Abstract
Point mutations in the mitochondrial (mt) tRNALeu(UUR) gene are responsible for mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS), a subgroup of mitochondrial encephalomyopathic diseases. We previously showed that mt tRNALeu(UUR) with an A3243G or T3271C mutation derived from patients with MELAS are deficient in a normal taurine-containing modification (τm5U; 5-taurinomethyluridine) at the anticodon wobble position. To examine decoding disorder of the mutant tRNA due to the wobble modification deficiency independent of the pathogenic point mutation itself, we used a molecular surgery technique to construct an mt tRNALeu(UUR) molecule lacking the taurine modification but without the pathogenic mutation. This “operated” mt tRNALeu(UUR) without the taurine modification showed severely reduced UUG translation but no decrease in UUA translation. We thus concluded that the UUG codon–specific translational defect of the mutant mt tRNAsLeu(UUR) is the primary cause of MELAS at the molecular level. This result could explain the complex I deficiency observed clinically in MELAS.
Mitochondrial (mt) DNA mutations are responsible for a wide spectrum of human diseases that are caused by mitochondrial dysfunction. Point mutations in the genes encoding mt tRNAs are found particularly frequently in mitochondrial diseases (MITOMAP: A Human Mitochondrial Genome Database, www.mitomap.org, 2004) (1). Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS), one of the major clinical subgroups of the mitochondrial encephalomyopathies, is caused by a single base replacement in the tRNALeu gene, which is responsible for the translation of the UUR (R = A or G) leucine codons (tRNALeu(UUR)) (2). The majority (80%) of MELAS patients possess an A to G transition at nucleotide position (np) 3243 (3, 4), whereas, in ≈10% of the patients, a T to C transition is observed at np 3271 (5). The mutation at np 3243 has also been observed in maternally inherited diabetes with deafness (6) and in progressive external ophthalmoplegia (7, 8). An A to G transition at np 8344 in the tRNALys gene is found in most patients with myoclonus epilepsy associated with ragged red fibers (MERRF) (9), another major clinical subgroup of the mitochondrial encephalomyopathies. Thus, clinical features depend on the tRNA species and/or positions of the mutations; however, the molecular mechanisms linking the locations of the mutations and their leading phenotypes are not fully understood.
Cybrid cell lines (10, 11), in which mutant mtDNA derived from patients was intercellularly transferred into human cells lacking mtDNA (ρ0 cells), were used to demonstrate that the above three mutations (A3243G, T3271C, and A8344G) are directly involved in mitochondrial dysfunction. In the case of MELAS, a decline in enzymatic activity and a decrease in protein synthesis were observed in cybrid cells containing a high ratio of mutated mtDNA (12–14). Several lines of studies proposed dysfunction of the mutant tRNAsLeu(UUR) as a possible outcome arising directly from the MELAS mutations, which would in turn cause a decrease in respiratory activity (15). However, there is as yet no conclusive evidence for a molecular mechanism of mitochondrial dysfunction induced by pathogenic point mutations.
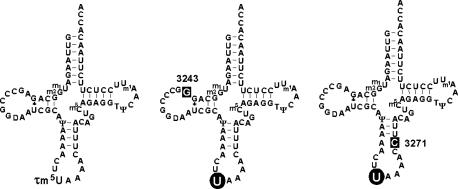
We have previously shown that, in cybrid cells possessing homoplasmic pathogenic mutations, the taurine-containing modified uridine (τm5U; 5-taurinomethyluridine) (16) that normally occurs at the anticodon wobble position of mt tRNALeu(UUR) remains unmodified in the mt tRNALeu(UUR) bearing the A3243G or T3271C mutation (Fig. 1) (17). This finding explains why these different point mutations are associated with the same clinical phenotype. In addition, we have shown by using cybrid cells from a patient with MERRF that the mutant mt tRNALys bearing the A8344G mutation also lacks the appropriate taurine-modification (τm5s2U; 5-taurinomethyl-2-thiouridine) (16, 18). These two types of mitochondrial diseases thus have in common the lack of taurine modification of their respective mutant tRNAs. Thus, the point mutations apparently function to hinder the biosynthesis of the wobble taurine modification of mt tRNAs. Because uridine modifications at the wobble position are responsible for precise and efficient codon recognition (19, 20), a considerable decoding disorder might be caused by the wobble modification deficiency. In the case of MERRF, we have previously shown that tRNALys with a mutation at np 8344 actually does lose translational activity for both of its cognate codons due to the wobble modification defect (21).
Fig. 1.
Cloverleaf structures of human mt tRNAsLeu(UUR) from WT cells (Left) and from MELAS cybrid cells with the A3243G mutation (Center) or T3271C mutation (Right). “U” on a round black background indicates unmodified wobble uridine. White letters on a square black background represent the respective point mutations. Symbols for the modifications are 5-taurinomethyluridine (τm5U), 1-methyladenosine (m1A), 1-methylguanosine (m1G), 2-methylguanosine (m2G), pseudouridine (Ψ), ribothymidine (T), dihydrouridine (D), and 5-methylcytidine (m5C) (17).
