of DNA content, Oliveira et al. (1992) and Shimabukuro-Dias et al. (2004) proposed the existence of five groups of species in Corydoras. However, the vast majority of studies in Corydoras is restricted to conventional analysis and little is known about location of the different types of rDNA, only in Corydoras paleatus (Jenyns, 1842) and Corydoras ehrhardti Steindachner, 1910 for 18S rDNA (Artoni et al. 2006) and Corydoras britskii (Nijssen & Isbrücker, 1983) for 18S and 5S rDNA (Takagui et al. 2014), making essential the development of studies with this approach to better understand the relationships between species of Corydoras.
Thus, the current paper presents the first cytogenetic description of Corydoras carlae, focusing on karyotype characterization, heterochromatin distribution pattern and location of 5S and 18S rDNA sites. Besides the new data for the species, this study also reveal for the first time the co-localization of 5S and major rDNA in Callichthyidae.
Materials and methods
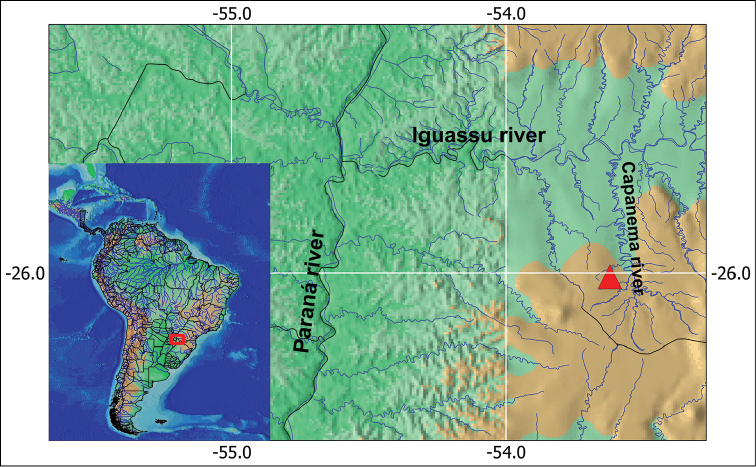
Ten individuals (four females and six males) of Corydoras carlae were sampled in the Florido River (26°00'32.60"S; 53°25'50.70"W), Paraná State, Brazil. A sub-tributary of left margin of Lower Iguassu River that flows into Capanema River, which flows immediately above of the Iguassu falls (Fig. 1). Voucher specimens were deposited in the fish collection of the (NUPELIA), Universidade Estadual de Maringá, Paraná, Brazil, as Corydoras carlae (NUP 17885).
Figure 1.
Localization of Florido River from the Iguassu River basin, where Corydoras carlae individuals were captured. Red triangle indicates the sampled point.
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals, approved by the Committee on the Ethics of Animal Experiments of the Universidade Estadual do Oeste do Paraná (License Number: Protocol 13/09 – CEEAAP/Unioeste). Before the evisceration process, the individuals were anesthetized by an overdose of clove oil (Griffiths 2000). Metaphase chromosomes were obtained from anterior kidney cells using the air-drying technique (Bertollo et al. 1978). Analysis of the C-positive heterochromatin (C-bands) followed the basic procedure of Sumner (1972), with some minor adaptations (Lui et al. 2012). The NORs were detected by means of silver nitrate staining (Ag-NORs), according to Howell and Black (1980). The chromosomes were classified as (m), (sm), and (st) according to their arm ratio (Levan et al. 1964). For the determination of the (FN), or number of chromosome arms, the m, sm and st chromosomes were considered as bearing two arms and the acrocentric chromosomes only one arm.
The localization of the 5S and 18S rDNA sites in the chromosomes was performed using the fluorescence in situ hybridization (FISH) method (Pinkel et al. 1986) with modifications (Margarido and Moreira-Filho 2008), with probes obtained from the fish species Leporinus elongatus Valenciennes, 1850 (Martins and Galetti Jr 1999) and Prochilodus argenteus Spix & Agassiz, 1829 (Hatanaka and Galetti Jr 2004), respectively. The probes were labelled through nick translation, with digoxigenin-11-dUTP (5S rDNA) and biotin-16-dUTP (18S rDNA) (Roche). Detection and amplification of the hybridization signal were made using avidin-FITC and anti-avidin biotin (Sigma) for probes labelled with biotin, and anti-digoxigenin rhodamine (Roche) for probes labelled with digoxigenin. Slides were counterstained with DAPI (50 µg/mL) and analysed in epifluorescence microscope (Olympus BX61). The images were captured using the software DP controller (Media Cybernetics).
