Abstract
The 37-kDa laminin receptor (37LRP or RPSA) is a remarkable, multifaceted protein that functions in processes ranging from matrix adhesion to ribosome biogenesis. Its ability to engage extracellular laminin is further thought to contribute to cellular migration and invasion. Most commonly associated with metastatic cancer, RPSA is also increasingly found to be important in other pathologies, including microbial infection, neurodegenerative disease and developmental malformations. Importantly, it is thought to have higher molecular weight forms, including a 67-kDa species (67LR), the expression of which is linked to strong laminin binding and metastatic behavior. The composition of these larger forms has remained elusive and controversial. Homo- and heterodimerization have been proposed as events capable of building the larger species from the monomeric 37-kDa precursor, but solid evidence is lacking. Here, we present data suggesting that higher molecular weight species require SUMOylation to form. We also comment on the difficulty of isolating larger RPSA species for unambiguous identification and demonstrate that cell lines stably expressing tagged RPSA for long periods of time fail to produce tagged higher molecular weight RPSA. It is possible that higher molecular weight species like 67LR are not derived from RPSA.
Keywords: 37 67 laminin receptor, RPSA, 67LR, 37LRP, Non-integrin laminin receptor, LAMR1, p40, SUMO
INTRODUCTION
The 37-kDa laminin receptor or ribosomal protein SA (RPSA) is an RPS2-family ribosomal subunit required for protein synthesis [1] and ribosome biogenesis [2]. In addition to its importance for translation, cellular viability and ribosomal RNA (rRNA) processing [3], it is thought to have a secondary function as a laminin receptor [4].
Laminins are a family of extracellular matrix proteins, long known for their involvement in cellular adhesion and migratory processes [5]. RPSA was discovered early in the search for proteins that mediate laminin signals thanks to its association with the related 67-kDa laminin receptor (67LR) [6]. This 67-kDa protein is presumed to be a higher molecular weight form of RPSA with a significantly greater affinity for laminin [7]. Understanding the relationship between RPSA and 67LR may be critical for unraveling the contribution of RPSA to laminin binding, tissue development [8, 9], and the malignant properties of cancer cells [10–16]. The importance of this relationship may also extend to microbial infection and neurodegeneration: RPSA has been implicated as a receptor for several pathogenic agents, such as bacteria [17, 18], viruses [19, 20] and the toxic amyloids of Alzheimer’s disease [21, 22] and prion encephalopathies [23, 24].
Because a precursor relationship is thought to exist between the 37-kDa RPSA and 67LR [7, 25], RPSA has the alternate name 37-kDa laminin receptor precursor protein (37LRP). Full protein sequencing of 67LR has not been reported, but partial sequencing identified a single peptide (MLAREVLR) that corresponds to amino acids 177–184 of RPSA [25]. Additionally, a number of antibodies raised against either 67LR or RPSA occasionally cross-react [25–27]. Although a relationship between RPSA and 67LR is generally accepted, there is no conclusive description of the transition. This issue has recently been reviewed in detail [14].
Attempts to demonstrate that 67LR is indeed derived from RPSA have generated two lines of evidence. The first describes the appearance of a faint 67-kDa signal coinciding with a decrease in the 37-kDa signal after immunoprecipitation of RPSA from radiolabeled cells [28]. A second report recognized that treatment of cells with cerulenin, a fatty acid synthase inhibitor, reduces the presence of 67LR with a subsequent increase in RPSA, suggesting acylation is required to maintain 67LR [29, 30].
Speculation about the nature of the 37- to 67-kDa transition has typically focused on a homo- or heterodimerization event. The evidence for either scenario is limited to dated amino acid composition analysis [30] and crossreactivity with lectin-targeted antibodies [29, 31]. Alternative splicing does not appear to be a satisfactory explanation given that a single mRNA transcript is generated from the RPSA gene [32].
Recently, fluorescence complementation was used to demonstrate that RPSA may be capable of homo- and heterodimerizing (with galectin-3) [33]. It is not clear if these dimers represent 67LR because they may not be stable under denaturing conditions, which 67LR is known to resist [30]. Dimerization is also contradicted by the failure of RPSA (albeit truncated) to self-associate in 2-hyrbid and pull-down systems [34]. Skepticism regarding whether 67LR is truly derived from RPSA is warranted, as a conclusive demonstration of the relationship is lacking.
Though post-translational modifications – specifically acylation – are apparently necessary for the 37 to 67 transition, they are considered insufficient because of their low molecular weight contributions [30]. However, there are modifications that can confer significant size to their targets. Ubiquitin-like proteins (UBLs) such as SUMO can add 8–20 kDa of mass to proteins and endure under denaturing conditions owing to their covalent attachment [35]. Importantly, ubiquitin has already been shown to modify RPSA as part of a laminin-dependent system to regulate the presence of 67LR at the cell membrane [36]. SUMOylation is especially interesting as it has crosstalk with the ubiquitin degradation system and can also affect protein trafficking and protein–protein interactions [37, 38]. Modification by UBLs may thus have significant explanatory power in describing the transition of RPSA to higher molecular weight species, the stability of 67LR, laminin binding, and its contribution to cell migration and metastasis.