In this study, we estimated the decoding disorder in mitochondrial translation arising from the deficient modification of the wobble taurine of mutant tRNALeu(UUR). It is necessary to discriminate the specific effect of the wobble modification deficiency from that of the different point mutations (A3243G or T3271C). We used the molecular surgery technique to construct an mt tRNALeu(UUR) molecule lacking the taurine modification but without the pathogenic mutation. By clarifying the decoding and translational activities of this “operated” tRNALeu(UUR), we were able to obtain clear experimental evidence that the mt tRNALeu(UUR) lacking the wobble taurine modification has a codon-specific deficit in translational ability.
Materials and Methods
Cybrid Cell Lines. The mutant cybrid cell lines, which were constructed by the intercellular transfer of MELAS patient mtDNA to ρ0 HeLa cells, have been described (17). The ML2-2-2 and ML5-1-13 cell lines exclusively contain mtDNA with the A3243G and T3271C point mutations, respectively. The cells were cultured in normal medium [Dulbecco's modified Eagle's medium/F-12 (1:1; GIBCO/BRL), supplemented with 10% FCS].
Large-Scale Purification of mt tRNALeu(UUR) from Human Placenta. Large-scale purification of mt tRNALeu(UUR) from human placenta was basically carried out according to methods described in the literature (16). A crude RNA fraction (7.8 g) was extracted from 27 kg of human placenta. The tRNA fraction (3.5 g) was obtained by anion exchange column chromatography by using DEAE-Sepharose fast flow (8 × 73 cm) with linear gradients of NaCl (200–500 mM) and MgCl2 (8–16 mM) in a buffer containing 20 mM Hepes-KOH (pH 7.5). The fractions enriched with the mitochondrial tRNAs were monitored by dot hybridization (22) and pooled. The mt tRNALeu(UUR) was purified from the fraction by using an improved solid-phase DNA probe method (23), which we have named “chaplet” column chromatography, and a synthetic DNA probe with 3′ biotin that is complementary to tRNALeu(UUR) (5′-GCGATTACCGGGCTCTGCCATCTTAA-3′). The isolated tRNALeu(UUR) was incubated at 65°C for 8 min and annealed at room temperature in buffer containing 50 mM Hepes-KOH (pH 7.5) and 10 mM MgCl2. Thereafter, 5 mM DTT and 0.3 unit/μl T4 polynucleotide kinase (Toyobo, Tokyo) were added to the mixture, and the dephosphorylation of the 3′ end of tRNALeu(UUR) was performed at 37°C for 1 h. Then, the 3′ end was repaired with mammalian mitochondrial CCA-adding enzyme (24) at 37°C for 1 h by adding 0.04 μg/μl CCA-adding enzyme, 1 mM ATP, 1 mM CTP, and 100 mM KCl as final concentrations. The repaired tRNALeu(UUR) was finally purified by 10% PAGE in a 40-cm gel containing 7 M urea. One hundred sixty micrograms of the purified tRNALeu(UUR) was obtained.
Mutant mt tRNAsLeu(UUR) bearing the A3243G or T3271C mutation were purified from the relevant cybrid cells as described (17).