Results
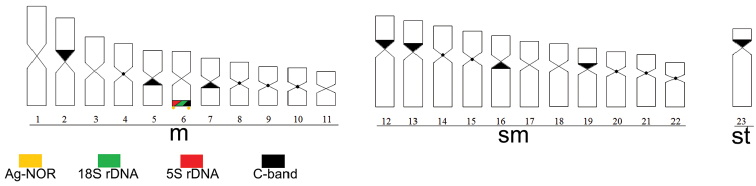
Corydoras carlae presented a modal diploid number of 46 chromosomes in males and females, and the karyotype contained 22 metacentric, 22 submetacentric and 2 subtelocentric chromosomes (22m+22sm+2st), yielding a FN of 92 in both sexes (Fig. 2a). The Ag-NORs was detected in the terminal position on the long arm of metacentric pair 6 (Box Fig. 2a). Positive C-band heterochromatins were detected in the centromeric and pericentromeric regions of nine and eight pairs, respectively, and coincident with the ribosomal sites (Fig. 2b). The double-FISH technique showed 5S rDNA cluster appears interspersed with 18S cistrons in the terminal portion of the long arm of pair 6 (Fig. 2c). Thus, featuring synteny and co-location of ribosomal genes in Corydoras carlae. The ideogram summarizes all markers on chromosomes of Corydoras carlae (Fig. 3).
Figure 2.
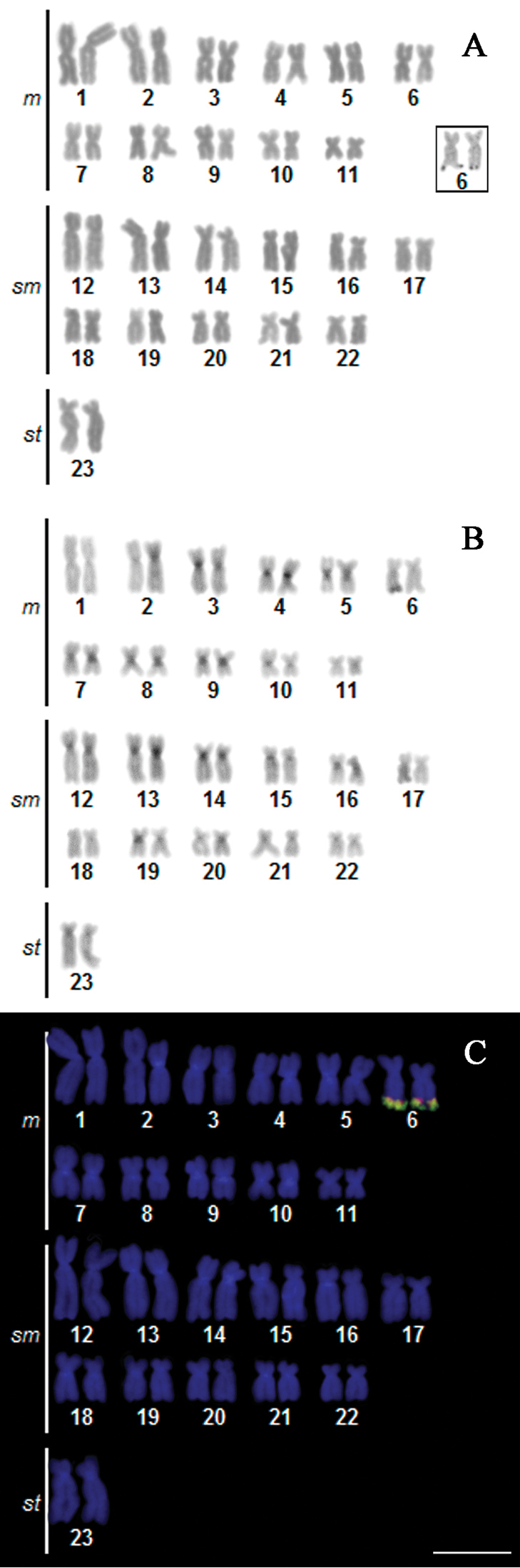
Karyotypes of Corydoras carlae stained with a Giemsa b C-banded and c after double FISH with 5S rDNA probes (red) and 18S rDNA (green). The NORs bearing chromosomes (pair 6) are boxed. Bar = 10 µm.