Here, we present evidence that higher molecular weight species of RPSA are built via SUMOylation. However, we also discuss the difficulty in directly identifying these larger forms and the failure of affinity-tagged RPSA to produce them.
MATERIALS AND METHODS
Cell culture, vectors and transfection
HT1080, HeLa and NIH 3T3 cells were obtained from ATCC and maintained according to supplier recommendations. All media were supplemented with 10% FBS, a 100 mg/ml penicillin–streptomycin cocktail and 0.5 mg/ml amphotericin B. RPSAFLAG stable cell lines were derived using pcDNA3.1v5/His vectors (Life Technologies) containing N- or C-terminally DYKDDDDK-tagged RPSA and neomycin resistance markers. Selection was with G418 sulfate (Corning). The cells were maintained under selection except for the 24 h prior to protein harvesting or transfection. The pcDNA3-HA-SUMO1, 2 and 3 vectors and their non-cleavable mutants [39] were donated by Miklos Békés of New York University.
Transfection was performed with Lipofectamine 2000 (Life Technologies). siRNA was transfected using DharmaFECT-4 (Dharmacon) at 50 nmol/l. Human and mouse UBC9-targeted siRNAs were from Santa Cruz Biotechnology. RPSA and RISC-free control (siGlo) siRNAs were from Dharmacon. For sustained knockdown (5–8 days), cells were re-transfected once 72 h after initial transfection.
Reagents and antibodies
Whole cell lysates were prepared with M-PER (Thermo Scientific) supplemented with 150 mM NaCl, 1 mM EDTA and 25–50 mM N-ethylmaleimide (NEM) in the presence of protease inhibitors (Roche). When indicated, 10 or 1 μM MG-132 (Sigma-Aldrich) was used to treat cells for 7 h or overnight, respectively. Cellular fractionation was performed with Qproteome (Qiagen) using one-tenth the recommended buffer volumes. The following antibodies were used: RPSA/Laminin-R H-141 (Santa Cruz Biotechnology); UBC9 D26F2 and HA-tag 6E2 (Cell Signaling Technology); and FLAG M2 (Sigma-Aldrich). RPSA antibody 4099-1 was produced as in [7].
In vitro SUMOylation
In vitro SUMOylation was performed with SUMOlink (Active Motif) and IV SUMOylation (Enzo Life Sciences) kits using recombinant RPSA (full length, 1–295) purified with nickel affinity chromatography and subsequent Superdex 75 (GE Healthcare) gel filtration as previously described [40]. 1 μg of target protein was used for the reactions.
Immunoprecipitation
Protein lysates were incubated with 10 μg of capturing antibody for 2 h at 4ºC. The mixture was added to Protein G DynaBeads (Life Technologies) and left overnight at 4ºC. After three washes with PBS+0.05% Tween-20, captured proteins were eluted with 0.1 M glycine (pH 3.5) or via boiling in protein sample buffer. For capture of FLAG- or HA-tagged proteins, FLAG M2 (Sigma-Aldrich) or HA (Thermo Scientific) antibody-conjugated magnetic beads were used as above with elution carried out using 150 ng/μl FLAG or 2 mg/ml HA peptides. Protein G DynaBeads (Life Technologies) loaded with normal mouse IgG were used as the negative controls.
Quantitative PCR
RNA from cell lines was extracted from cells using an RNAeasy kit (Qiagen). 1 μg was used for reverse transcription (iScript, BioRad). Quantitative real-time PCR was performed with a BioRad iCycler as described [1]. FLAG-RPSA was detected using the following primers: GCCCTCTGTGCCTATTCAG–CFlag-F; CTTTACTTATCGTCGTCATCCTTG–CFlag-R; GACTACAAGGACGACGATGACAAG–NFlag-F; CTTCTCCCAGGTCCTCTTGAG–NFlag-R
RESULTS
Higher molecular weight RPSA species
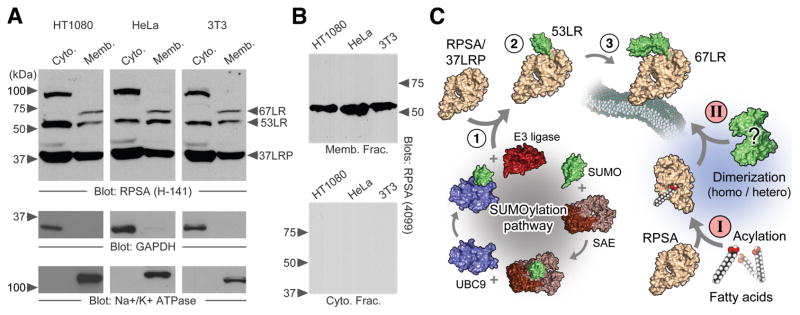
In probing lysates of mammalian cells for RPSA, a 37-kDa signal on immunoblots was regularly observed, corresponding to the monomeric form. Two antibodies raised against RPSA, H-141 and 4099-1, also detect higher molecular weight (HMW) forms in multiple cell lines (Fig. 1A and B). These HMW species include 67LR and a ~53-kDa product we now refer to as 53LR, which represents a previously reported [41] but infrequently discussed HMW form of RPSA. It is present in both the cytosol and the membrane, unlike 67LR, which was detected exclusively in the membrane fractions. It is not clear if 53LR and 67LR have been conflated with each other throughout the literature, but they appear to be distinct proteins.
