Abstract Abstract
The diploid number 2n = 30 is a presumed synapomorphy of Dendropsophus Fitzinger, 1843, although a noticeable variation in the number of biarmed/telocentric chromosomes is observed in this genus. Such a variation suggests that several chromosomal rearrangements took place after the evolutionary origin of the hypothetical ancestral 30-chromosome karyotype; however, the inferred rearrangements remain unknown. Distinct numbers of telocentric chromosomes are found in the two most cytogenetically studied species groups of Dendropsophus. In contrast, all three species of the Dendropsophus marmoratus (Laurenti, 1768) group that are already karyotyped presented five pairs of telocentric chromosomes. In this study, we analyzed cytogenetically three additional species of this group to investigate if the number of telocentric chromosomes in this group is not as variable as in other Dendropsophus groups. We described the karyotypes of Dendropsophus seniculus (Cope, 1868), Dendropsophus soaresi (Caramaschi & Jim, 1983) and Dendropsophus novaisi (Bokermann, 1968) based on Giemsa staining, C-banding, silver impregnation and in situ hybridization with telomeric probes. Dendropsophus seniculus, Dendropsophus soaresi and Dendropsophus novaisi presented five pairs of telocentric chromosomes, as did the remaining species of the group previously karyotyped. Though the species of this group show a high degree of karyotypic similarity, Dendropsophus soaresi was unique in presenting large blocks of het-ITSs (heterochromatic internal telomeric sequences) in the majority of the centromeres. Although the ITSs have been interpreted as evidence of ancestral chromosomal fusions and inversions, the het-ITSs detected in the karyotype of Dendropsophus soaresi could not be explained as direct remnants of ancestral chromosomal rearrangements because no evidence of chromosomal changes emerged from the comparison of the karyotypes of all of the species of the Dendropsophus marmoratus group.
Keywords: Chromosomes, Anura, telomeric sequence
Introduction
Faivovich et al. (2005) resurrected the genus Dendropsophus Fitzinger, 1843 to accommodate all Neotropical hylid species known or suspected to have a diploid chromosome number 2n = 30. This cytogenetic character state was later confirmed as a synapomorphy for this genus by Suárez et al. (2013) after the description of a 2n = 24 karyotype for Xenohyla Izecksohn, 1998, the sister genus of Dendropsophus (see Faivovich et al. 2005, Pyron and Wiens 2011, Duellman et al. 2016). Based on preliminary data, Bogart (1973) hypothesized that centric fission events may have been involved in the origin of an ancestral 30-chromosome karyotype, which is a hypothesis that was also considered by Suárez et al. (2013). However, the chromosomes that are putatively involved in these events have not yet been recognized, and this hypothesis remains to be validated.
Although all of the Dendropsophus species karyotyped to date show 2n = 30 (see review in Catroli and Kasahara 2009, Medeiros et al. 2013, Suárez et al. 2013, Oliveira et al. 2016), a noticeable variation in the number of biarmed/telocentric chromosomes is observed among them, suggesting that several chromosomal rearrangements took place after the evolutionary origin of the hypothetical ancestral 30-chromosome karyotype. Karyotypes with only biarmed chromosomes [as in Dendropsophus minutus (Peters, 1872) (Gruber et al. 2005) and Dendropsophus leali (Bokermann, 1964) (Bogart 1973)] and karyotypes with up to five pairs of telocentric/subtelocentric chromosomes [as in Dendropsophus labialis (Peters, 1863) (Bogart 1973), Dendropsophus sanborni (Schmidt, 1944) and Dendropsophus jimi (Napoli & Caramaschi, 1999) (Medeiros et al. 2013)] may be observed. However, the chromosomes and events involved in these rearrangements also remain undiscovered because most Dendropsophus species karyotypes are not yet described, and few chromosomal markers are available for the known karyotypes, preventing reliable hypotheses of chromosome homeology.
Of the nine species groups recognized in Dendropsophus (for a review of the Dendropsophus groups, see Faivovich et al. 2005), the Dendropsophus microcephalus (Cope, 1886) group is the most species-rich (currently with 40 species—Frost 2016) and the most studied cytogenetically (17 species karyotyped—review by Catroli and Kasahara 2009, Medeiros et al. 2013, Oliveira et al. 2016). It is noteworthy that karyotypes without any telocentric chromosome (in Dendropsophus leali—Bogart 1973) and with one [as in Dendropsophus bipunctatus (Spix, 1824)—Bogart 1973], two [as in Dendropsophus phlebodes (Steineger, 1906)—Kaiser et al. 1996], three [as in Dendropsophus cruzi (Pombal & Bastos, 1998)—Gruber et al. 2005], four [as in Dendropsophus nanus (Boulenger, 1889)—Medeiros et al. 2003] or five (as in Dendropsophus jimi—Medeiros et al. 2013) telocentric chromosome pairs are observed in this group. Karyotypes with distinct numbers of telocentric chromosomes were also found in the Dendropsophus leucophyllatus (Beireis, 1783) group (Bogart 1973, Kaiser et al. 1996, Gruber et al. 2005), which currently has 11 species (see Frost 2016) and is the second most cytogenetically studied species group of Dendropsophus (four of the named species are karyotyped). In contrast, all of the three species of the Dendropsophus marmoratus group that are already karyotyped (i.e. Dendropsophus marmoratus, Dendropsophus melanargyreus and Dendropsophus nahdereri) present five pairs of telocentric chromosomes (Bogart 1973, Gruber et al. 2005, Suárez et al. 2013).