Figure 3.
Ideogram of Corydoras carlae, showing the heterochromatin, Ag-NORs, 18S and 5S rDNA distribution pattern.
Discussion
Cytogenetic studies have classified the species of the genus Corydoras into five groups according to their karyotype composition (Oliveira et al. 1992, Shimabukuro-Dias et al. 2004). Corydoras carlae has been included in group 4 (2n = 40-52 chromosomes, with many metacentric and submetacentric chromosomes). Considering our results, three species of this group occurring in the Iguassu River basin were cytogenetically analyzed: Corydoras carlae (2n=46, 22m+22sm+2st), collected in the Lower Iguassu River; Corydoras paleatus (2n=44, 20m+24sm) collected in the Upper Iguassu River (Oliveira et al. 1993), and Corydoras paleatus and Corydoras ehrhardti (2n=44, 18m+26sm), collected in the Upper Tibagi River (Artoni et al. 2006).
Individuals of Corydoras carlae analyzed here probably do not co-occur with Corydoras paleatus from Upper Iguassu River, since the lower portion is characterized by numerous waterfalls which gave rise to several reservoirs (Baumgartner et al. 2012). Therefore, the geographic isolation of Corydoras carlae may have facilitated the establishment of this numerical and structural karyotypic variation, as also observed in different populations of Glanidium ribeiroi Haseman, 1911 along the Iguassu River basin (Lui et al. 2015). Thus, the lack of gene flow among Corydoras species in the Iguassu River basin could favor different changes in each sample, supposedly resulting in speciation.
The number and position of NORs in Corydoras species are quite variable and almost all information pertaining to the characterization of NORs in this species is based on silver nitrate impregnation (Table 1). These data show that most species have simple NORs located in the terminal position, as in the case of Corydoras carlae. However, not all species have this pattern, as in the case of Corydoras simulatus Weitzman & Nijssen, 1970 with interstitial NORs (Oliveira et al. 1992), as well as Corydoras britskii (Takagui et al. 2014), Corydoras simulatus, Corydoras sp. Galheiro river, Corydoras flaveolus Ihering, 1911 and Corydoras metae Eigenmann, 1914 (Oliveira et al. 1992), which exhibits a systems of multiple NORs. According to Oliveira and Gosztonyi (2000), the condition of simple Ag-NORs in terminal location is the possible basal condition for Siluriformes. Thus, Corydoras carlae and other species presenting simple Ag-NORs in terminal location seem to maintain this basal condition.
Table 1.
Ag-NORs, major and minor ribosomal genes sites distribution in Callichthyidae. The 45S and 5S columns report the number of chromosomes bearing the cistrons and its location.
| Family Callichthyidae | Locality | 2n | Ag-NOR | 45S | 5S | Note | References |
|---|---|---|---|---|---|---|---|
| Subfamily Corydoradinae | |||||||
| Corydoras carlae | Florido River/Paraná State, Brazil | 46 | simple | 2, q terminal | 2, q terminal | Synteny, Co-localization | Present study |
| Corydoras britskii | Miranda River/ Mato Grosso do Sul State, Brazil | 90 | multiple | 3, p terminal | 2 p, interstitial | Non-Synteny | Takagui et al. (2014) |
| Corydoras paleatus | Tibagi River/Paraná State, Brazil | 44 | simple | 3, q terminal | ------- | --------- | Artoni et al. (2006) |
| Corydoras ehrhardti | Tibagi River/Paraná State, Brazil | 44 | simple | 2, q terminal | ------- | --------- | Artoni et al. (2006) |
| Corydoras sodalis | from aquarium | 74 | simple | --------- | --------- | --------- | Shimabukuro-Dias et al. 2004 |