Molecular Surgery to Construct the Artificial mt tRNALeu(UUR) with an Unmodified Uridine at Position 34. Molecular surgery was basically carried out according to methods described in the literature (25). The hammerhead ribozyme was designed to cleave mt tRNALeu(UUR) obtained from human placenta at the anticodon wobble position (between positions 34 and 35). The ribozyme sequence 5′-GGGACUGUAAAGUUUUCUGAUGAGCCGAAAGGCGAAAGUUUUAUGCG-3′ was transcribed in vitro by using T7 RNA polymerase. Eighty micrograms of the purified mt tRNALeu(UUR) was digested at 37°C for 3 h in a reaction mixture (300 μl) containing 50 mM Tris·HCl (pH 8.0), 25 mM MgCl2, 240 μg of the hammerhead ribozyme, and 240 μg each of two synthetic DNA probes complementary to the top part of tRNALeu(UUR), 5′-GCGATTACCGGGCTCTGCCATCTTAA-3′ and 5′-TGTTAAGAAGAGGAATTGAACCTC-3′. The synthetic DNA probes are designed to destabilize the tRNA to enhance the accessibility of the ribozyme. The resulting 5′ and 3′ half fragments were separated by 10% PAGE in a gel containing 7 M urea. The purified 5′ half fragment (18 μg) was treated with 0.1 M HCl at 0°C for 3 h to cleave the 2′,3′ cyclic phosphate of the 3′ end and then dephosphor ylated with bacterial alkaline phosphatase (Takara). The τm5U34 at the 3′ end of the 5′ half fragment was removed by periodate oxidation (26). After dephosphorylation, the truncated 5′ half fragment was ligated with pUp (kindly provided by K. Nishikawa, Gifu University, Gifu, Japan) by using T4 RNA ligase (Takara) at 11°C for 15 h in a reaction mixture containing 50 mM Tris·HCl (pH 7.5), 15 mM MgCl2, 3.5 mM DTT, 15 μg/ml BSA, 5% PEG 6000, 300 μM ATP, 15 μM of the 5′ half fragment, 45 μM pUp, and 2.5 units/μl T4 RNA ligase. The ligated 5′ half fragment and the 3′ half fragment were incubated at 68°C for 7 min in a reaction mixture consisting of 50 mM Hepes-KOH, 15 mM MgCl2, and 10 μM of each fragment and then annealed at room temperature. Phosphorylation of the 5′ end of the 3′ half fragment and dephosphorylation of the 3′ end of the 5′ half fragment were performed simultaneously at 37°C for 30 min by adding 3.5 mM DTT, 15 μg/ml BSA, 300 μM ATP, and 0.5 unit/μl T4 polynucleotide kinase (Toyobo). Thereafter, 1.25 units/μl T4 RNA ligase (Takara) was added to the mixture, and the ligation was performed at 37°C for 30 min. The reconstituted tRNA was purified by 12% PAGE in a gel containing 7 M urea. In the end, 5.2 μg of the tRNA bearing an unmodified uridine at the wobble position was obtained. The sequence of the resultant tRNALeu(UUR) was confirmed by Donis-Keller's method (27).
In Vitro Mitochondrial Translation. The in vitro translation assay was carried out according to methods described in the literature (28). Briefly, the aminoacyl-tRNALeu(UUR) was prepared at 37°C for 10 min in a reaction mixture (50 μl) consisting of 50 mM Tris·HCl (pH 8.0), 15 mM MgCl2, 2 mM ATP, 3.3 μM[3H]l-leucine (1.85 MBq/mmol, American Radiolabeled Chemicals, St. Louis), 9 pmol tRNALeu(UUR), and 20 μg of human mt leucyl-tRNA synthetase. The leucylation level of WT and the operated tRNAsLeu(UUR) achieved to be 88% and 87%, respectively. Although it is known that leucylation is affected by the MELAS mutation, MELAS tRNA with the 3243 or 3271 mutation was respectively leucylated to be 48% and 80% in the presence of an excess amount of mt leucyl-tRNA synthetase. Poly(UUA)30, Poly(UUG)30 and Poly(UUC)30 were synthesized in vitro by using T7 RNA polymerase. The reaction mixture (20 μl) contained 50 mM Tris·HCl (pH 8.6), 15 mM MgCl2, 5 mM KCl, 1 mM DTT, 0.5 mM spermine, 2.5 mM phosphoenolpyruvate, 2.5 units/ml pyruvate kinase, 0.5 mM GTP, 12 pmol mt EF-Tu, 8 pmol mt EF-G, 2 pmol mt ribosome, 4 μg of mRNA [Poly-(UUA)30, Poly(UUG)30 and Poly(UUC)30], and 0.1 pmol [3H]Leu-tRNALeu(UUR). The mixture was incubated at 37°C for 15 min, and the radioactivity of the polymerized amino acids was measured by liquid scintillation counting as described (29, 30).
Ribosomal A-Site Binding. The A-site binding was carried out according to methods described in the literature (31) with slight modifications. The mRNA containing A-site UUA or UUG codon was synthesized in vitro by using T7 RNA polymerase to create the following sequence (A-site codon underlined): 5′-GGGUUAACUUUAAGUAAGGAGGUAUACUAUGUURUAACUGCAGAAAAAA-3′. First, we filled the ribosomal P-site with initiator tRNA in a mixture (10 μl) consisting of 5 pmol Escherichia coli 70S ribosome, 2 μg of mRNA, 19 pmol E. coli initiator tRNAfMet, 50 mM Tris·HCl (pH 7.5), 6.5 mM MgCl2, 60 mM KCl, 1 mM DTT, and 0.5 mM spermine, which was incubated at 37°C for 17 min. Thereafter, four different amounts (0.25, 0.5, 0.75, and 1 pmol) of either the WT or operated mt tRNALeu(UUR) in a mixture (10 μl) consisting of 50 mM Tris·HCl (pH 7.5), 6.5 mM MgCl2, 60 mM KCl, 1 mM DTT, and 0.5 mM spermine were added to the ribosomal mixture, and a nonenzymatic binding reaction was performed at 37°C for 12 min. The amount of tRNA bound was quantified as described in the literature (32).