Chromosomal sites composed of telomeric repeats localized apart from the telomeres, also known as interstitial or intrachromosomal telomeric sequences (ITSs) or repeats (ITRs), have been detected in several animals (Meyne et al. 1990, Nanda et al. 2002, Rovatsos et al. 2015, Schmid and Steinlein 2016) and plants (Tek and Jiang 2004, He et al. 2013). Based on the genomic location and sequence organization, especially in the number of telomeric repeats, Ruiz-Herrera et al. (2008) classified the ITSs in short ITSs (s-ITSs) and heterochromatic ITS (het-ITS). The s-ITSs [called short interstitial telomeres, short ITs, by Azzalin at al. (2001)] are short stretches of telomeric hexamers distributed at internal chromosomal positions, presumably present in all vertebrate species, whereas het-ITSs are large blocks of telomeric-like repeats localized mainly in centromeric and pericentromeric regions (Ruiz-Herrera et al. 2008). The s-ITSs probably originated from the insertion of telomeric repeats during the repair of DNA double-strand breaks, as was originally proposed by Nergadze et al. (2004, 2007). On the other hand, the het-ITSs have been widely considered to be remnants of ancestral chromosomal rearrangements as fusions (e.g., Lee et al. 1993, Slijepcevic 1998, Ropiquet et al. 2010, Paço et al. 2013, Young et al. 2013) and inversions (e.g., Farré et al. 2009, Ocalewicz et al. 2013, Paço et al. 2013). Recently, Schmid and Steinlein (2016) proposed an additional category of ITS, named euchromatic-ITSs (eu-ITSs), to accommodate the large ITSs that are not revealed as heterochromatic sites by C-banding or staining with base-specific fluorochromes.
The Dendropsophus marmoratus group currently includes eight species, i.e., Dendropsophus marmoratus, Dendropsophus acreanus (Bokermann, 1964), Dendropsophus dutrai (Gomes & Peixoto, 1996), Dendropsophus melanargyreus, Dendropsophus nahdereri, Dendropsophus novaisi, Dendropsophus seniculus and Dendropsophus soaresi (Faivovich et al. 2005). Some adult and larval morphological synapomorphies of this species group may be recognized (Faivovich et al. 2005); however, its cladistic proximity with other Dendropsophus species groups as well as the internal phylogenetic relationships of this group remain unclear (Faivovich et al. 2005; Pyron and Wiens 2011, Fouquet et al. 2011, Medeiros et al. 2013). To date, up to three of the eight species of the Dendropsophus marmoratus group have been included in phylogenetic analysis (Fouquet et al. 2011).
In this study, we analyzed cytogenetically three additional species of the Dendropsophus marmoratus group to investigate if the number of telocentric chromosomes in this group is not as variable as in other Dendropsophus groups. Because karyotypic variation in number of telocentric chromosomes may result from rearrangements involving telomeric sequences (review in Ruiz-Herrera et al. 2008), we included here the mapping of telomeric sequences in the karyotypes of two of the analyzed species. Additionally, we provided the nucleotide sequence of a fragment of the 16S ribosomal RNA gene of one exemplar for each of the species that were analyzed cytogenetically to yield a reliable association of the chromosomal data set with a DNA data set that has been remarkably useful for taxonomic and phylogenetic studies of anurans.
Material and methods
Specimens
Four male exemplars of Dendropsophus seniculus from Ribeirão Grande, state of São Paulo, Brazil, nine Dendropsophus soaresi males from Barreiras, state of Bahia, Brazil and one female of Dendropsophus novaisi from Jequié, state of Bahia, Brazil were analyzed cytogenetically. The specimens were collected under a permit issued by the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA) (#32483), and deposited at the amphibian collection of the Museu de Zoologia “Prof. Adão José Cardoso” at the Institute of Biology – University of Campinas, Campinas, Brazil, under the accession numbers ZUEC 17225–17228 (Dendropsophus seniculus), ZUEC 16867–16875 (Dendropsophus soaresi) and ZUEC 17858 (Dendropsophus novaisi).
Cytogenetic analyses
Animals were injected intraperitoneally with 2% colchicine (Sigma – Aldrich; 0.02 mL per 1 g body weight) for an “in vivo” treatment that lasted at least 4 hours. The animals were deeply anesthetized with lidocaine gel 2% and their intestines were removed and used for obtaining chromosomal preparations according to the method of King and Rofe (1976). Chromosomes were conventionally stained with 10% Giemsa and sequentially submitted to C-banding (Sumner 1972) and silver staining by the Ag-NOR method (Howell and Black 1980).
To localize telomeric sequences, the karyotypes were in situ hybridized with the probe (CCCTAA)3 (PNA – Peptid Nucleic Acid TelC-Cy3; PNA Bio Inc.), following the manufacturer’s instructions.
Mitochondrial DNA sequences
Samples of genomic DNA were obtained from Dendropsophus seniculus (ZUEC 17225), Dendropsophus soaresi (ZUEC 16867) and Dendropsophus novaisi (ZUEC 17858) following the procedure reported by Medeiros et al. (2013). A fragment of approximately 1300 bp of the 16S ribosomal RNA gene was PCR-amplified using the primers 12L13(L) (Feller and Hedges 1998) and 16Sbr(H) (Palumbi et al. 1991). The amplified products were purified with the GFX PCR and Gel Band DNA purification Kit (GE Healthcare) and directly sequenced in an automatic DNA ABI/Prism sequencer (Applied Biosystems) using BigDye Terminator kits (Applied Biosystems) and the primers 12L13 (Feller and Hedges 1998), TitusI(H) (Titus 1992), Hedges16L2a (Hedges 1994), Hedges16H10 (Hedges 1994), 16Sar(L) (Palumbi et al. 1991) and 16Sbr(H) (Palumbi et al. 1991). DNA sequences were aligned using ClustalW option implemented in BioEdit v. 7.2.5 (Hall 1999) and compared to each other and to the 16S rDNA sequence of Dendropsophus seniculus available at GenBank (AY843666).