| Corydoras arcuatus | Tabatinga River/frontier Brazil and Peru | 46 | simple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras trilineatus | Caripi River/Pará State, Brazil | 46 | simple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras schwartzi | Negro River/Amazonas State, Brazil | 46 | simple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras cf. simulatus | Colombia | 62 | simple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras sp. Caripi River | Caripi River/Pará State, Brazil | 60 | simple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras reticulatus | Negro River/Amazonas State, Brazil | 74 | simple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras aff. punctatus Negro River | Negro River/Amazonas State, Brazil | 102 | simple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras simulatus | Colombia | 62 | multiple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras sp. Galheiro River | Galheiro River/Minas Gerais State, Brazil | 84 | multiple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras flaveolus | Alambari River/São Paulo State, Brazil | 58 | multiple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Corydoras metae | Colombia | 92 | multiple | --------- | --------- | --------- | Oliveira et al. 1992 |
| Subfamily Callichthyinae | |||||||
| Hoplosternum littorale | Contas River/Bahia State, Brazil | 60 | simple | 2, p terminal | 4, p terminal | Non-Synteny | Almeida et al. (2012) |
| Callichthys callichthys | Paraná River/Paraná State, Brazil | 56 | simple | 2-3, p terminal e interstitial | 7-9, p interstitial and terminal | Synteny, Adjacent regions | Konerat et al. (2014) |
| Hoplosternum littorale | Coastal River/São Paulo State Brazil | 60 | simple | 2, p terminal | 4 p terminal | Non-Synteny | Pazza et al. (2005) |
| Callichthys callichthys | Contas River/Bahia State, Brazil | 54 | multiple | 7, p terminal, 5, q terminal, 1 p interstitial | 8-12, p interstitial and terminal | Non-Synteny | Almeida et al. (2013) |
| Lepthoplosternum pectorale | Paraná River/Paraná State, Brazil | 64 | multiple | 10, p terminal; 2, q terminal | 6, p terminal | Non-Synteny | Konerat et al. (2014) |
Despite exhibiting wide variation on the diploid number, chromosome morphology and location of NORs, Corydoras species share a heterochromatin distribution pattern very similar, preferably centromeric and pericentomeric, and in most cases, associated to NORs. In Corydoras carlae, this pattern was also observed, with heterochromatic blocks also displayed in many chromosomes. Corydoras britskii from Miranda River also showed large amount of pericentromeric heterochromatin, but with terminal heterochromatic blocks (Takagui et al. 2014), which were not observed in this study.
The mapping of 18S rDNA and 5S rDNA are scarce in Callichthyidae, being known only for some species (Table 1). Concerning the genus Corydoras, Corydoras carlae exhibited only one chromosome pair bearing 18S rDNA sites, as well as Corydoras ehrhardti (Artoni et al. 2006), confirming the system of simple NORs evidenced by silver impregnation for both species. FISH with rDNA probes has helped detect the presence of inactive NORs, as in the case of Corydoras paleatus (Artoni et al. 2006), which presented multiple NORs sites after 18S-FISH, while the silver impregnation had detected only simple NORs. Thus, studies with 18S-FISH can be useful for better evaluating the pattern distribution of the NORs in Corydoras.
In Corydoras, data on the location and number of 5S rDNA cistrons had only been described for Corydoras britskii, for which was detected interstitially in a pair of subtelocentric chromosomes (Takagui et al. 2014). In Corydoras carlae, the 5S rDNA was displayed at terminal position on the long arm of the metacentric pair 6. The presence of one chromosome pair bearing 5S rDNA is a common feature in different families of Siluriformes (Swarça et al. 2009), although multiple loci of 5S rDNA have been observed in Callichthyinae (Table 1). Inter– and intra–individual numerical and position variations of 5S rDNA cistrons have been observed in Callichthyidae and seem to represent a species-specific marker.