Figure 1.
The higher molecular weight species of RPSA. A – SDS-PAGE of cytosolic and membrane extracts from HT1080, HeLa and NIH 3T3 cells. The monomeric 37-kDa RPSA is indicated along with higher molecular weight species. GAPDH and the Na+/K+-ATPase are shown as cytosolic and membrane markers, respectively. H-141 antibody is used to detect RPSA. B – As above with RPSA 4099-1 antibody. C – Two hypothetical pathways leading to the construction of 67LR from a 37-kDa RPSA precursor (37LRP). Progressive SUMOylation (1-2-3) and fatty acid acylation (I) followed by either a homo- or heterodimerization event (II) are depicted.
Previously reported larger HMW species (~100-kDa) were also detected [42]. Intermediate and larger species have also been described [41–48]. These species persist in the presence of SDS, 2-mercaptoethanol, DTT and denaturing temperatures.
Sustained use of RPSA-targeted siRNA ablates RPSA and all HMW species within 48–72 h (data not shown). This observation does not in itself support the precursor relationship, owing to knockdown of RPSA causing global decreases in protein synthesis [1].
Fig. 1C depicts two possible scenarios to explain the transition of RPSA to HMW species: homo/heterodimerization and progressive modification by SUMO (SUMO2 and 3 have the ability to form poly-SUMO chains).
The SUMOylation pathway is necessary for 53LR production
The apparent quantized laddering of RPSA species (i.e., 37–53–67–100, Fig. 1A) and their stability under reducing and denaturing conditions (e.g., SDS-PAGE) led us to consider UBL additions as a plausible explanation for the transition of the 37-kDa monomer to the HMW forms.
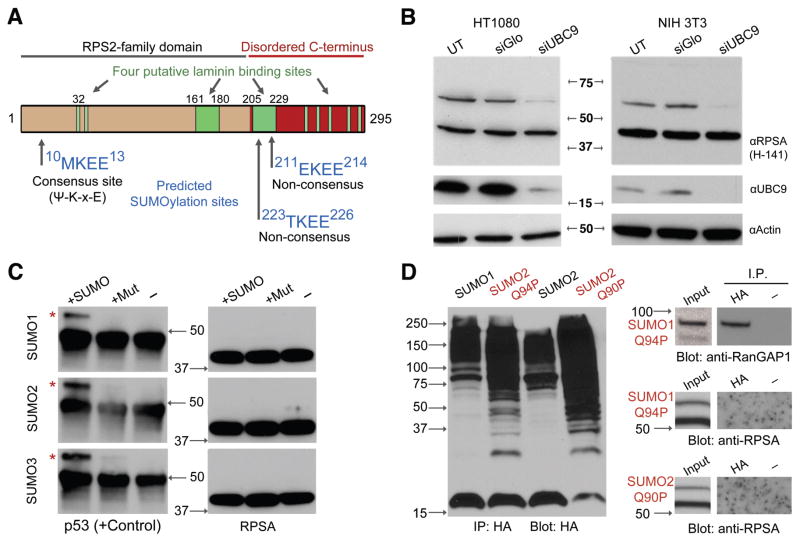
Because the molecular weights of SUMO proteins approximately correspond to the correct size intervals, we inspected the RPSA sequence for SUMOylation sites using a consensus sequence search [49]. One canonical consensus site is found near the N-terminus of RPSA (Lys-11) in addition to two non-consensus sites in proximity to a putative laminin-binding region (Fig. 2A).
Figure 2.
The RPSA transition to 53LR requires SUMOylation. A – Prediction of SUMOylation sites based on the RPSA primary sequence (amino acids 1–295). High probability target motifs are shown along with the location of four reported laminin binding sites (green). Ψ indicates a hydrophobic residue, x is any residue. B – Knockdown of UBC9 to disrupt SUMOylation. UT, mock transfected. siGlo, control siRNA. C – In vitro SUMOylation assay using recombinant p53 or RPSA. Normal (SUMO) or non-conjugating mutants (Mut) of SUMO1, 2 or 3 were used. Red asterisks indicate SUMOylated forms. D – Capture of HA-tagged SUMO proteins from HT1080 cells to immunoprecipitate target proteins. RanGAP1 is shown as a positive control. Q94P and Q90P indicate “uncleavable” mutant SUMO proteins. Anti-RPSA blots are shown overexposed to indicate capture failure.
Surveying raw data from a number of proteomics screens, we noted that RPSA had been previously detected as being modified by SUMO1, 2, 3 and 4 [37, 50–52]. To confirm the SUMOylation of RPSA, siRNA was used to ablate UBC9/UBE2I, the SUMO E2 conjugating enzyme (Fig. 1C). In HT1080, HeLa and NIH 3T3 cells, reduction of UBC9 caused a marked decrease in HMW RPSA species (Fig. 2B). Specifically, 53LR was eliminated between four and five days of sustained knockdown while RPSA/37LRP remained at normal levels. We consistently failed to detect 67LR in these experiments, and suspect that its expression levels (or its preservation) are condition sensitive.