Results
Cytogenetic analyses
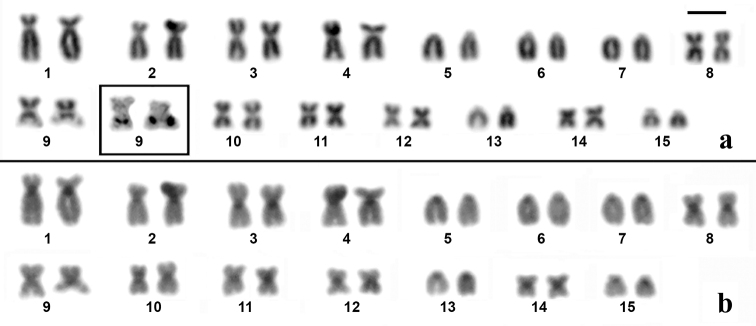
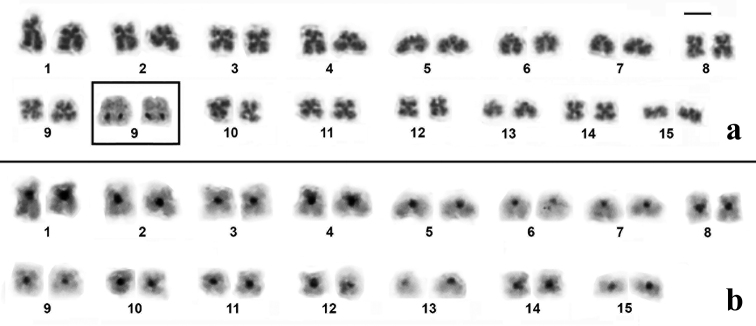
The karyotypes of Dendropsophus seniculus, Dendropsophus soaresi and Dendropsophus novaisi were very similar and presented three pairs (pairs 1, 2 and 4) of submetacentric chromosomes, seven pairs (pairs 3, 8–12 and 14) of metacentric chromosomes and five pairs (pairs 5–7, 13 and 15) of telocentric chromosomes (Figures 1–3). The nucleolus organizer region (NOR) was detected by silver staining in the long arm of chromosome 9 of the three species (insets in Figures 1–3). C-banding only detected the centromeric region of all the chromosomes of Dendropsophus seniculus (Figure 1b), Dendropsophus soaresi (Figure 2b) and Dendropsophus novaisi (Figure 3b).
Figure 1.
Karyotype of Dendropsophus seniculus stained with Giemsa (a) and C-banded (b). In the inset in (a), the NOR-bearing chromosome pair 9 after silver staining. Bar = 5 µm.
Figure 3.
Karyotype of Dendropsophus novaisi stained with Giemsa (a) and C-banded (b). In the inset in (a), the NOR-bearing chromosome pair 9 after silver staining. Bar = 5 µm.
Figure 2.
Karyotype of Dendropsophus soaresi stained with Giemsa (a) and C-banded (b). In the inset in (a), the NOR-bearing chromosome pair 9 after silver staining. Bar = 5 µm.
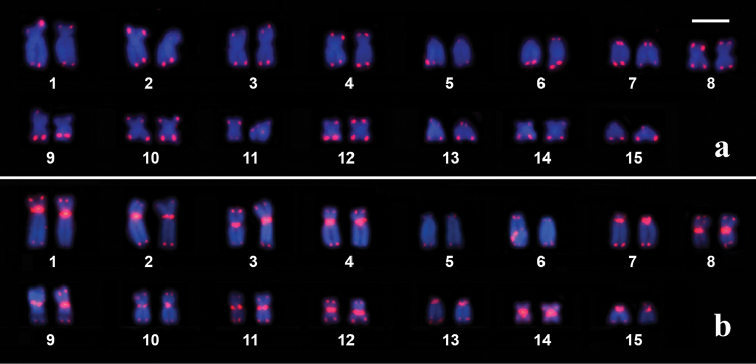
In situ hybridization detected telomeric sequences in all of the telomeres of Dendropsophus seniculus and Dendropsophus soaresi (Figure 4). Additionally, interstitial telomeric sequences (ITSs) were detected in the centromeres of the chromosomes of Dendropsophus soaresi, except in two of the five pairs of telocentric chromosomes (pairs 5 and 6) (Figure 4b).
Figure 4.
Karyotypes of Dendropsophus seniculus (a) and Dendropsophus soaresi (b) hybridized with telomeric probe.
Mitochondrial DNA sequences
The nucleotide sequence (1312 bp) (see Suppl. material 1) of the 16S rDNA of the specimen of Dendropsophus seniculus that we analyzed was highly similar (99.6%) to the corresponding sequence available at GenBank (AY843666; Faivovich et al. 2005) from a specimen of Dendropsophus seniculus from Angra dos Reis, Rio de Janeiro State, Brazil. The sequences obtained from Dendropsophus soaresi (1314 bp) and Dendropsophus novaisi (1310 bp) (see Suppl. material 1) were 86.4% similar to each other, and 88.4% and 90.7% (average value) similar to the sequences of Dendropsophus seniculus, respectively.
Discussion
Karyotypic comparisons
The three species analyzed showed karyotypes composed of five pairs of telocentric chromosomes, similarly to the other three species of the Dendropsophus marmoratus group previously studied cytogenetically [i.e. Dendropsophus marmoratus (Bogart 1973, Suárez et al. 2013), Dendropsophus melanargyreus (Suárez et al. 2013) and Dendropsophus nahdereri (Gruber et al. 2005)]. The interspecific morphological conservation of the karyotypes of the species of the Dendropsophus marmoratus group contrasts with the variation found in other Dendropsophus groups. The Dendropsophus microcephalus group, for instance, includes species with zero to five pairs of telocentric chromosomes (see Bogart 1973, Kaiser et al. 1996, Gruber et al. 2005, Medeiros et al. 2003). Variation in the number of telocentric chromosomes could also be found in the Dendropsophus leucophyllatus group, although only four named species of this group have been karyotyped (see Bogart 1973, Kaiser et al. 1996, Gruber et al. 2005).