Furthermore, 5S rDNA cluster appears interspersed with 18S cistrons, featuring synteny and co-location of ribosomal genes in Corydoras carlae. The synteny is an unusual feature in fish, and such condition could influence an unwanted translocation of 5S sequences within 45S clusters, which could probably occur if these clusters were maintained linked in the same chromosome area (Martins and Galetti Jr 1999). This may explain why most vertebrates have these sequences on different chromosomes. Interestingly, all the possible syntenic conditions have been found in fishes, both sets of genes in distinct and disjoint chromosomal regions, as observed in Parodon nasus Kner, 1859 cited as Parodon tortuosus (Vicente et al. 2001) and Astyanax paranae Eigenmann, 1914 cited as Astyanax scabripinnis (Mantovani et al. 2005), or in adjacent regions, as in Triportheus nematurus (Kner, 1858) (Diniz et al. 2009), Mugil incilis Hancock, 1830 (Hett et al. 2011), Kronichthys lacerta, Isbrueckerichthys duseni, Parotocinclus maculicauda, Trichomycterus sp. (Ziemniczak et al. 2012) and Callichthys callichthys (Konerat et al. 2014), or the 5S rDNA interspersed along the clusters of 45S rDNA (co-localization), as in Astyanax altiparanae Garutti & Britski, 2000, Astyanax lacustris (Lütken, 1875), Astyanax fasciatus (Cuvier, 1819), Astyanax schubarti Britski, 1964 and Astyanax paranae cited as Astyanax scabripinnis (Almeida-Toledo et al. 2002), Solea senegalensis Kaup, 1858 (Cross et al. 2006), Bryconamericus cf. iheringii (Piscor et al. 2013) and Corydoras carlae (present study).
Despite little studies about mapping of rDNA genes in Callichthyidae, the majority of the species share the condition of non-synteny between the 5S rDNA and 45S rDNA. Therefore, this condition of co-localization of ribosomal genes in Corydoras carlae seems to represent a marker for this species.
Acknowledgments
The authors thank the Ineo/Gerpel, specially Poliana, Guido, Angélica and Werike by assistance in sample collection. The authors are also grateful to Dr. Weferson Júnio da Graça and MSc. Luiz Tencatt for taxonomic identification of the specimens. We would also like to express our gratitude to Brazilian agency Fundação Araucária for scholarship to first author.
Citation
Rocha RH, Baumgärtner L, Paiz LM, Margarido VP, Fernandes CA, Gubiani ÉA (2016) An uncommon co-localization of rDNA 5S with major rDNA clusters in Callichthyidae (Siluriformes): a report case in Corydoras carlae Nijssen & Isbrücker, 1983. Comparative Cytogenetics 10(4): 603–613. doi: 10.3897/CompCytogen.v10i4.9507
References
- Almeida JS, Affonso PRAM, Dias AL. (2012) Remarkable karyotypic homogeneity in a widespread tropical fish species: Hoplosternum littorale (Siluriformes, Callichthyidae). Journal of Fish Biology 81: 1415–1421. doi: 10.1111/j.1095-8649.2012.03387.x [DOI] [PubMed] [Google Scholar]
- Almeida JS, Affonso PRAM, Diniz D, Carneiro PLS, Dias AL. (2013) Chromosomal variation in the tropical armoured catfish Callichthys callichthys (Siluriformes, Callichthyidae): implications for conservation and taxonomy in a species complex from a Brazilian hotspot. Zebrafish 10: 451–458. doi: 10.1089/zeb.2013.0885 [DOI] [PubMed] [Google Scholar]
- Almeida-Toledo LF, Ozouf-Costaz C, Foresti F, Bonillo C, Porto-Foresti F, Daniel-Silva MFZ. (2002) Conservation of the 5S-bearing chromosome pair and co-localization with major rDNA clusters in five species of Astyanax (Pisces, Characidae). Cytogenetic and Genome Research 97: 229–233. doi: 10.1159/000066609 [DOI] [PubMed] [Google Scholar]
- Artoni RF, Terêncio ML, Vicari MR, Matiello MCA, Cestari MM, Bertollo LAC. (2006) Cytogenetics of two sympatric Corydoras species (Pisces, Siluriformes, Callichthyidae) of Southern Brazil. Brazilian Journal of Biology 66: 191–198. http://www.scielo.br/pdf/bjb/v66n1b/a02v661b.pdf [DOI] [PubMed] [Google Scholar]
- Baumgartner G, Pavanelli CS, Baumgartner D, Bifi AG, Debona T, Frana VA. (2012) Peixes do baixo rio Iguaçu. EDUEM, Maringá, 203 pp. doi: 10.7476/9788576285861 [Google Scholar]
- Bertollo LAC, Takahashi CS, Moreira-Filho O. (1978) Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Brazilian Journal of Genetics 1: 103–120. [Google Scholar]
- Cross I, Merlo A, Manchado M, Infante C, Cañavate JP, Rebordinos L. (2006) Cytogenetic characterization of the sole Solea senegalensis (Teleostei: Pleuronectiformes: Soleidae): Ag-NOR, (GATA)n, (TTAGGG)n and ribosomal genes by one-color and two-color FISH. Genetica 128: 253–259. doi: 10.1007/s10709-005-5928-9 [DOI] [PubMed] [Google Scholar]
- Diniz D, Laudicina A, Bertollo LAC. (2009) Chromosomal location of 18S and 5S rDNA sites in Triportheus fish species (Characiformes, Characidae). Genetics and Molecular Biology 32: 37–41. doi: 10.1590/S1415-47572009005000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschmeyer WN, Fong JD. (2016) Catalog of Fishes – Species by Family/Subfamily. http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp
- Galetti PM, Jr, Martins C. (2004) Contribuição da hibridização in situ para o conhecimento dos cromossomos de peixes. In: Guerra M. (Ed.) FISH: conceitos e aplicações na Citogenética. Sociedade Brasileira de Genética, Ribeirão Preto, 61–88.
