Abstract
There is controversy concerning the role of genetic factors in species extinctions. Many authors have asserted that species are usually driven to extinction before genetic factors have time to impact them, but few studies have seriously addressed this issue. If this assertion is true, there will be little difference in genetic diversity between threatened and taxonomically related nonthreatened species. We compared average heterozygosities in 170 threatened taxa with those in taxonomically related nonthreatened taxa in a comprehensive metaanalysis. Heterozygosity was lower in threatened taxa in 77% of comparisons, a highly significant departure from the predictions of the no genetic impact hypothesis. Heterozygosity was on average 35% lower (median 40%) in threatened taxa than in related nonthreatened ones. These differences in heterozygosity indicate lowered evolutionary potential, compromised reproductive fitness, and elevated extinction risk in the wild. Independent evidence from stochastic computer projections has demonstrated that inbreeding depression elevates extinction risk for threatened species in natural habitats when all other threatening processes are included in the models. Thus, most taxa are not driven to extinction before genetic factors affect them adversely.
There is controversy about the impact of genetic factors on extinction risk for threatened species and populations in nature (1). Species population sizes are reduced by habitat loss, overexploitation, impact of introduced species, and pollution until they reach a point where stochastic factors further elevate extinction risk (2). Stochastic factors encompass demographic, environmental, and genetic stochasticity and natural catastrophes. Threatened species typically have small and/or declining populations, such that inbreeding and loss of genetic diversity are unavoidable. In random mating populations, neutral genetic variation is lost and inbreeding accumulates, as follows:
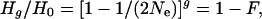 |
1 |
where Hg is the heterozygosity at generation g, H0 the initial heterozygosity, Ne the long-term effective population size, and F the inbreeding coefficient (1). Inbreeding reduces reproduction and survival in essentially all well studied species (1, 3), reduced population heterozygosity is associated with reduced population reproductive fitness (4), and inbreeding depression increases extinction risk (5). Further, loss of genetic diversity reduces the ability of populations to evolve to cope with environmental change (1, 6). Thus, reduced heterozygosity is a marker of populations with reduced reproductive fitness and an elevated risk of future extinction caused by genetic factors, irrespective of the cause of the initial decline.
However, in an influential review, Lande (7) argued that “demography may usually be of more immediate importance than population genetics in determining the minimum viable size of wild populations.” This argument has been widely interpreted to mean that ecological and demographic factors would typically drive threatened populations to extinction before genetic factors had time to impact them adversely (8-14). Although Lande (15) subsequently modified his views, it was not a retraction of the “no genetic impact” scenario, but a consequence of his later view that mutational accumulation contributes substantially to extinction risk.
Two studies have reported adverse genetic impacts on extinction risk in populations in the wild (16, 17), and the generality of the no genetic impact hypothesis (7-14) has been questioned (1). Inbreeding and reduced genetic diversity were associated with elevated extinction risk in wild butterfly populations (16), and extinction rates were markedly higher in populations of the plant Clarkia pulchella with higher versus lower inbreeding (17). Do these studies indicate that genetic factors usually contribute to extinctions, or are they special cases? It is critical to resolve this issue, so that threatened taxa can be managed appropriately.
Evaluating comprehensively the role of genetics in extinction for a diversity of taxa by experiments on wild populations would be an enormous task and quite impractical in the short term. Furthermore, conservation biology is a crisis discipline where it is not reasonable or practicable to wait for data collection before making decisions (18). However, a comparison of published data on genetic diversity in threatened and related nonthreatened taxa will provide an overall perspective, as threatened taxa are considered to be on the path to extinction. If the no genetic impact hypothesis (7-14) is correct there should be little difference in genetic diversity between threatened and taxonomically related nonthreatened taxa. Conversely, if most threatened taxa do indeed show less genetic diversity than related nonthreatened taxa, then this is strong evidence that genetic factors are adversely impacting these taxa.
Methods
We carried out a comprehensive metaanalysis to examine this hypothesis by using the internationally recognized IUCN-The World Conservation Union Red List threatened categorization system (19) that comprises critically endangered, endangered, and vulnerable taxa and applied it to identify threatened species and subspecies and taxonomically related nonthreatened taxa. Additional analyses were done on other IUCN-listed categories of extinct, extinct in the wild, lower risk, and data deficient. Generally, pairs of taxa were from the same genus or family, but some were at the class level. Analyses were done on percentage difference in heterozygosity between threatened and the nearest related nonthreatened species or group of species, based on data for allozymes, microsatellites, and minisatellites (paired comparisons only involved the same markers). Data from listed species were paired with data of the same type (either expected or observed heterozygosity, and either from allozymes or microsatellites or minisatellites) from the most closely related nonlisted species or the weighted (according to sample size) average of the most closely related species. Generally, pairs were from the same genus or family, but some were at the class level. If expected Hardy-Weinberg heterozygosity and observed heterozygosity were both available, the expected was used as it is least affected by the size of the sample (20). If allozyme and microsatellite pairs were available for the same taxon, the combined weighted (measure × no. sampled × no. of loci tested) average was used. Table 2, which is published as supporting information on the PNAS web site, contains full details of the threatened taxa, the related nonthreatened taxa, and the sources of data.