Notably, the presence and detection of 67LR appear to fluctuate dramatically even under consistent conditions while 53LR is more reliably detected. We were not able to isolate a variable that leads to consistent 67LR expression, leading to great difficulty in its identification. Cell confluency, placement on laminin, frequency of passage, age of culture, cell cycle stage, and frequency of media refreshment were not factors in 67LR expression in our hands.
HMW RPSA species are not readily immunoprecipitated
Hoping to determine the identities of 53LR and 67LR unambiguously, we attempted to immunoprecipitate the protein for purification and mass spectrometry. Unfortunately, we were unable to purify or detect HMW RPSA species through standard immunoblotting or with 2D electrophoresis following immunoprecipitation (data not shown). Antibody recognition of the HMW species may rely on epitopes only exposed following denaturation. Alternatively, RPSA antibodies may not be suitable for immunoprecipitation.
In vitro SUMOylation does not produce HMW RPSA species
To confirm SUMOylation and determine the site of modification, recombinant purified RPSA was used for in vitro SUMOylation assays in the presence of E1 and E2 activating and conjugating enzymes. Interestingly, SUMOylation events leading to HMW RPSA were not detected despite the production of SUMO-modified control proteins p53 (Fig. 2C) and RanGAP1 (data not shown). This may suggest that an E3 ligase or other factor (e.g. post-translational modification) is required for the transition event.
Immunoprecipitation of HA-SUMO does not capture HMW RPSA
Because knockdown of UBC9 provides only an indirect assessment of RPSA SUMOylation status, and because HMW species could not be purified, we attempted to capture SUMOylated proteins and probe for RPSA. To achieve this, HT1080 and NIH 3T3 cells were transfected with HA-tagged SUMO1 or SUMO2 or 3 proteins. We also tested “uncleavable” HA-SUMO mutants (Q94P and Q90P) designed to resist removal by SUMO-specific proteases in order to stabilize SUMO additions to their targets [39]. HA-SUMO was immunoprecipitated with anti-HA magnetic beads and the immunoblots were probed for RPSA or control SUMO targets. No HMW RPSA was detected (Fig. 2D).
It may be that HMW species do not survive the immunoprecipitation process, as SUMO modifications are known to be fragile. The use of NEM to dissuade protease activity did not alleviate the issue. It is also conceivable that the HMW RPSA content is too low to adequately compete in the affinity capture.
Exogenous FLAG-tagged RPSA does not produce HMW species
Reasoning that difficulty in immunoprecipitating HMW RPSA species may have at least in part been due to antibody quality and low levels, we attempted to capture HMW species through exogenous expression of RPSA labeled with a FLAG-tag. This would simultaneously provide strong evidence of a relationship between RPSA and the HMW species.
Numerous examples in the literature indicate that affinity-tagged RPSA constructs do not produce detectable HMW forms [e.g., 28, 33, 53]. This observation is often attributed to the need for secondary factors that may not be sufficiently abundant to allow the transition [28, 30]. To combat this issue, stable cell lines were generated to allow tagged RPSA ample time to integrate into the endogenous population undergoing transition events.
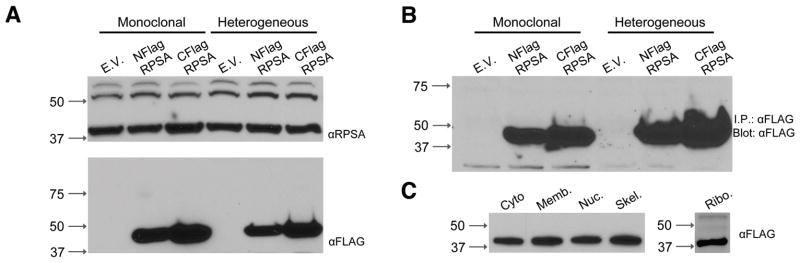
No trace of tagged HMW forms was found in HT1080RPSA-FLAG stable cell lines, whether monoclonally or heterogeneously selected (Fig. 3A). Indeed, periodic testing from 24–48 h after transfection and up to 8 weeks of continuous culture did not yield HMW signals in anti-FLAG immunoblots, despite the presence of HMW forms when using anti-RPSA antibodies. RPSA constructs with either N- or C-terminal FLAG-tags were used to guard against the possibility that the location of the tag could interfere with the HMW transition or its detection.
Figure 3.
FLAG-tagged RPSA does not produce higher molecular weight RPSA species. A – Membrane extracts of HT1080 cells stably expressing N- or C-terminally FLAG-tagged RPSA (NFlag or CFlag respectively). Monoclonal and heterogeneous stable cell lines were examined. Blots are representative of stable expression for 1–8 weeks (4 weeks shown). E.V. – vector control. B – Anti-FLAG immunoprecipitation of membrane extracts from stable HT1080RPSA-FLAG cells. C – Subcellular fractionation demonstrating normal localization of tagged RPSA in all expected fractions: cytoplasmic, membrane, nuclear, cytoskeletal and ribosome fractions.
The use of the proteasome inhibitor MG-132 did not enhance HMW levels or enable detection of FLAG-HMW forms. To examine the possibility that the FLAG-RPSA content was too low to compete for the transition event(s) with the endogenous protein, we performed qPCR to quantitate exogenous expression. RPSA-FLAG was found to be expressed at approximately 5–8% of the endogenous RPSA level in stable RPSA-FLAG cell lines.