- Griffiths SP. (2000) The use of clove oil as an anaesthetic and method for sampling intertidal rockpool fishes. Journal of Fish Biology 57: 1453–1464. doi: 10.1006/jfbi.2000.1406 [Google Scholar]
- Hatanaka T, Galetti PM., Jr (2004) Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus, Agassiz, 1829 (Characiformes, Prochilodontidae). Genetica 122: 239–244. doi: 10.1007/s10709-004-2039-y [DOI] [PubMed] [Google Scholar]
- Hett AS, Nirchio M, Oliveira C, Siccha ZR, Rossi AR, Sola L. (2011) Karyotype characterization of Mugil incilis Hancock, 1830 (Mugiliformes: Mugilidae), including a description of an unusual co-localization of major and minor ribosomal genes in the family. Neotropical Ichthyology 9: 107–112. http://www.scielo.br/pdf/ni/v9n1/aop0511.pdf [Google Scholar]
- Howell WM, Black DA. (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 36: 1014–1015. doi: 10.1007/BF01953855 [DOI] [PubMed] [Google Scholar]
- Konerat JT, Bueno V, Martins-Santos IC, Margarido VP. (2014) Karyotypic diversity and chromosome evolution in the armored catfishes Callichthyinae (Siluriformes, Callichthyidae). Caryologia 67: 140–148. doi: 10.1080/00087114.2014.931635 [Google Scholar]
- Levan A, Fredga K, Sandberg AA. (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52: 201–220. doi: 10.1111/j.1601 5223.1964.tb01953.x [Google Scholar]
- Lui RL, Blanco DR, Moreira-Filho O, Margarido VP. (2012) Propidium iodide for making heterochromatin more evident in the C-banding technique. Biotechnic & Histochemistry 87: 433–438. doi: 10.3897/CompCytogen.v7i1.4368 [DOI] [PubMed] [Google Scholar]
- Lui RL, Blanco DR, Traldi JB, Margarido VP, Moreira-Filho O. (2015) Karyotypic variation of Glanidium ribeiroi Haseman, 1911 (Siluriformes, Auchenipteridae) along the Iguazu river basin. Brazilian Journal of Biology 75: 215–221. doi: 10.1590/1519-6984.10714 [DOI] [PubMed] [Google Scholar]
- Mantovani M, Abel LDS, Moreira-Filho O. (2005) Conserved 5S and variable 45S rDNA chromosomal localization revealed by fish in Astyanax scabripinnis (Pisces, Characidae). Genetica 123: 211–216. doi: 10.1007/s10709-2281-3 [DOI] [PubMed] [Google Scholar]
- Margarido VP, Moreira-Filho O. (2008) Karyotypic differentiation through chromosome fusion and number reduction in Imparfinis hollandi (Ostariophysi, Heptapteridae). Genetics and Molecular Biology 31: 235–238. http://www.scielo.br/pdf/gmb/v31n1s0/12.pdf [Google Scholar]
- Martins C, Galetti PM., Jr (1999) Chromosome localization of 5S rDNA genes in Leporinus (Anostomidae, Characiformes). Chromosome Research 7: 363–365. doi: 10.1023/A:1009216030316 [DOI] [PubMed] [Google Scholar]
- Oliveira CL, Toledo FDA, Filho SDAT. (1990) Comparative cytogenetic analysis of three cytotypes of Corydoras nattereri (Pisces, Siluriformes, Callichthyidae). Cytologia 21–26. doi: 10.1508/cytologia.55.21
- Oliveira C, Almeida-Toledo LF, Mori L, Toledo-Filho AS. (1993) Cytogenetic and DNA content studies on armoured catfishes of the genus Corydoras (Pisces, Siluriformes, Callichthyidae) from the southeast coast of Brazil. Brazilian Journal of Genetics 16: 617–629. http://repositorio.unesp.br/bitstream/handle/11449/37711/WOSA1993MC77000008.pdf?sequence=1&isAllowed=y [Google Scholar]