Major Taxa. Major taxa were delineated according to the amount of data obtained. Thus, plants were subdivided into gymnosperms and angiosperms. As there were more data, animals were categorized as invertebrates (no further subdivision) and vertebrates, which were further subdivided into poikilotherms (fish, amphibia, and reptiles), homeotherms, birds, and mammals.
Data Analyses. The null hypothesis was that the genetic diversity of threatened taxa does not differ from that of nonthreatened taxa. The alternative hypothesis was that threatened taxa have less genetic diversity than comparable nonthreatened taxa. Thus, the statistical tests were one-tailed.
The common metric used was the percentage difference in heterozygosity. When a nonthreatened taxon had zero heterozygosity, this percentage was infinite. In simulations using the data set, we found that the use of only the nonthreatened taxon as the divisor gave biased estimates of the true differences, but the use of the larger of the threatened or nonthreatened heterozygosities as the divisor gave unbiased estimates of the true differences. Consequently, we used the larger measure of heterozygosity of each pair as the denominator [100 × (nonthreatened - threatened)/nonthreatened or 100 × (nonthreatened - threatened)/threatened]. The larger heterozygosity is more likely to represent the former heterozygosity for the taxon. Percentage difference in heterozygosity has a firm theoretical and conservation basis and is also interpretable as the effective inbreeding coefficient (1). The use of nonthreatened heterozygosities as the divisor throughout does not alter the conclusions.
Because the data are not normally distributed, nonparametric Wilcoxon's signed rank tests were performed on the difference in heterozygosity of each threatened taxon compared with the most closely related available taxon or taxa not included in the IUCN Red Lists. We tested for differences among the different Red List categories (critically endangered, endangered, vulnerable, lower risk, and data deficient) by using Kruskal-Wallis tests. For this test, data on extinct plus extinct in the wild (combined), lower-risk, and data-deficient taxa were added. The distribution of listed species with lower heterozygosity than closely related nonlisted species among major taxa and among IUCN Red List categories were investigated by using contingency χ2 and Kruskal-Wallis tests.
As publication bias (file drawer effect) may affect the conclusions of metaanalyses (21), we tested for bias by regressing percentage difference between threatened and nonthreatened taxa on W (sample size × no. of loci) separately for allozymes and microsatellites, based on Palmer's recommendation (22).
Another test of the file drawer effect involved adding negative percentage differences to the data set until the results of the Wilcoxon's signed rank test became nonsignificant. This number indicates the minimum number of data points that must have remained unpublished for the results to be nonsignificant and can be compared with the number analyzed to indicate the existence or potential magnitude of this confounding problem.
All statistical analyses were carried out by using the minitab statistical package (release 13, Minitab, State College, PA).
Results
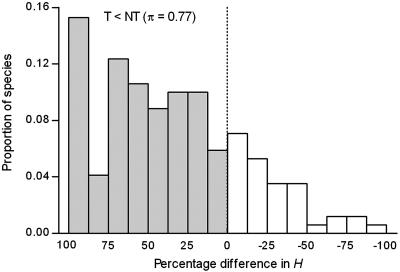
Overall, 77% of the 170 threatened taxa had lower heterozygosity than related nonthreatened taxa (Ht < Hnt), a highly significant deviation from equality as predicted by the no genetic impact hypothesis (7-14) (Table 1). Differences were significant for both allozyme and microsatellite data, and the two did not differ (see Supporting Text, which is published as supporting information on the PNAS web site). The distribution of percentage differences in heterozygosity is shown in Fig. 1, the median difference being 40% (mean, 35%).