Finally, we immunoprecipitated FLAG-RPSA using anti-FLAG magnetic beads to enrich the exogenous RPSA population. No HMW signal was detected (Fig. 3B). The FLAG-tagged proteins could rescue the functional defects associated with siRNA-mediated knockdown of RPSA [53] and localized to all known RPSA subcellular fractions (i.e. cytosol, ribosomes, membrane, cytoskeleton and nucleus; Fig. 3C).
The possibility that 53LR and 67LR may not be related to RPSA must be considered in light of this evidence. Concrete verification of the HMW species and their identity will be critical for moving forward in the study of this protein and its functions.
DISCUSSION
This report highlights the difficulties associated with identifying the higher molecular weight species of RPSA. Indeed, given that affinity-tagged RPSA does not yield HMW bands, it is necessary to consider the possibility that the relationship with RPSA (also called the 37-kDa laminin receptor or 37LRP) has been misinterpreted.
Multiple reports using tagged RPSA constructs would seem to confirm this observation (i.e., only the 37-kDa monomer is detected), but only one explicitly comments on this specific issue [28, 30]. Often cited data from Montuori et al. [54] regarding a polyhistidine-labeled RPSA yielding a 67-kDa species were not published. The entity purified from bacterial expression is also exclusively the monomeric 37-kDa form [40].
The use of stable cell lines in this study would seem to have resolved the issue of a yet-unknown required secondary factor that is rate-limiting for the transition. The dimerization hypothesis (Fig. 1C) has been the leading explanatory model, recently receiving support from investigators demonstrating homo- and heterodimerization in cells [33]. Even in this study, transiently expressed YFP-RPSA fusion proteins fail to show stable dimers in immunoblots. It is unclear if otherwise modified (e.g., SUMO) HMW species are produced. It is unusual that an interaction strong enough to resist the denaturing and reducing conditions of SDS-PAGE should prove to be so elusive.
The matter is further complicated by the existence of proteins potentially contributing artifactual signals. For example, galactosidase beta-1 (GLB1), a laminin/elastin-binding molecule, shares epitopes with RPSA and is natively 67 kDa [55]. Additionally, there exist so called laminin-binding lectins, high molecular weight multimers that denature into subunits of ~70 kDa [56].
Unfortunately, we were unable to adequately demonstrate the SUMOylation status of RPSA. Nonetheless, SUMOylation as an explanatory paradigm does deserve attention because of its implications. Specifically, it may factor into a dynamic regulatory mechanism controlling a cell’s ability to bind laminin.
It is tempting to speculate that SUMO modification interacts with the already described ubiquitin/proteasome regulation of 67LR [36]. If poly-SUMOylation indeed builds HMW species, it may be targeting RPSA for ubiquitination and subsequent degradation via the SUMO-targeted ubiquitin ligase (STUbL) pathway [57]. This may explain the difficulty in HMW capture.
We did not observe stabilization of HMW forms with a proteasome inhibitor, confirming the results of a large-scale proteomics study examining increases in SUMOylated proteins following the use of proteasome inhibitors (i.e., SUMO-RPSA was not increased) [37]. It is also possible that SUMO modification guards against proteasome degradation, as SUMO has been known to act antagonistically to ubiquitination by competitive blockage of the modification site [38]. Interestingly, this possibility allows for stable HMW RPSA proteins to exist at the membrane. Modulation of protein–protein interactions is another known role of SUMO modification [38]. Since 67LR is thought to have higher affinity for laminin than the monomer, SUMOylation conceivably affects the binding of laminin, especially considering the presence of SUMOylation sites within a potential laminin-binding region (Fig. 2A).
Alternatively, only 53LR may be a SUMOylated species while 67LR may have a distinct genesis, such as a dimer or unrelated protein. An underlying dilemma for the SUMO/ubiquitin hypothesis is the difficulty in explaining how an extracellular protein could be dynamically subjected to modification by UBLs. A transmembrane domain within RPSA is now properly considered implausible given cytosolic solubility and the examination of the crystal structure [14, 40].
Our failure to confirm SUMOylation despite some compelling evidence is unfortunate while supporting evidence of the RPSA/67LR precursor relationship remains decidedly lacking. Several recommendations emerge for investigators working on detecting 67LR: (1) clearly indicate the molecular weight of all species being discussed; (2) explicitly comment when HMW species are/are not produced using tagged proteins; (3) publish conditions under which HMW species can be reliably detected; and (4) attempt to purify and sequence the HMW forms. Definitive identification is crucial for advancing our understanding of RPSA and its potential contributions to laminin binding, migration, metastasis and numerous other functions.
Acknowledgments
This work was supported by US Public Health grant CA100687 from the National Cancer Institute, National Institutes of Health and Department of Health and Human Services as well as Institutional National Research Service Award T32-CA009161 (Levy). Protein structure graphics were generated using the PyMOL Molecular Graphics System, Schrödinger, LLC.