According to the estimated dates of divergence provided by Duellman et al. (2016), the Dendropsophus marmoratus, Dendropsophus microcephalus and Dendropsophus leucophyllatus groups arose at similar times in the mid-Miocene (17.0, 17.2 and 18.7 Mya, respectively). Therefore, differential time for divergence does not justify the different levels of karyotypic variation observed among the three aforementioned species groups. Further analyses of the chromosomal rearrangements involved in the karyotypic variations in Dendropsophus combined with phylogeographic studies are still necessary to elucidate about the high conservation in the number of telocentric chromosomes in the Dendropsophus marmoratus group.
With respect to the number and relative size of the telocentric chromosomes, the karyotypes of the species of the Dendropsophus marmoratus group are similar to that of Dendropsophus labialis (Bogart 1973), a species included in the Dendropsophus labialis group. This morphological similarity suggests that the telocentric chromosomes of these karyotypes could be homeologous, although a better characterization of these chromosomes is fundamental to test this hypothesis. According to the most comprehensive phylogenetic analysis of Dendropsophus (Duellman et al. 2016) and assuming the telocentric chromosomes of the species of the Dendropsophus marmoratus group and Dendropsophus labialis are homeologous, it is possible to hypothesize that this karyotype configuration is plesiomorphic with respect to those constituted by other numbers and/or relative sizes of the telocentric chromosomes. However, internal relationships within Dendropsophus are consistently poorly supported and small taxonomic additions cause huge impacts (e.g., Fouquet et al. 2015, Duellman et al. 2016).
Five pairs of telocentric chromosomes were also observed in the karyotypes of Dendropsophus jimi and Dendropsophus sanborni (Medeiros et al. 2013), which are species that belong to the Dendropsophus microcephalus group. In these karyotypes, however, the telocentric pairs were classified as pairs 5, 6, 12, 13 and 15, whereas the telocentric chromosomes of the karyotypes of Dendropsophus labialis and the species of the Dendropsophus marmoratus group are numbered as pairs 5, 6, 7, 13 and 15. Because only a few chromosomal markers are available for a comparison of these karyotypes, it is still not possible to determine if the telocentric chromosomes of all of these species are homeologous. Therefore, we cannot discard the possibility that chromosomes 12 of Dendropsophus jimi and Dendropsophus sanborni are homeologous to chromosomes 7 of Dendropsophus labilais and the species of the Dendropsophus marmoratus group, although these chromosomes differ by the presence of NOR in chromosomes 12 of Dendropsophus jimi and Dendropsophus sanborni (Medeiros et al. 2013).
The similarities among the karyotypes of the species of the Dendropsophus marmoratus group are not restricted to the number of telocentric chromosomes. Dendropsophus seniculus, Dendropsophus soaresi and Dendropsophus novaisi also share with Dendropsophus marmoratus and Dendropsophus melanargyreus the location of the NOR at a distal site of the long arm of chromosome 9, which differs from Dendropsophus nahdereri, whose NOR is located on the short arm of the submetacentric chromosome 1 (Gruber et al. 2005).
C-banding did not reveal any differential band that could be considered exclusive to the karyotypes of Dendropsophus seniculus, Dendropsophus soaresi or Dendropsophus novaisi, since only the centromeric regions were detected by this technique (present work). Conspicuous non-centromeric C-bands were also absent in the karyotypes of Dendropsophus marmoratus and Dendropsophus melanargyreus, the other two species of the Dendropsophus marmoratus group whose karyotypes were already C-banded, although Suárez et al. (2013) reported the presence of some distal and interstitial C-bands in those karyotypes.
Despite the high similarity of the karyotypes of the species of the Dendropsophus marmoratus group with respect to the number and morphology of the chromosomes, C-banding pattern and location of NOR (except for Dendropsophus nahdereri), the karyotype of Dendropsophus soaresi stands out because of the presence of internal telomeric sequences in addition to the terminal telomeric sequences.
Interstitial telomeric sequences
Large and short ITSs are likely to play a role in karyotypic evolution. Several studies support the hypothesis that, in addition to possibly representing relics of chromosomal changes, the het-ITSs may themselves induce chromosome breakage and subsequent chromosomal rearrangements (reviewed in Ruiz-Herrera et al. 2008 and Bolzán 2012). Similarly, experimental and associative studies have also suggested the involvement of s-ITSs with genomic instability or chromosomal hot spots of recombination (Aksenova et al. 2013, Wood et al. 2015).
The het-ITSs detected in the present study in the karyotype of Dendropsophus soaresi cannot be explained as direct remnants of ancestral chromosomal rearrangements because no evidence of chromosomal changes has emerged from the comparison of the karyotypes of all species of the Dendropsophus marmoratus group already known (Bogart 1973, Gruber et al. 2005, Suárez et al. 2013, present work). Although it is very similar to the karyotypes of the other species of the group, the karyotype of Dendropsophus soaresi is unique in presenting large blocks of centromeric ITSs because the karyotypes of Dendropsophus seniculus (present study), Dendropsophus melanargyreus and Dendropsophus marmoratus (Suárez et al. 2013) showed only telomeric sites hybridized with telomeric probes.