Table 1. Percentages of threatened taxa with lower heterozygosity than taxonomically related nonthreatened taxa (Ht < Hnt) in a range of major taxa and the magnitudes of those differences.
| Taxon | Ht < Hnt, % | Median difference, % | Mean difference, % | n | P |
|---|---|---|---|---|---|
| All | 77 | 40 | 35 | 170 | <0.0005 |
| Animals | 78 | 38 | 35 | 134 | <0.0005 |
| Vertebrates | 78 | 35 | 35 | 129 | <0.0005 |
| Homeotherms | 81 | 43 | 40 | 94 | <0.0005 |
| Mammals | 84 | 46 | 42 | 63 | <0.0005 |
| Birds | 74 | 40 | 35 | 31 | 0.001 |
| Poikilotherms | 69 | 26 | 20 | 35 | 0.001 |
| Invertebrates | 80 | 67 | 37 | 5 | 0.140 |
| Plants | 75 | 57 | 38 | 36 | <0.0005 |
| Angiosperms | 81 | 58 | 40 | 21 | 0.005 |
| Gymnosperms | 67 | 51 | 35 | 15 | 0.012 |
n, Number of threatened taxa; P, probabilities based on Wilcoxon's signed rank tests.
Fig. 1.
Distribution of percentage differences in heterozygosity (H) between threatened (T) and taxonomically related nonthreatened taxa (NT). π is the proportion of taxa for which T < NT, indicated by the shaded bars.
The proportions of threatened taxa with Ht < Hnt did not differ among major taxa (χ2 = 5.4, df = 5, P = 0.366). All major taxa with sufficient sample sizes showed a significant majority of threatened taxa with Ht < Hnt (Table 1). The average magnitudes of the differences were also similar across taxa.
There were no indications of selective reporting bias in the data set. Regressions of percentage difference in heterozygosity between threatened and nonthreatened taxa on W (sample size × no. of loci) were nonsignificant for both allozymes (b = 0.003, t = 1.60, df = 122, P = 0.11, and r2 = 1.3%) and microsatellites (b = -0.006, t = -1.40, df = 49, P = 0.17, and r2 = 1.9%). Further details and additional analyses are given in Supporting Text and Table 3, which is published as supporting information on the PNAS web site.
Discussion
A significant majority of threatened taxa in all major taxa with more than five data points showed lower genetic diversity than that in taxonomically related nonthreatened taxa. This finding is in conflict with the predictions of the no genetic impact hypothesis (7-14). Our results also refute the prediction that threatened mammals will show a difference in heterozygosity of <5% (23). The median and mean differences were 40% and 35%, respectively, vastly greater than the prediction. Prior studies in plants have noted lowered genetic diversity in rare versus common species and negative associations between genetic diversity and range size (24, 25), but did not connect these with endangerment and the no genetic impact hypothesis.
Taxa currently showing no adverse genetic impacts may still experience genetic impacts before extinction. For example, vulnerable taxa have approximately a 10% probability of extinction within 100 years (19), ample time for genetic impacts.
Are the reduced genetic diversities we found of sufficient magnitude to reflect adverse genetic impacts and elevated extinction risks? We were unable to determine whether genetic factors have contributed to the current threatened status of the taxa in our study. However, reduced genetic diversity is a marker indicating that their reproductive fitness is already compromised and that their subsequent extinction risk is elevated. Each of the essential links between reduced genetic diversity and subsequent extinction risk has been verified. First, reduced genetic diversity has been shown to reduce times to extinction under changing environments (1, 6). Second, from Eq. 1 the difference in heterozygosity is a measure of the inbreeding coefficient of a taxon. As loss of reproductive fitness is related to the inbreeding coefficient, a positive correlation between heterozygosity and population fitness is predicted and has been verified (4). Inbreeding depression has been shown to increase extinction risk in laboratory and wild populations (1, 16, 17, 26-28). The 40% median percentage reduction in genetic diversity between threatened and nonthreatened taxa corresponds to an inbreeding coefficient where deliberately inbred laboratory populations show elevated extinction risks (1, 29, 30), and inbreeding depression has greater impact in more stressful natural environments than in benign captive environments (3, 27, 28).
Third, computer projections demonstrate that inbreeding depression adversely affects the extinction risk of threatened species in the wild even when all other demographic, environmental, and catastrophic factors are operating (5). Computer projections using data for 20 threatened species showed 24-31% reductions in median times to extinction when inbreeding depression for juvenile survival was included in the models, compared to simulations where inbreeding depression was omitted. This result is conservative as inbreeding depression of only 3.14 diploid lethal equivalents for juvenile survival was applied, whereas actual levels in the wild are ≈12 lethal equivalents spread over the full life cycle (31). With the latter level of inbreeding depression, there is a 78% projected reduction in median time to extinction (unpublished data). In addition, small natural populations of a topminnow fish, a greater prairie chicken, and a Swedish adder all have declined in numbers, in part because of inbreeding, and recovered after outbreeding (32-34). Thus, our results refute the view that species are typically driven to extinction before genetic factors have time to impact them.