Abbreviations used
- 37LRP
37-kDa laminin receptor precursor
- 53LR
53-kDa laminin receptor
- 67LR
67-kDa laminin receptor
- HMW
higher molecular weight
- RPSA
ribosomal protein SA
- SUMO
small ubiquitin-like modifier
- UBL
ubiquitin-like protein
Footnotes
The original publication is available www.springerlink.com. DOI: https://doi.org/10.1515/cmble-2015-0031
References
- 1.Scheiman J, Tseng JC, Zheng Y, Meruelo D. Multiple functions of the 37/67-kd laminin receptor make it a suitable target for novel cancer gene therapy. Mol Ther. 2010;18:63–74. doi: 10.1038/mt.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demianova M, Formosa TG, Ellis SR. Yeast proteins related to the p40/laminin receptor precursor are essential components of the 40 S ribosomal subunit. J Biol Chem. 1996;271:11383–11391. doi: 10.1074/jbc.271.19.11383. [DOI] [PubMed] [Google Scholar]
- 3.O’Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol. 2010;190:853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardini E, Pesole G, Tagliabue E, Magnifico A, Castronovo V, Sobel ME, Colnaghi MI, Menard S. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol Biol Evol. 1998;15:1017–1025. doi: 10.1093/oxfordjournals.molbev.a026000. [DOI] [PubMed] [Google Scholar]
- 5.Miner JH. Laminins and their roles in mammals. Microsc Res Tech. 2008;71:349–356. doi: 10.1002/jemt.20563. [DOI] [PubMed] [Google Scholar]
- 6.Rao NC, Barsky SH, Terranova VP, Liotta LA. Isolation of a tumor cell laminin receptor. Biochem Biophys Res Commun. 1983;111:804–808. doi: 10.1016/0006-291x(83)91370-0. [DOI] [PubMed] [Google Scholar]
- 7.Rao CN, Castronovo V, Schmitt MC, Wewer UM, Claysmith AP, Liotta LA, Sobel ME. Evidence for a precursor of the high-affinity metastasis-associated murine laminin receptor. Biochemistry (Mosc) 1989;28:7476–7486. doi: 10.1021/bi00444a047. [DOI] [PubMed] [Google Scholar]
- 8.Hara K, Satoh K, Ide H. Apical ectodermal ridge-dependent expression of the chick 67 kDa laminin binding protein gene (cLbp) in developing limb bud. Zoolog Sci. 1997;14:969–978. doi: 10.2108/zsj.14.969. [DOI] [PubMed] [Google Scholar]
- 9.Bolze A, Mahlaoui N, Byun M, Turner B, Trede N, Ellis SR, Abhyankar A, Itan Y, Patin E, Brebner S, Sackstein P, Puel A, Picard C, Abel L, Quintana-Murci L, Faust SN, Williams AP, Baretto R, Duddridge M, Kini U, Pollard AJ, Gaud C, Frange P, Orbach D, Emile JF, Stephan JL, Sorensen R, Plebani A, Hammarstrom L, Conley ME, Selleri L, Casanova JL. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science. 2013;340:976–978. doi: 10.1126/science.1234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menard S, Tagliabue E, Colnaghi MI. The 67 kDa laminin receptor as a prognostic factor in human cancer. Breast Cancer Res Treat. 1998;52:137–145. doi: 10.1023/a:1006171403765. [DOI] [PubMed] [Google Scholar]
- 11.Ardini E, Sporchia B, Pollegioni L, Modugno M, Ghirelli C, Castiglioni F, Tagliabue E, Menard S. Identification of a novel function for 67-kDa laminin receptor: increase in laminin degradation rate and release of motility fragments. Cancer Res. 2002;62:1321–1325. [PubMed] [Google Scholar]
- 12.Chetty C, Khumalo T, Da Costa Dias B, Reusch U, Knackmuss S, Little M, Weiss SF. Anti-LRP/LR specific antibody IgG1-iS18 impedes adhesion and invasion of liver cancer cells. PLoS One. 2014;9:e96268. doi: 10.1371/journal.pone.0096268.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liotta LA, Rao NC, Barsky SH, Bryant G. The laminin receptor and basement membrane dissolution: role in tumour metastasis. Ciba Found Symp. 1984;108:146–162. doi: 10.1002/9780470720899.ch10. [DOI] [PubMed] [Google Scholar]
- 14.DiGiacomo V, Meruelo D. Looking into laminin receptor: critical discussion regarding the non-integrin 37/67-kDa laminin receptor/RPSA protein. Biol Rev Camb Philos Soc. 2015 doi: 10.1111/brv.12170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanjuan X, Fernandez PL, Miquel R, Munoz J, Castronovo V, Menard S, Palacin A, Cardesa A, Campo E. Overexpression of the 67-kD laminin receptor correlates with tumour progression in human colorectal carcinoma. J Pathol. 1996;179:376–380. doi: 10.1002/(SICI)1096-9896(199608)179:4<376::AID-PATH591>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 16.Viacava P, Naccarato AG, Collecchi P, Menard S, Castronovo V, Bevilacqua G. The spectrum of 67-kD laminin receptor expression in breast carcinoma progression. J Pathol. 1997;182:36–44. doi: 10.1002/(SICI)1096-9896(199705)182:1<36::AID-PATH802>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 17.Chung JW, Hong SJ, Kim KJ, Goti D, Stins MF, Shin S, Dawson VL, Dawson TM, Kim KS. 37-kDa laminin receptor precursor modulates cytotoxic necrotizing factor 1-mediated RhoA activation and bacterial uptake. J Biol Chem. 2003;278:16857–16862. doi: 10.1074/jbc.M301028200. [DOI] [PubMed] [Google Scholar]