- Oliveira C, Almeida‐Toledo LF, Mori L, Toledo‐Filho AS. (1992) Extensive chromosomal rearrangements and nuclear DNA content changes in the evolution of the armoured catfishes genus Corydoras (Pisces, Siluriformes, Callichthyidae). Journal of Fish Biology 40: 419–431. doi: 10.1111/j.1095-8649.1992.tb02587.x [Google Scholar]
- Oliveira C, Gosztonyi AE. (2000) A cytogenetic study of Diplomystes mesembrinus (Teleostei, Siluriformes, Diplomystidae) with a discussion of chromosome evolution in siluriformes. Caryologia 53: 31–37. doi: 10.1080/00087114.2000.10589178 [Google Scholar]
- Pazza R, Kavalco KF, Almeida-Toledo LF, Bertollo LAC. (2005) Hoplosternum littorale (Teleostei, Callichthyidae) from a Coastal River basin in Brazil - Cytogenetic analysis and gene mapping of 5S and 18S rDNA. Caryologia 58: 339–344. doi: 10.1080/00087114.2005.10589472 [Google Scholar]
- Pinkel D, Straume T, Gray JW. (1986) Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proceedings of the National Academy of Sciences 83: 2934–2938. http://www.pnas.org/content/83/9/2934.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscor D, Ribacinko-Piscor DB, Fernandes CA, Partise-Maltempi PP. (2013) Cytogenetic analysis in three Bryconamericus species (Characiformes, Characidae): first description of the 5S rDNA-bearing chromosome pairs in the genus. Molecular Cytogenetics 6: 13. doi: 10.1186/1755-8166-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro-Dias CK, Oliveira C, Foresti F. (2004) Cytogenetic analysis of five species of the subfamily Corydoradinae (Teleostei: Siluriformes: Callichthyidae). Genetics and Molecular Biology 27: 549–554. doi: 10.1590/S1415-47572004000400014 [Google Scholar]
- Sumner AT. (1972) A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 75: 304–306. doi: 10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Swarça AC, Fenocchio AS, Cestari MM, Dias AL. (2009) Localization and characterization of the 5S rDNA bearing chromosome in two Steindachneridion species by means of different cytogenetic techniques. Cytologia 74: 323–327. doi: 10.1508/cytologia.74.323 [Google Scholar]
- Takagui FH, Venturelli NB, Sampaio TR, Dias AL, Giuliano-Caetano L. (2014) Cytogenetic study in Corydoras britskii (Siluriformes: Callichthyidae), using different chromosomal banding and fluorescence hybridization in situ with rDNA probes. Ichthyological Research 61: 201–206. doi: 10.1007/s10228-014-0392-0 [Google Scholar]
- Turner BJ, Diffot N, Rasch EM. (1992) The callichthyid catfish Corydoras aeneus is an unresolved diploid-tetraploid sibling complex. Ichthyological Exploration Freshwaters 3: 17–23. [Google Scholar]
- Vicente VE, Jesus CM, Moreira-Filho O. (2001) Chromosomal localization of 5S and 18S rRNA genes in three Parodon species (Pisces, Parodontidae). Caryologia 54: 365–69. doi: 10.1080/00087114.2001.10589247 [Google Scholar]
- Ziemniczak K, Barros AV, Rosa KO, Nogaroto V, Almeida MC, Cestari MM, Moreira-Filho O, Artoni RF, Vicari MR. (2012) Comparative cytogenetics of Loricariidae (Actinopterygii, Siluriformes): emphasis in Neoplecostominae and Hypoptopomatinae. Italian Journal of Zoology 79: 492–501. doi: 10.1080/11250003.2012.676677 [Google Scholar]