It is not possible given current knowledge to answer with precision the question of when the genetic effects of lowered diversity are of sufficient magnitude that they must be directly managed. The answer will depend on the inbreeding coefficient and thus on effective population size and number of generations, as indicated by Eq. 1. Inbreeding levels where impacts will be important will be somewhat lower when the prior rate of inbreeding is lower and the potential for purging higher (1). It is also likely to vary among species, particularly in relation to their population growth rate (5). With rapid environmental change, the levels of inbreeding and loss of genetic diversity where adverse genetic impacts are expected will be lower than for stable environments (1, 27, 28). Estimated times to extinction for different-sized housefly populations in a benign captive environment approximated the effective sizes in generations (1).
Why does the no genetic impact hypothesis (7-14) not apply to most threatened taxa? There are four factors where subsequent information has changed perceptions since Lande's 1988 paper (7) in ways that would have led to underestimates of the impact of genetic factors. First, ratios of effective population size to census size have subsequently been found to average 0.11 (35), much lower than assumed in 1988. For example, in 1991 Mace and Lande (36) assumed that the ratio was 0.2-0.5. Thus, inbreeding and loss of genetic diversity occur at a substantially greater rate than Lande would have assumed. Second, impacts of interactions between genetic and other stochastic factors may have been underestimated. Fluctuations in population size caused by environmental stochasticity and catastrophes reduce the effective population size and increase the rates of inbreeding and loss of genetic diversity (37). Third, information on inbreeding depression for the full life cycle in natural environments was very limited in 1988, so its impacts are likely to have been underestimated, based on data from captive populations. The most prominent data at that time reported 3.14 lethal equivalents for juvenile mortality in captive mammals (38), whereas the full impact of inbreeding depression in the wild has more recently been reported as almost 4 times that (31). Fourth, Lande (7) considered that natural selection was highly effective in purging deleterious alleles under slow rates of inbreeding, but purging has subsequently been found to have relatively small effects (1, 30, 39).
In conclusion, most threatened taxa have lower genetic diversity than closely related nonthreatened taxa, indicating reduced reproductive fitness and elevated extinction risks. Consequently, our results are not compatible with the hypothesis that most species are driven to extinction before genetic factors impact them.
Acknowledgments
We thank F. Allendorf, J. Barker, A. Beattie, L. Beheregaray, K. Belov, M. Burgman, T. Caro, D. Colgan, R. Crozier, M. Eldridge, P. England, M. Festa-Bianchet, S. Haig, I. Hanski, D. Hartl, A. Hoffmann, C. Johnson, L. Lim, L. Mills, F. Nicholas, K. Paige, J. Pemberton, S. Pimm, R. Primack, M. Shaffer, M. Soulé, R. Vrijenhoek, R. Waples, E. Wilson, and T. Young for helpful comments.
Author contributions: D.S., B.W.B., and R.F. designed research; D.S. and R.F. performed research; D.S., B.W.B., and R.F. analyzed data; and D.S., B.W.B., and R.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ht, average heterozygosity in threatened taxa; Hnt, average heterozygosity in taxonomically related nonthreatened taxa.
References
- 1.Frankham, R., Ballou, J. D. & Briscoe, D. A. (2002) Introduction to Conservation Genetics (Cambridge Univ. Press, Cambridge, U.K).
- 2.Shaffer, M. L. (1981) BioScience 31, 131-134. [Google Scholar]
- 3.Crnokrak, P. & Roff, D. A. (1999) Heredity 83, 260-270. [DOI] [PubMed] [Google Scholar]
- 4.Reed, D. H. & Frankham, R. (2003) Conserv. Biol. 17, 230-237. [Google Scholar]
- 5.Brook, B. W., Tonkyn, D. W., O'Grady, J. J. & Frankham, R. (June 27, 2002) Conserv. Ecol. 6, www.consecol.org/vol6/iss1/art16.
- 6.Frankham, R., Lees, K., Montgomery, M. E., England, P. R., Lowe, E. & Briscoe, D. A. (1999) Anim. Conserv. 2, 255-260. [Google Scholar]
- 7.Lande, R. (1988) Science 241, 1455-1460. [DOI] [PubMed] [Google Scholar]
- 8.Pimm, S. L. (1991) The Balance of Nature: Ecological Issues in the Conservation of Species and Communities (Univ. of Chicago Press, Chicago).
- 9.Young, T. P. (1991) Nature 352, 10. [Google Scholar]
- 10.Wilson, E. O. (1992) The Diversity of Life (Harvard Univ. Press, Cambridge, MA).