Results
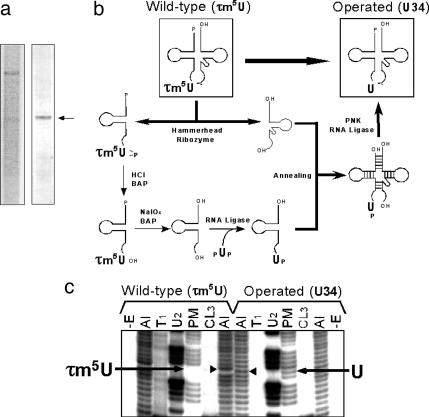
Construction of an mt tRNALeu(UUR) Variant with an Unmodified Wobble Uridine Using the Molecular Surgery Technique. To investigate the effect of the wobble modification deficiency on the translational activity of tRNA independently of the effect of the pathogenic point mutation itself, we operated on the human native mt tRNALeu(UUR) using the molecular surgery technique (25) to construct an artificial mt tRNALeu(UUR) that has a completely normal sequence, with all of the normal modified bases except the τm5U at the wobble position. For this purpose, a large amount of mt tRNALeu(UUR) (160 μg) was isolated from 27 kg of human placenta (Fig. 2a) by using a solid-phase DNA probe method as described in Materials and Methods. The purified mt tRNALeu(UUR) was cut in half at the wobble position by the hammerhead ribozyme (Fig. 2b). τm5U in the 5′ half fragment was removed by periodate oxidation and replaced with an unmodified uridine by enzymatic ligation. Then the 5′ fragment was re-ligated to the 3′ half fragment to construct an mt tRNALeu(UUR) lacking the τm5U modification (Fig. 2b; for details, see Materials and Methods). The sequence of the resultant tRNA was confirmed by Donis-Keller's method (27) (Fig. 2c). As shown in Fig. 2c, the wobble base of the operated tRNA is cleaved by RNase PhyM (A- or U-specific) to give a discrete band, whereas that of the WT tRNA is resistant to digestion. An unusual upward shift in the alkaline ladders due to the τm5U modification of the WT tRNA became normal in the ladder of the operated tRNA. Otherwise, we could see no differences in the digestion patterns between these two tRNAs. These results suggest that the resultant operated tRNA had an unmodified wobble uridine but no changes in other modifications.
Fig. 2.
Construction of mt tRNALeu(UUR) with an unmodified uridine at position 34 using the molecular surgery technique. (a) Purification of mt tRNALeu(UUR) from human placenta. (Left) Denaturing gel electrophoresis of the total RNA fraction extracted from human placenta. (Right) Purified human mt tRNALeu(UUR) (indicated by an arrow). (b) Schematic depiction of the molecular surgery procedure used to construct mt tRNALeu(UUR) with an unmodified wobble uridine. The details of each step are described in Materials and Methods. BAP and PNK represent bacterial alkaline phosphatase and polynucleotide kinase, respectively. (c) RNA sequence ladders for the 5′ end-labeled WT and operated tRNALeu(UUR) were obtained by the Donis-Keller method (27). –E, without enzyme; Al, treatment with alkali; T1, RNase T1 (specific for G); U2, RNase U2 (for A > G); PM, RNase Phy M (for A and U); and CL3, RNase CL3 (for C). Arrows indicate the bands that were cleaved by RNase PhyM digestion. There is no band for τm5U34 in the WT tRNA because the modification prevents PhyM digestion, but there is a cleaved band for U34 in the operated tRNA. Arrowheads show the wobble positions in the alkaline ladders. The band corresponding to τm5U34 is shifted up due to its bulky substituent, whereas U34 shows a normal band in the ladder.
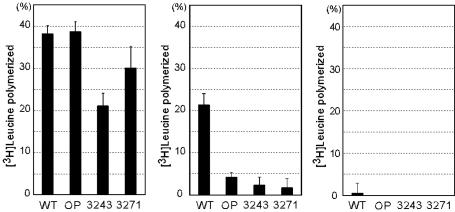
UUG Codon-Specific Translational Defect of mt tRNAsLeu(UUR) Lacking the Wobble Modification in an in Vitro Mitochondrial Translation System. We examined whether the mt tRNALeu(UUR) lacking the wobble modification could function in the translation process. The translational activity was measured by using an in vitro mitochondrial translation system (28). We used an mRNA transcribed in vitro that has 30 triplet repeats for the leucine codons UUA and UUG, as well as UUC as a negative control. As shown in Fig. 3, the WT mt tRNALeu(UUR) was efficient in decoding both the UUA and UUG codons and showed no activity with the UUC noncognate codon. The MELAS mutant tRNAsLeu(UUR) purified from the relevant mutant cybrid cells were also examined. These mutant tRNAs, which not only possess the MELAS A3243G or T3271C mutation but also lack the wobble modification (Fig. 1), showed a considerable reduction in UUA decoding as well as a severe reduction in UUG decoding. In the case of the operated tRNALeu(UUR), whose wobble modification had been surgically removed, no appreciable reduction was observed in UUA decoding, but a severe reduction was observed in UUG decoding.