The occurrence of het-ITSs at the majority of the centromeres of the karyotype of Dendropsophus soaresi is also remarkable and suggests the expansion and homogenization of telomeric sequences throughout the repetitive elements that compose these centromeric regions. Repetitive DNA, such as centromeric satellite DNA, is expected to expand in the genome and evolve in concert by a series of mechanisms, including unequal crossing-over, gene conversion, rolling circle replication and reinsertion, and transposon-mediated exchange (see Dover 1982, 1986 and the review by Plohl et al. 2008). The telomeric repeats present in heterochromatic sites should be subject to the same evolutionary forces (see Ruiz-Herrera et al. 2008). In contrast, the absence of het-ITS in the centromere of two chromosome pairs (telocentric chromosome pairs 5 and 6) of Dendropsophus soaresi suggests that these centromeres do not evolve in concert with the remaining centromeric regions of the genome. The reason for such differential behavior is intriguing and remains unknown.
Similar to observations of Dendropsophus soaresi, large blocks of centromeric/pericentromeric ITSs that were widely distributed throughout the genome were previously found in four other hylid species [i.e. Aplastodiscus albofrenatus (Lutz, 1924), Aplastodiscus arildae (Cruz & Peixoto, 1987) and Aplastodiscus eugenioi (Carvalho-e-Silva & Carvalho-e-Silva, 2005)—Carvalho et al. 2009, Gruber et al. 2012a; Hypsiboas faber (Wied-Neuwied, 1821)—Schmid and Steinlein 2016]. In the hylid Itapotihyla langsdorffii (Duméril & Bibron, 1841), ITSs were also observed in several centromeres (Gruber et al. 2012b), but in this case the het-ITSs are not as large as those previously mentioned. In addition to the aforementioned hylids, other fifteen hylid species showed ITSs in their karyotypes (Meyne et al. 1990, Wiley et al. 1992, Suárez et al. 2013, Mattos et al. 2014, Bruschi et al. 2014, Schmid and Steinlein 2016), which suggests that the appearance of this type of sequence is recurrent in the Hylidae family. Only the centromeric ITS found in chromosome 3 of Scarthyla goinorum (Bokermann, 1962) was clearly interpreted as a remnant of a chromosomal fusion that in that case could respond to the reduced chromosome number observed in this species (Suárez et al. 2013). The insertion of telomeric repeats during the repair of double-strand breaks in DNA as a phenomenon putatively involved in the origin of ITS in Hylidae remains unexplored.
It is worth noticing that in the sample of metaphases analyzed in this paper, large signals of the telomeric probe were detected at a subterminal non-heterochromatic site of some chromosomes of Dendropsophus seniculus (Figure 4a, at the long arm of the right homologous of chromosome 9). This hybridization pattern resembles that pattern interpreted by Wood et al. (2014, 2015) as cytological evidence of the occurrence of t-loops formed between telomere and s-ITS. However, studies designed to search for s-ITSs in hylid karyotypes have not yet been performed, and the prevalence of ITSs in Hylidae remains an intriguing question to be assessed in further studies.
Association between cytogenetic data and 16S rDNA sequences
The high similarity between the 16S rDNA sequence of Dendropsophus seniculus we provided and that previously obtained by Faivovich et al. (2005) enables a reliable association between the cytogenetic data shown here and the analyses hitherto conducted with the previously available sequence, including the studies of Fouquet et al. (2015) and Duellman et al. (2016). On the other hand, the nucleotide sequences obtained here from Dendropsophus soaresi and Dendropsophus novaisi were the first report of 16S rDNA sequences for these species.
Dendropsophus systematics are in flux and even comprehensive datasets are unable to provide a stable historical hypothesis (Fouquet et al. 2011; Peloso et al. 2016). The association between the cytogenetic dataset and 16S rDNA sequences may be very helpful in future analyses, especially because the species-level taxonomy of Dendropsophus has been subject to several changes. A number of Dendropsophus species has been described in the last few years (Rivera-Correia and Orrico 2013, Ortega-Andrade and Ron 2013, Orrico et al. 2014, Fouquet et al. 2015, Peloso et al. 2016) as well as species synonymizaton has been proposed (Guarnizo et al. 2012, Orrico et al. 2013). Therefore, a reliable association between different sets of data is fundamental for further integrative studies.
Conclusion
All of the karyotypes found in the Dendropsophus marmoratus group to date showed five pairs of telocentric chromosomes and were also similar in the location of NORs (except for the Dendropsophus nahdereri karyotype, described by Gruber et al. 2005) and C-banding pattern. Because of this karyotypic conservatism, the het-ITSs present in the majority of the centromeres of the karyotype of Dendropsophus soaresi may not be interpreted as direct remnants of ancestral chromosomal rearrangements.
Acknowledgments
The authors are thankful to Daniel Pacheco Bruschi for help in producing the cytogenetic data from Dendropsophus novaisi. This study was supported by the Brazilian agencies São Paulo Research Foundation (FAPESP) and Fundo de Apoio ao Ensino, à Pesquisa e Extensão (FAEPEX) da Universidade Estadual de Campinas.
Citation
Teixeira LSR, Seger KG, Targueta CP, Orrico VGD, Lourenço LB (2016) Comparative cytogenetics of tree frogs of the Dendropsophus marmoratus (Laurenti, 1768) group: conserved karyotypes and interstitial telomeric sequences. Comparative Cytogenetics 10(4): 753–767. https://doi.org/10.3897/CompCytogen.v10i4.9972
Supplementary materials
16S rDNA sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Lívia S. R. Teixeira, Karin Regina Seger, Cíntia Pelegrineti Targueta, Victor G. Dill Orrico, Luciana Bolsoni Lourenço
Data type: Tif file
Explanation note: Alignment of 16S rDNA fragments obtained from specimens of Dendropsophus seniculus, Dendropsophus novaisi and Dendropsophus soaresi that were used in cytogenetic analyses. GenBank accession numbers: KY053469 (Dendropsophus seniculus), KY053470 (Dendropsophus novaisi), and KY053471 (Dendropsophus soaresi).