- 11.Caro, T. M. & Laurenson, M. K. (1994) Science 263, 485-486. [DOI] [PubMed] [Google Scholar]
- 12.Caughley, G. (1994) J. Anim. Ecol. 63, 215-244. [Google Scholar]
- 13.Dobson, A. P. (1999) in Genetics and the Extinction of Species: DNA and the Conservation of Biodiversity, eds. Landweber, L. F. & Dobson, A. P. (Princeton Univ. Press, Princeton), pp. xiii-xviii.
- 14.Elgar, M. A. & Clode, D. (2001) Conserv. Biol. 15, 284-286. [Google Scholar]
- 15.Lande, R. (1995) Conserv. Biol. 9, 782-791. [Google Scholar]
- 16.Saccheri, I., Kuussaari, M., Kankare, M., Vikman, P., Fortelius, W. & Hanski, I. (1998) Nature 392, 491-494. [Google Scholar]
- 17.Newman, D. & Pilson, D. (1997) Evolution (Lawrence, Kans.) 51, 354-362. [DOI] [PubMed] [Google Scholar]
- 18.Soulé, M. E. (1985) BioScience 35, 727-734. [Google Scholar]
- 19.Species Survival Commission, IUCN-The World Conservation Union (2000) The 2000 Red List of Threatened Species (IUCN-The World Conservation Union, Gland, Switzerland).
- 20.Nei, M. (1978) Genetics 89, 583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Møller, A. P. & Jennions, M. D. (2001) Trends Ecol. Evol. 16, 580-586. [DOI] [PubMed] [Google Scholar]
- 22.Palmer, A. R. (2000) Annu. Rev. Ecol. Syst. 31, 441-480. [Google Scholar]
- 23.Amos, W. & Balmford, A. (2001) Heredity 87, 257-265. [DOI] [PubMed] [Google Scholar]
- 24.Karron, J. D. (1991) in Genetics and Conservation of Rare Plants, eds. Falk, D. A. & Holsinger, K. E. (Oxford Univ. Press, New York), pp. 87-98.
- 25.Hamrick, J. L. & Godt, M. J. W. (1989) in Plant Population Genetics, Breeding, and Genetic Resources, eds. Brown, A. H. D., Clegg, M. T., Kahler, A. L. & Weir, B. S. (Sinauer, Sunderland, MA), pp. 43-63.
- 26.Bijlsma, R., Bundgaard, J. & Van Putten, W. F. (1999) J. Evol. Biol. 12, 1125-1137. [Google Scholar]
- 27.Bijlsma, R., Bundgaard, J. & Boerema, A. C. (2000) J. Evol. Biol. 13, 502-514. [Google Scholar]
- 28.Reed, D. H., Briscoe, D. A. & Frankham, R. (2002) Conserv. Genet. 3, 301-307. [Google Scholar]
- 29.Frankham, R. (1998) Conserv. Biol. 12, 665-675. [Google Scholar]
- 30.Reed, D. H., Lowe, E., Briscoe, D. A. & Frankham, R. (2003) Conserv. Genet. 4, 405-410. [Google Scholar]
- 31.Keller, L. F. (1998) Evolution (Lawrence, Kans.) 52, 240-250. [Google Scholar]
- 32.Vrijenhoek, R. C. (1994) in Conservation Genetics, eds. Loeschcke, V., Tomiuk, J. & Jain, S. K. (Birkhäuser, Basel), pp. 37-53.
- 33.Westemeier, R. L., Brawn, J. D., Simpson, S. A., Esker, T. L., Jansen, R. W., Walk, J. W., Kershner, E. L., Bouzat, J. L. & Paige, K. N. (1998) Science 282, 1695-1698. [DOI] [PubMed] [Google Scholar]
- 34.Madsen, T., Shine, R., Olsson, M. & Wittzell, H. (1999) Nature 402, 34-35. [Google Scholar]
- 35.Frankham, R. (1995) Genet. Res. 66, 95-107. [Google Scholar]
- 36.Mace, G. M. & Lande, R. (1991) Conserv. Biol. 5, 148-157. [Google Scholar]
- 37.van Noordwijk, A. J. (1994) in Conservation Genetics, eds. Loeschcke, V., Tomiuk, J. & Jain, S. K. (Birkhäuser, Basel), pp. 131-146.
- 38.Ralls, K., Ballou, J. D. & Templeton, A. (1988) Conserv. Biol. 2, 185-193. [Google Scholar]
- 39.Byers, D. L. & Waller, D. M. (1999) Annu. Rev. Ecol. Syst. 30, 479-513. [DOI] [PubMed] [Google Scholar]