Fig. 3.
Translational activity of the operated tRNALeu(UUR) without the wobble modification and of the MELAS mutant tRNAsLeu(UUR). In vitro mitochondrial translation of test mRNAs containing the UUA (Left), UUG (Center), or UUC (Right) (negative control) codons was performed with WT tRNALeu(UUR)(WT), operated tRNALeu(UUR) with an unmodified wobble uridine (OP), and two MELAS mutant tRNAsLeu(UUR) that bear the A3243G (3243) or U3271C (3271) mutations and an unmodified wobble uridine. The radioactivity of the [3H]leucyl-tRNA input to the reaction mixture was defined arbitrarily as 100. The averages of three independent experiments with SD values are shown.
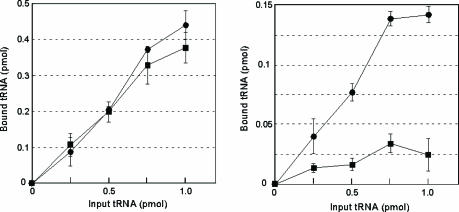
Defective Binding of the mt tRNALeu(UUR) Without the Taurine Modification to the UUG Codon on the Ribosomal A-Site. To confirm that the wobble modification is responsible for UUG decoding, we carried out a ribosomal A-site-binding experiment. Native mt tRNALeu(UUR) efficiently bound to both the UUA and UUG codons whereas the operated tRNA bearing an unmodified wobble uridine showed strong binding to the UUA codon but weak binding affinity for the UUG codon (Fig. 4). This finding suggests that the UUG codon-specific translational defect of mt tRNALeu(UUR) lacking wobble modification is caused by an inability to form codon-anticodon base pairs on the ribosomal A-site. From these results, we conclude that the modified wobble uridine plays a functional role in the decoding of the UUG codon by stabilizing the U:G wobble base pairing on the ribosomal A-site.
Fig. 4.
Ability of the operated tRNALeu(UUR) without the wobble modification to bind to UUR codons. Shown is binding of the WT tRNALeu(UUR) (filled circles) and the operated tRNALeu(UUR) that lacks the wobble modification (filled squares) to ribosomal A-sites containing UUA (Left) or UUG (Right). Three independent experiments were performed, and the average values are plotted with SD values.
Discussion
Many studies have implicated pathogenic point mutations in tRNAsLeu(UUR) in the dysfunctions associated with MELAS mutations (15, 33). These dysfunctions include impaired termination (34), impaired pre-tRNA processing (35), decreased stability and aminoacylation (36), and abnormal conformation (37), and commonly lead to a decreased steady-state level of the normal aminoacylated tRNA, which in turn leads to reduced protein synthesis. Some of these biochemical tRNA analyses were performed by using an unmodified tRNA transcribed in vitro, because it is difficult to obtain a sufficient number of native mt tRNAs with the MELAS mutation for a series of analyses. However, tRNAs mature through posttranscriptional modifications and do not function without these modifications. Indeed, human mt tRNALeu(UUR) (78 bases in length) has nine modified bases (11.5% of total bases). In particular, wobble modification plays a crucial role in decoding the genetic code. Our observations suggest that the wobble taurine modification deficiency in MELAS and MERRF should be considered one of the major molecular pathogeneses of these diseases. Although a biochemical study using unmodified tRNAs is a powerful approach for studying mitochondrial diseases in some cases, our observations in the mutant tRNAs from MELAS and MERRF prompted us to carefully estimate the defect of the single posttranscriptional modification using fully modified native tRNAs.
A number of analyses have suggested that the extent of the deficiency in protein synthesis did not seem to parallel the decline in the enzymatic activity of the respiratory complexes in the case of MELAS (12–14, 35). Thus, a quantitative decrease in functional mt tRNALeu(UUR) alone seems insufficient to be the direct cause of the mitochondrial dysfunction, although the mitochondrial dysfunction in MELAS arises from multiple causes. The facts prompted us to consider that some qualitative differences could be involved in the molecular pathogenesis of MELAS, including the wobble modification deficiency.