- 18.Orihuela CJ, Mahdavi J, Thornton J, Mann B, Wooldridge KG, Abouseada N, Oldfield NJ, Self T, Ala’Aldeen DA, Tuomanen EI. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J Clin Invest. 2009;119:1638–1646. doi: 10.1172/JCI36759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig GV, Kondig JP, Smith JF. A putative receptor for Venezuelan equine encephalitis virus from mosquito cells. J Virol. 1996;70:5592–5599. doi: 10.1128/jvi.70.8.5592-5599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Da Costa Dias B, Jovanovic K, Gonsalves D, Moodley K, Reusch U, Knackmuss S, Penny C, Weinberg MS, Little M, Weiss SF. Anti-LRP/LR specific antibody IgG1-iS18 and knock-down of LRP/LR by shRNAs rescue cells from Abeta42 induced cytotoxicity. Science Reports. 2013;3:2702. doi: 10.1038/srep02702.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Da Costa Dias B, Jovanovic K, Gonsalves D, Moodley K, Reusch U, Knackmuss S, Weinberg MS, Little M, Weiss SF. The 37kDa/67kDa laminin receptor acts as a receptor for Abeta42 internalization. Sci Rep. 2014;4:5556. doi: 10.1038/srep05556.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gauczynski S, Peyrin JM, Haik S, Leucht C, Hundt C, Rieger R, Krasemann S, Deslys JP, Dormont D, Lasmezas CI, Weiss S. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rieger R, Edenhofer F, Lasmezas CI, Weiss S. The human 37-kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat Med. 1997;3:1383–1388. doi: 10.1038/nm1297-1383. [DOI] [PubMed] [Google Scholar]
- 25.Wewer UM, Liotta LA, Jaye M, Ricca GA, Drohan WN, Claysmith AP, Rao CN, Wirth P, Coligan JE, Albrechtsen R, et al. Altered levels of laminin receptor mRNA in various human carcinoma cells that have different abilities to bind laminin. Proc Natl Acad Sci USA. 1986;83:7137–7141. doi: 10.1073/pnas.83.19.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castronovo V, Taraboletti G, Sobel ME. Functional domains of the 67-kDa laminin receptor precursor. J Biol Chem. 1991;266:20440–20446. [PubMed] [Google Scholar]
- 27.Buto S, Ghirelli C, Aiello P, Tagliabue E, Ardini E, Magnifico A, Montuori N, Sobel ME, Colnaghi MI, Menard S. Production and characterization of monoclonal antibodies directed against the laminin receptor precursor. Int J Biol Markers. 1997;12:1–5. doi: 10.1177/172460089701200101. [DOI] [PubMed] [Google Scholar]
- 28.Castronovo V, Claysmith AP, Barker KT, Cioce V, Krutzsch HC, Sobel ME. Biosynthesis of the 67 kDa high affinity laminin receptor. Biochem Biophys Res Commun. 1991;177:177–183. doi: 10.1016/0006-291x(91)91965-f. [DOI] [PubMed] [Google Scholar]
- 29.Buto S, Tagliabue E, Ardini E, Magnifico A, Ghirelli C, van den Brule F, Castronovo V, Colnaghi MI, Sobel ME, Menard S. Formation of the 67-kDa laminin receptor by acylation of the precursor. J Cell Biochem. 1998;69:244–251. doi: 10.1002/(sici)1097-4644(19980601)69:3<244::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 30.Landowski TH, Dratz EA, Starkey JR. Studies of the structure of the metastasis-associated 67 kDa laminin binding protein: fatty acid acylation and evidence supporting dimerization of the 32 kDa gene product to form the mature protein. Biochemistry (Mosc) 1995;34:11276–11287. doi: 10.1021/bi00035a037. [DOI] [PubMed] [Google Scholar]
- 31.Castronovo V, Luyten F, van den Brule F, Sobel ME. Identification of a 14-kDa laminin binding protein (HLBP14) in human melanoma cells that is identical to the 14-kDa galactoside binding lectin. Arch Biochem Biophys. 1992;297:132–138. doi: 10.1016/0003-9861(92)90650-l. [DOI] [PubMed] [Google Scholar]
- 32.Satoh K, Narumi K, Isemura M, Sakai T, Abe T, Matsushima K, Okuda K, Motomiya M. Increased expression of the 67kDa-laminin receptor gene in human small cell lung cancer. Biochem Biophys Res Commun. 1992;182:746–752. doi: 10.1016/0006-291x(92)91795-r. [DOI] [PubMed] [Google Scholar]
- 33.Alqahtani F, Mahdavi J, Wheldon LM, Vassey M, Pirinccioglu N, Royer PJ, Qarani SM, Morroll S, Stoof J, Holliday ND, Teo SY, Oldfield NJ, Wooldridge KG, Ala’Aldeen DA. Deciphering the complex three-way interaction between the non-integrin laminin receptor, galectin-3 and Neisseria meningitidis. Open Biol. 2014;4 doi: 10.1098/rsob.140053.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hundt C, Peyrin JM, Haik S, Gauczynski S, Leucht C, Rieger R, Riley ML, Deslys JP, Dormont D, Lasmezas CI, Weiss S. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 2001;20:5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 36.Kim DG, Choi JW, Lee JY, Kim H, Oh YS, Lee JW, Tak YK, Song JM, Razin E, Yun SH, Kim S. Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 2012;26:4142–4159. doi: 10.1096/fj.12-207639. [DOI] [PubMed] [Google Scholar]