References
- Aksenova AY, Greenwell PW, Dominska M, Shishkin AA, Kim JC, Petes TD, Mirkina SM. (2013) Genome rearrangements caused by interstitial telomeric sequences in yeast. PNAS 110: 19866–19871. https://doi.org/10.1073/pnas.1319313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Nergadze SG, Giulotto E. (2001) Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma 110: 75–82. https://doi.org/10.1007/s004120100135 [DOI] [PubMed] [Google Scholar]
- Bogart JP. (1973) Evolution of anuran karyotypes. In: Vial JL. (Ed.) Evolutionary Biology of the Anurans: Contemporary Research on Major Problems. University of Missouri Press, Columbia, 337–349. [Google Scholar]
- Bolzán AD. (2012) Chromosomal aberrations involving telomeres and interstitial telomeric sequences. Mutagenesis 27: 1–15. https://doi.org/10.1093/mutage/ger052 [DOI] [PubMed] [Google Scholar]
- Bruschi DP, Rivera M, Lima AP, Zúñiga AB, Recco-Pimentel SM. (2014) Interstitial Telomeric Sequences (ITS) and major rDNA mapping reveal insights into the karyotypical evolution of Neotropical leaf frogs species (Phyllomedusa, Hylidae, Anura). Molecular Cytogenetics 7: 22 https://doi.org/10.1186/1755-8166-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catroli GF, Kasahara S. (2009) Cytogenetic data on species of the family Hylidae (Amphibia, Anura): results and perspectives. Publ. UEPG. Ciências Biológicas e da Saúde 15: 67–86. [Google Scholar]
- Carvalho KA, Garcia PCA, Recco-Pimentel SM. (2009) NOR Dispersion, Telomeric Sequence Detection in Centromeric Regions and Meiotic Multivalent Configurations in Species of the Aplastodiscus albofrenatus Group (Anura, Hylidae). Cytogenetic Genome Research 126: 359–367. https://doi.org/10.1159/000264179 [DOI] [PubMed] [Google Scholar]
- Dover GA. (1982) Molecular drive: a cohesive mode of species evolution. Nature 299: 111–117. https://doi.org/10.1038/299111a0 [DOI] [PubMed] [Google Scholar]
- Dover GA. (1986) Molecular drive in multigene families: how biological novelties arise, spread and are assimilated. Trends Genet 122: 159–165. https://doi.org/10.1016/0168-9525(86)90211-8 [Google Scholar]
- Duellman WE, Marion AB, Hedges SB. (2016) Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa 4101: 1–109. https://doi.org/10.11646/zootaxa.4104.1.1 [DOI] [PubMed] [Google Scholar]
- Faivovich J, Haddad CFB, Garcia PCA, Frost DR, Campbell JA, Wheeler WC. (2005) Systematic review of the frog family Hylidae, with special reference to Hylinae: phylogenetic analysis and taxonomic revision. Bulletin of the American Museum of Natural History 294: 1–240. https://doi.org/10.1206/0003-0090(2005)294[0001:SROTFF]2.0.CO;2 [Google Scholar]
- Farré M, Ponsà M, Bosch M. (2009) Interstitial telomeric sequences (ITSs) are not located at the exact evolutionary breakpoints in primates. Cytogenetic and Genome Research 124: 128–131. https://doi.org/10.1159/000207517 [DOI] [PubMed] [Google Scholar]
- Feller A, Hedges SB. (1998) Molecular evidence for the early history of living amphibians. Molecular Phylogenetics and Evolution 9: 509–516. https://doi.org/10.1006/mpev.1998.0500 [DOI] [PubMed] [Google Scholar]
- Fouquet A, Noonan B, Blanc M, Orrico VGD. (2011) Phylogenetic position of Dendropsophus gaucheri (Lescure and Marty 2000) highlights the need for an in-depth investigation of the phylogenetic relationships of Dendropsophus (Anura: Hylidae). Zootaxa 3035: 59–67. https://doi.org/10.11646/zootaxa.4052.1.2 [Google Scholar]
- Fouquet A, Orrico VGD, Ernst R, Blanc M, Martinez Q, Vacher JP, Rodrigues MT, Ouboter PE, Jairam R, Ron S. (2015) A new Dendropsophus Fitzinger, 1843 (Anura: Hylidae) of the parviceps group from the lowlands of the Guiana Shield. Zootaxa 4052: 39–64. [DOI] [PubMed] [Google Scholar]
- Frost DR. (2016) Amphibian species of the world: an online reference. Version 6.0 American Museum of Natural History, New York, USA: http://research.amnh.org/herpetology/amphibia/index.html [1st July, 2016] [Google Scholar]
- Guarnizo CE, Escallón C, Cannatella DC, Amésquita A. (2012) Congruence between acoustic traits and genealogical history reveals a new species of Dendropsophus (Anura: Hylidae) in the high Andes of Colombia. Herpetologica 68: 523–540. https://doi.org/10.1655/HERPETOLOGICA-D-10-00038 [Google Scholar]
- Gruber SL, Haddad CFB, Kasahara S. (2005) Evaluating the karyotypic diversity in species of Hyla (Anura; Hylidae) with 2n=30 chromosomes based on the analysis of ten species. Folia Biologica (Praha) 51: 68–75. [PubMed] [Google Scholar]
- Gruber SL, Zina J, Narimatsu H, Haddad CFB, Kasahara S. (2012a) Comparative karyotype analysis and chromosome evolution in the genus Aplastodiscus (Cophomantini, Hylinae, Hylidae). BMC Genetics 13: 28 https://doi.org/10.1186/1471-2156-13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber SL, Haddad CFB, Kasahara S. (2012b) Karyotype analysis of seven species of the tribe Lophiohylini (Hylinae, Hylidae, Anura), with conventional and molecular cytogenetic techniques. Comparative Cytogenetics 6: 409–423. https://doi.org/10.3897/compcytogen.v6i4.3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- He L, Liu J, Torres GA, Zhang H, Jiang J, Xie C. (2013) Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Research 21: 5–13. https://doi.org/10.1007/s10577-012-9332-x [DOI] [PubMed] [Google Scholar]