In this study, we successfully clarified the critical effect of the wobble modification deficiency in mt tRNALeu(UUR) on UUG codon-specific translation by using an in vitro mitochondrial translation system and a ribosome A-site-binding assay. Although the aminoacylation level of four tRNAsLeu(UUR) put into the translation system was not equal (48–88%; see Materials and Methods), the difference in aminoacylation level and presence of deacylated tRNA could not influence the translational activity, because it is likely that almost all leucylated tRNA is trapped by EF-Tu to form the ternary complex (aminoacyl-tRNA/EF-Tu/GTP) under the existence of overabundant EF-Tu and that the ribosomal A-site exclusively binds the ternary complex (38). In the case of the nonenzymatic ribosomal A-site-binding experiment, the aminoacylation level of the two aminoacylated tRNAs was almost equal (88% for WT and 87% for operated), so it hardly became a problem. We used E. coli ribosome instead of the human mitochondrial counterpart in the binding assay, because the precise condition for the assay using E. coli ribosome is completely established and it has been shown that mt tRNA enables translation to proceed on E. coli ribosome and, conversely, that E. coli tRNA dose likewise on mt ribosome (39, 40).
Our results demonstrate two major points. First, the severe reduction in UUG decoding by the MELAS mutant tRNAs can be mainly attributed to the lack of the wobble modification. Many studies have unraveled the contribution of the position 5-substituted wobble uridines to decoding, including restricting decoding by a modified uridine with a methylene carbon directly bonded to the C5 position of uracil ring (xm5U) or expanding decoding by a modified uridine with an oxygen atom directly bonded to the C5 position (xo5U) (19, 20, 41, 42). In this study, we concluded that the taurine modification at the C5 position of wobble uridine (τm5U) in mt tRNALeu(UUR) plays a crucial role in the decoding of the UUG codon by stabilizing the U:G wobble base pairing. Second, because there was a considerable reduction in UUA decoding when the point mutations were present but not when they were absent, the MELAS point mutations themselves impose a certain negative effect on translation. Thus, we could estimate the negative effect of the respective pathogenic point mutations (A3243G and U3271C) on decoding the cognate UUA codon, by comparison with the result of the operated tRNA lacking the wobble modification. The negative effect might arise from the fragile relaxed structure caused by the mutations, because the A3243G mutation could disrupt the potential tertiary interaction U8-A14-A21 (43–45), and the T3271C mutation destabilizes the anticodon stem (46). The MELAS tRNALeu(UUR) with the A3243G mutation showed a more severe reduction in UUA decoding than the tRNALeu(UUR) with the T3271C mutation. This result is consistent with translational activities of MELAS cybrid cells with these point mutations (12, 14).
We have noticed a specific bias of leucine codon usage in 13 proteins encoded by human mtDNA genes (Table 1). For example, despite the minor usage of the UUG codon in most of the proteins, the ND6 gene, which is a component of respiratory chain complex I (NADH-coenzyme Q reductase), contains eight UUG codons that constitute 42.1% of the total leucine codons and 4.6% of the total codons in ND6. It has been reported that the translational activity of ND6 in cybrid cells is specifically and markedly reduced without a decrease in total mitochondrial protein synthesis when the A3243G or T3271C mtDNA levels are increased (13, 14). Furthermore, a point mutation (A14453G) in the structural gene for ND6 was found to be associated with severe MELAS syndrome (47). Considering the UUG codon–specific translational defect described in this study, these facts support the idea that MELAS patients experience a translational depression of ND6. This idea nicely explains why a specific reduction of complex I activity is characteristic of MELAS patients (48, 49). These results indicate that the UUG codon–specific translational defect caused by defective wobble taurine modification is primarily responsible for the molecular pathogenesis of MELAS. Additionally, our study suggested that the point mutation itself, in particular the A3243G mutation, contributes to the tRNALeu(UUR) translational defect to a considerable extent. Thus, the level of decoding disorder for each MELAS mutant tRNA should vary with the effect of each pathogenic point mutation.
Table 1.
Leucine codon usage for 13 protein genes encoded by mtDNA (52)
| Gene | No. of amino acids | No. of CUN codons | No. of UUA codons | No. of UUG codons | UUG codons/Leu codons, % | UUG codons/total codons, % |
|---|---|---|---|---|---|---|
| ND1 | 318 | 57 | 5 | 1 | 1.6 | 0.3 |
| ND2 | 347 | 55 | 8 | 1 | 1.6 | 0.3 |
| ND3 | 115 | 18 | 10 | 1 | 3.4 | 0.9 |
| ND4 | 459 | 87 | 8 | 1 | 1.0 | 0.2 |
| ND4L | 98 | 22 | 1 | 0 | 0.0 | 0.0 |
| ND5 | 603 | 95 | 7 | 2 | 1.9 | 0.3 |
| ND6 | 174 | 3 | 8 | 8 | 42.1 | 4.6 |
| Cyt b | 378 | 55 | 7 | 2 | 3.1 | 0.5 |
| COI | 513 | 55 | 7 | 0 | 0.0 | 0.0 |
| COII | 227 | 28 | 4 | 1 | 3.0 | 0.4 |
| COIII | 260 | 31 | 3 | 0 | 0.0 | 0.0 |
| A6 | 226 | 39 | 4 | 1 | 2.3 | 0.4 |
| A8 | 68 | 8 | 1 | 1 | 10.0 | 1.5 |
The UUG codon ratio (%) to total leucine codons and to total amino acids (total codons) is shown for each gene. A6 and A8 represent subunits 6 and 8 for ATPase, respectively.