- 37.Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, Andersen JS, Vertegaal AC. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics. 2008;7:2107–2122. doi: 10.1074/mcp.M800025-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bekes M, Prudden J, Srikumar T, Raught B, Boddy MN, Salvesen GS. The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J Biol Chem. 2011;286:10238–10247. doi: 10.1074/jbc.M110.205153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jamieson KV, Wu J, Hubbard SR, Meruelo D. Crystal structure of the human laminin receptor precursor. J Biol Chem. 2008;283:3002–3005. doi: 10.1074/jbc.C700206200. [DOI] [PubMed] [Google Scholar]
- 41.Tandon NN, Holland EA, Kralisz U, Kleinman HK, Robey FA, Jamieson GA. Interaction of human platelets with laminin and identification of the 67 kDa laminin receptor on platelets. Biochem J. 1991;274(Pt 2):535–542. doi: 10.1042/bj2740535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weeks BS, Kopp JB, Horikoshi S, Cannon FB, Garrett M, Kleinman HK, Klotman PE. Adult and fetal human mesangial cells interact with specific laminin domains. Am J Physiol. 1991;261:F688–695. doi: 10.1152/ajprenal.1991.261.4.F688. [DOI] [PubMed] [Google Scholar]
- 43.Simoneau S, Haik S, Leucht C, Dormont D, Deslys JP, Weiss S, Lasmezas C. Different isoforms of the non-integrin laminin receptor are present in mouse brain and bind PrP. Biol Chem. 2003;384:243–246. doi: 10.1515/BC.2003.027. [DOI] [PubMed] [Google Scholar]
- 44.Clement B, Segui-Real B, Savagner P, Kleinman HK, Yamada Y. Hepatocyte attachment to laminin is mediated through multiple receptors. J Cell Biol. 1990;110:185–192. doi: 10.1083/jcb.110.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis CM, Papadopoulos V, Jia MC, Yamada Y, Kleinman HK, Dym M. Identification and partial characterization of laminin binding proteins in immature rat Sertoli cells. Exp Cell Res. 1991;193:262–273. doi: 10.1016/0014-4827(91)90095-c. [DOI] [PubMed] [Google Scholar]
- 46.Douville PJ, Harvey WJ, Carbonetto S. Isolation and partial characterization of high affinity laminin receptors in neural cells. J Biol Chem. 1988;263:14964–14969. [PubMed] [Google Scholar]
- 47.Kleinman HK, Ogle RC, Cannon FB, Little CD, Sweeney TM, Luckenbill-Edds L. Laminin receptors for neurite formation. Proc Natl Acad Sci USA. 1988;85:1282–1286. doi: 10.1073/pnas.85.4.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercurio AM, Shaw LM. Macrophage interactions with laminin: PMA selectively induces the adherence and spreading of mouse macrophages on a laminin substratum. J Cell Biol. 1988;107:1873–1880. doi: 10.1083/jcb.107.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- 50.Blomster HA, Hietakangas V, Wu J, Kouvonen P, Hautaniemi S, Sistonen L. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol Cell Proteomics. 2009;8:1382–1390. doi: 10.1074/mcp.M800551-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282.. [DOI] [PubMed] [Google Scholar]
- 52.Guo D, Han J, Adam BL, Colburn NH, Wang MH, Dong Z, Eizirik DL, She JX, Wang CY. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem Biophys Res Commun. 2005;337:1308–1318. doi: 10.1016/j.bbrc.2005.09.191. [DOI] [PubMed] [Google Scholar]
- 53.Scheiman J, Jamieson KV, Ziello J, Tseng JC, Meruelo D. Extraribosomal functions associated with the C terminus of the 37/67 kDa laminin receptor are required for maintaining cell viability. Cell Death & Disease. 2010;1:e42. doi: 10.1038/cddis.2010.19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montuori N, Siri J, Simmons T, Gillet C, Senterre G, Wrathall L, Sobel ME. Precursor-product relationship between a 37-kDa polypeptide and the 67-kDa laminin receptor. FASEB. 1995;9:A539. [Google Scholar]
- 55.Mecham RP, Hinek A, Griffin GL, Senior RM, Liotta LA. The elastin receptor shows structural and functional similarities to the 67-kDa tumor cell laminin receptor. J Biol Chem. 1989;264:16652–16657. [PubMed] [Google Scholar]
- 56.Bao ZZ, Muschler J, Horwitz AF. LBL, a novel, developmentally regulated, laminin-binding lectin. J Biol Chem. 1992;267:4974–4980. [PubMed] [Google Scholar]
- 57.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]