- Hedges SB. (1994) Molecular evidence for the origin of birds. Proceedings of the National Academy of Sciences 91: 2621–2624. https://doi.org/10.1073/pnas.91.7.2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell WM, Black DA. (1980) Controlled silver staining of nucleolar organizer regions with a protective colloidal developer a 1-step method. Experientia 36: 1014–1015. https://doi.org/10.1007/BF01953855 [DOI] [PubMed] [Google Scholar]
- Kaiser H, Mais C, Bolãnos F, Steinlein C, Feichtinger W, Schmid M. (1996) Chromosomal, investigation of three Costa Rican frogs from the 30-chromosome radiation of Hyla with the description of a unique geographic variation in nucleolus organizer regions. Genetica 98: 95–102. https://doi.org/10.1007/BF00120223 [Google Scholar]
- King M, Rofe R. (1976) Karyotypic variation in the Australian gekko Phyllodactylus marmoratus (Gray) (Gekkonidae: Reptilia). Chromosoma 54: 75–87. https://doi.org/10.1007/BF00331835 [DOI] [PubMed] [Google Scholar]
- Lee C, Sasi R, Lin CC. (1993) Interstitial localization of telomeric DNA sequences in the Indian muntjac chromosomes: further evidence for tandem chromosome fusions in the karyotypic evolution of the Asian muntjacs. Cytogenetics and Cell Genetics 63: 156–159. https://doi.org/10.1159/000133525 [DOI] [PubMed] [Google Scholar]
- Mattos TL, Coelho AC, Schneider CH, Telles DOC, Menin M, Gross MC. (2014) Karyotypic diversity in seven Amazonian anurans in the genus Hypsiboas (family Hylidae). BMC Genetics 15: 43. https://doi.org/10.1186/1471-2156-15-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros LR, Rossa-Feres DC, Recco-Pimentel SM. (2003) Chromosomal differentiation of Hyla nana and Hyla sanborni (Anura, Hylidae) with a description of NOR polymorphism in H. nana. Journal of Heredity 94: 149–154. https://doi.org/10.1093/jhered/esg019 [DOI] [PubMed] [Google Scholar]
- Medeiros LC, Lourenço LB, Rossa-Feres DC, Lima AP, Andrade GV, Giaretta AA, Egito GTBT, Recco-Pimentel SM. (2013) Comparative cytogenetic analysis of some species of the Dendropsophus microcephalus group (Anura, Hylidae) in the light of phylogenetic inferences. BMC Genetics 14: 59 https://doi.org/10.1186/1471-2156-14-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyne J, Baker RJ, Hobart HH, Hsu TC, Ryder OA, Ward OG, Wiley JE, Wurster-Hill DH, Yates TL, Moyzis RK. (1990) Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 99: 3–10. https://doi.org/10.1007/BF01737283 [DOI] [PubMed] [Google Scholar]
- Nanda I, Schrama D, Feichtinger W, Haaf T, Schartl M, Schmid M. (2002) Distribution of telomeric (TTAGGG)n sequences in avian chromosomes. Chromosoma 111: 215–227. https://doi.org/10.1007/s00412-002-0206-4 [DOI] [PubMed] [Google Scholar]
- Nergadze SG, Rocchi M, Azzalin CM, Mondello C, Giulotto E. (2004) Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Research 14: 1704–1710. https://doi.org/10.1101/gr.2778904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze SG, Santagostino MA, Salzano A, Mondello C, Giulotto E. (2007) Contribution of telomerase RNA retrotranscription to DNA double-strand break repair during mammalian genome evolution. Genome Biology 8: R260 https://doi.org/10.1186/gb-2007-8-12-r260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocalewicz K, Furgala-Selezniow G, Szmyt M, Lisboa R, Kucinski M, Lejk AM, Jankun M. (2013) Pericentromeric location of the telomeric DNA sequences on the European grayling chromosomes. Genetica 141: 409–416. https://doi.org/10.1007/s10709-013-9740-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IS, Noleto RB, Oliveira AKC, Toledo LF, Cestari MM. (2016) Comparative cytogenetic analysis of four species of Dendropsophus (Hylinae) from the Brazilian Atlantic forest. Journal of Genetics 95: 349–355. https://doi.org/10.1007/s12041-016-0645-y [DOI] [PubMed] [Google Scholar]
- Orrico VGD, Duellman WE, Souza MB, Haddad CFB. (2013) The taxonomic status of Dendropsophus allenorum and D. timbeba (Anura: Hylidae). Journal of Herpetology 46: 615–618. https://doi.org/10.1670/12-208 [Google Scholar]
- Orrico VGD, Peloso PLV, Sturaro MJ, Silva-Filho HF, Neckel-Oliveira S, Gordo M, Faivovich J, Haddad CFB. (2014) A new “Bat-Voiced” species of Dendropsophus Fitzinger, 1843 (Anura, Hylidae) from the Amazon Basin, Brazil. Zootaxa 3881: 341–361. https://doi.org/10.11646/zootaxa.3881.4.3 [DOI] [PubMed] [Google Scholar]
- Ortega-Andrade HM, Ron SR. (2013) A new species of small tree frog, genus Dendropsophus (Anura: Hylidae) from the eastern Amazon lowlands of Ecuador. Zootaxa 3652: 163–178. https://doi.org/10.11646/zootaxa.3652.1.6 [DOI] [PubMed] [Google Scholar]
- Paço A, Chaves R, Vieira-da-Silva A, Adega F. (2013) The involvement of repetitive sequences in the remodelling of karyotypes: The Phodopus genomes (Rodentia, Cricetidae). Micron 46: 27–34. https://doi.org/10.1016/j.micron.2012.11.010 [DOI] [PubMed] [Google Scholar]