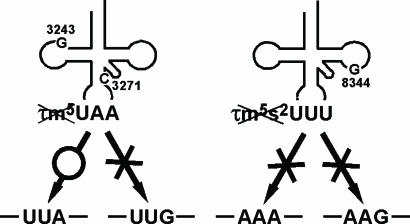
We previously examined the translational ability of the mutant mt tRNALys from MERRF patients that bears the A8344G mutation. This analysis showed that tRNALys lacking the τm5s2U-modification are incapable of translating both cognate codons AAA and AAG. This result is due to a complete loss of codon–anticodon pairing on the ribosome (21), because the 2-thio modification of the wobble base is known to be critical for decoding AAR codons (32). This result explains why MERRF patients show a marked defect in whole mitochondrial translation (21, 50, 51). Thus, the different symptoms exhibited by MELAS and MERRF patients may be explained by the fact that the mutant tRNAs lacking the wobble modification in these patients show a distinct pattern of codon recognition (Fig. 5).
Fig. 5.
Possible mechanism of molecular pathogenesis caused by the wobble modification deficiency of the mutant tRNAs in MELAS (Left) and MERRF (Right) patients. Pathogenic point mutation (A3243G or U3271C) in mutant tRNALeu(UUR) from MELAS patients causes a τm5U-modification deficiency, which results in a UUG codon–specific translational defect. The MERRF 8344 mutation in tRNALys causes a τm5s2U-modification deficiency that results in a translational defect for both cognate codons (AAA and AAG) (21).
In conclusion, our study has unraveled the essential molecular mechanism causing mitochondrial dysfunction in MELAS pathology. A point mutation at nucleotide position 3243 or 3271 in the mtDNA results in a taurine modification deficiency at the anticodon wobble position of the mutant tRNALeu(UUR), which subsequently causes a UUG-codon–specific translational defect, possibly leading to a translational depression of ND6. This defective codon-specific translation indicates that the deficiency in taurine modification could be a key to the expression of clinical phenotypes in mitochondrial diseases.
Acknowledgments
We are grateful to Drs. T. Ohtsuki, T. Hanada, and Takeo Suzuki for technical advice and helpful suggestions. This work was supported by grants-in-aid for scientific research on priority areas from the Ministry of Education, Science, Sports, and Culture of Japan (to T.S. and K.W.), by a Japan Society for the Promotion of Science Fellowship for Japanese Junior Scientists (to Y.K.), by a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to T.S.), and by Human Frontier Science Program Grant RG0349 (to T.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MELAS, mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes; MERRF, myoclonus epilepsy associated with ragged red fibers; np, nucleotide position; mt, mitochondrial; τm5U 5-taurinomethyluridine; τm5s2U, 5-taurinomethyl-2-thiouridine.
References
- 1.Schon, E. A., Bonilla, E. & DiMauro, S. (1997) J. Bioenerg. Biomembr. 29, 131–149. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi, Y., Momoi, M. Y., Tominaga, K., Shimoizumi, H., Nihei, K., Yanagisawa, M., Kagawa, Y. & Ohta, S. (1991) Am. J. Hum. Genet. 49, 590–599. [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi, Y., Momoi, M. Y., Tominaga, K., Momoi, T., Nihei, K., Yanagisawa, M., Kagawa, Y. & Ohta, S. (1990) Biochem. Biophys. Res. Commun. 173, 816–822. [DOI] [PubMed] [Google Scholar]
- 4.Goto, Y., Nonaka, I. & Horai, S. (1990) Nature 348, 651–653. [DOI] [PubMed] [Google Scholar]
- 5.Goto, Y., Nonaka, I. & Horai, S. (1991) Biochim. Biophys. Acta 1097, 238–240. [DOI] [PubMed] [Google Scholar]
- 6.van den Ouweland, J. M., Lemkes, H. H., Ruitenbeek, W., Sandkuijl, L. A., de Vijlder, M. F., Struyvenberg, P. A., van de Kamp, J. J. & Maassen, J. A. (1992) Nat. Genet. 1, 368–371. [DOI] [PubMed] [Google Scholar]
- 7.Johns, D. R. & Hurko, O. (1991) Lancet 337, 927–928. [DOI] [PubMed] [Google Scholar]
- 8.Moraes, C. T., Ciacci, F., Silvestri, G., Shanske, S., Sciacco, M., Hirano, M., Schon, E. A., Bonilla, E. & DiMauro, S. (1993) Neuromuscul. Disord. 3, 43–50. [DOI] [PubMed] [Google Scholar]