- Palumbi SR, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. (1991) The simple fool’s guide to PCR, Version 2.0. Privately published document compiled by Palumbi SR. Special Publication of Department of Zoology, University of Hawaii, Honolulu, USA, 44 pp. [Google Scholar]
- Peloso PLV, Orrico VGD, Haddad CFB, Lima-Filho GR, Sturaro MJ. (2016) A new species of clown tree frog Dendropsophus leucophyllatus species group, from Amazonia (Anura, Hylidae). South American Journal of Herpetology 11: 66–80. https://doi.org/10.2994/SAJH-D-16-00003.1 [Google Scholar]
- Plohl M, Luchetti A, Meštrović N, Mantovani B. (2008) Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409: 72–82. https://doi.org/10.1016/j.gene.2007.11.013 [DOI] [PubMed] [Google Scholar]
- Pyron RA, Wiens JJ. (2011) A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Molecular Phylogenetics and Evolution 61: 543–583. https://doi.org/10.1016/j.ympev.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Rivera-Correa M, Orrico VGD. (2013) Description and phylogenetic relationships of a new species of treefrog of the Dendropsophus leucophyllatus group (Anura: Hylidae) from the Amazon basin of Colombia and with an exceptional color pattern. Zootaxa 3686: 447–460. https://doi.org/10.11646/zootaxa.3686.4.3 [DOI] [PubMed] [Google Scholar]
- Ropiquet A, Hassanin A, Pagacova E, Gerbault-Seureau M, Cernohorska H, Kubickova S, Bonillo C, Rubes J, Robinson TJ. (2010) A paradox revealed: karyotype evolution in the four-horned antelope occurs by tandem fusion (Mammalia, Bovidae, Tetracerus quadricornis). Chromosome Research 18: 277–286. https://doi.org/10.1007/s10577-010-9115-1 [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Kratochvíl L, Altmanová M, Pokorná MJ. (2015) Interstitial telomeric motifs in squamate reptiles: when the exceptions outnumber the rule. PLoS ONE 10: e0134985 https://doi.org/10.1371/journal.pone.0134985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Nergadze SG, Santagostino M, Giulotto E. (2008) Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenetic and Genome Research 122: 219–228. https://doi.org/10.1159/000167807 [DOI] [PubMed] [Google Scholar]
- Schmid M, Steinelin C. (2016) Chromosome Banding in Amphibia. XXXIV. Intrachromosomal Telomeric DNA Sequences in Anura. Cytogenetic and Genome Research 148: 211–226. https://doi.org/10.1159/000446298 [DOI] [PubMed] [Google Scholar]
- Slijepcevic P. (1998) Telomeres and mechanisms of Robertsonian fusion. Chromosoma 107: 136–140. https://doi.org/10.1007/s004120050289 [DOI] [PubMed] [Google Scholar]
- Suárez P, Cardozo D, Baldo D, Pereyra MO, Faivovich J, Orrico VGD, Catroli GF, Grabiele M, Bernarde PS, Nagamachi CY, Haddad CFB, Pieczarka JC. (2013) Chromosome evolution in Dendropsophini (Amphibia, Anura, Hylinae). Cytogenetic and Genome Research 141: 295–308. https://doi.org/10.1159/000354997 [DOI] [PubMed] [Google Scholar]
- Sumner AT. (1972) A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 75: 304–306. https://doi.org/10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Tek AL, Jiang J. (2004) The centromeric regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similar sequence. Chromosoma 113: 77–83. https://doi.org/10.1007/s00412-004-0297-1 [DOI] [PubMed] [Google Scholar]
- Titus TA, Larson A. (1996) Molecular phylogenetics of desmognathine salamanders (Caudata: Plethodontidae): a reevaluation of evolution in ecology, life history, and morphology. Systematic Biology 45: 229–238. https://doi.org/10.1093/sysbio/45.4.451 [Google Scholar]
- Wiley JE, Meyne J, Little ML, Stout JC. (1992) Interstitial hybridization sites of the (TTAGGG)n telomeric sequence on the chromosomes of some North American hylid frogs. Cytogenetics and Cell Genetics 61: 55–57. https://doi.org/10.1159/000133368 [DOI] [PubMed] [Google Scholar]
- Wood AM, Rendtlew Danielsen JM, Lucas CA, Rice EL, Scalzo D, Shimi T, Goldman RD, Smith ED, Le Beau MM, Kosak ST. (2014) TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nature Communications 5: 5467 https://doi.org/10.1038/ncomms6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, Laster K, Rice EL, Kosak ST. (2015) A beginning of the end: new insights into the functional organization of telomeres. Nucleus 6: 172–178. https://doi.org/10.1080/19491034.2015.1048407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MJ, O’Meally D, Sarre SD, Georges A, Ezaz T. (2013) Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Research 21: 361–374. https://doi.org/10.1007/s10577-013-9362-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
16S rDNA sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Lívia S. R. Teixeira, Karin Regina Seger, Cíntia Pelegrineti Targueta, Victor G. Dill Orrico, Luciana Bolsoni Lourenço
Data type: Tif file
Explanation note: Alignment of 16S rDNA fragments obtained from specimens of Dendropsophus seniculus, Dendropsophus novaisi and Dendropsophus soaresi that were used in cytogenetic analyses. GenBank accession numbers: KY053469 (Dendropsophus seniculus), KY053470 (Dendropsophus novaisi), and KY053471 (Dendropsophus soaresi).