- 9.Shoffner, J. M., Lott, M. T., Lezza, A. M., Seibel, P., Ballinger, S. W. & Wallace, D. C. (1990) Cell 61, 931–937. [DOI] [PubMed] [Google Scholar]
- 10.King, M. P. & Attardi, G. (1989) Science 246, 500–503. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, J., Ohta, S., Kikuchi, A., Takemitsu, M., Goto, Y. & Nonaka, I. (1991) Proc. Natl. Acad. Sci. USA 88, 10614–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomyn, A., Martinuzzi, A., Yoneda, M., Daga, A., Hurko, O., Johns, D., Lai, S. T., Nonaka, I., Angelini, C. & Attardi, G. (1992) Proc. Natl. Acad. Sci. USA 89, 4221–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbar, D. R., Moonie, P. A., Zeviani, M. & Holt, I. J. (1996) Hum. Mol. Genet. 5, 123–129. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, J., Ohta, S., Takai, D., Miyabayashi, S., Sakuta, R., Goto, Y. & Nonaka, I. (1993) Biochem. Biophys. Res. Commun. 197, 1049–1055. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs, H. T. (2003) Hum. Mol. Genet. 12, Spec. No. 2, R293–R301. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki, T., Suzuki, T., Wada, T., Saigo, K. & Watanabe, K. (2002) EMBO J. 21, 6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yasukawa, T., Suzuki, T., Suzuki, T., Ueda, T., Ohta, S. & Watanabe, K. (2000) J. Biol. Chem. 275, 4251–4257. [DOI] [PubMed] [Google Scholar]
- 18.Yasukawa, T., Suzuki, T., Ishii, N., Ueda, T., Ohta, S. & Watanabe, K. (2000) FEBS Lett. 467, 175–178. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama, S. & Nishimura, S. (1995) in tRNA: Structure, Biosynthesis and Function, eds. Söll, D. & Rajbandary, U. L. (Am. Soc. Microbiol., Washington, DC), pp. 207–223.
- 20.Bjork, G. R. (1995) in tRNA: Structure, Biosynthesis and Function, eds. Söll, D. & Rajbandary, U. L. (Am. Soc. Microbiol, Washington, DC), pp. 165–205.
- 21.Yasukawa, T., Suzuki, T., Ishii, N., Ohta, S. & Watanabe, K. (2001) EMBO J. 20, 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokogawa, T., Kumazawa, Y., Miura, K. & Watanabe, K. (1989) Nucleic Acids Res. 17, 2623–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko, T., Suzuki, T., Kapushoc, S. T., Rubio, M. A., Ghazvini, J., Watanabe, K., Simpson, L. & Suzuki, T. (2003) EMBO J. 22, 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaike, T., Suzuki, T., Tomari, Y., Takemoto-Hori, C., Negayama, F., Watanabe, K. & Ueda, T. (2001) J. Biol. Chem. 276, 40041–40049. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, T., Ueda, T. & Watanabe, K. (1997) EMBO J. 16, 1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keith, G. & Gilham, P. T. (1974) Biochemistry 13, 3601–3606. [DOI] [PubMed] [Google Scholar]
- 27.Donis-Keller, H. (1980) Nucleic Acids Res. 8, 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanada, T., Suzuki, T., Yokogawa, T., Takemoto-Hori, C., Sprinzl, M. & Watanabe, K. (2001) Genes Cells 6, 1019–1030. [DOI] [PubMed] [Google Scholar]
- 29.Gradner, R. S., Wahba, A. J., Basilio, C., Miller, R. S., Lengyel, P. & Speyer, J. F. (1962) Proc. Natl. Acad. Sci. USA 48, 2087–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravel, J. M. & Shorey, R. L. A. (1971) Methods Enzymol. 20, 306–316. [Google Scholar]
- 31.Ogle, J. M., Murphy, F. V., Tarry, M. J. & Ramakrishnan, V. (2002) Cell 111, 721–732. [DOI] [PubMed] [Google Scholar]
- 32.Ashraf, S. S., Sochacka, E., Cain, R., Guenther, R., Malkiewicz, A. & Agris, P. F. (1999) RNA 5, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasukawa, T., Suzuki, T., Ohta, S. & Watanabe, K. (2002) Mitochondrion 2, 129–141. [DOI] [PubMed] [Google Scholar]
- 34.Hess, J. F., Parisi, M. A., Bennett, J. L. & Clayton, D. A. (1991) Nature 351, 236–239. [DOI] [PubMed] [Google Scholar]
- 35.Flierl, A., Reichmann, H. & Seibel, P. (1997) J. Biol. Chem. 272, 27189–27196. [DOI] [PubMed] [Google Scholar]
- 36.Chomyn, A., Enriquez, J. A., Micol, V., Fernandez-Silva, P. & Attardi, G. (2000) J. Biol. Chem. 275, 19198–19209. [DOI] [PubMed] [Google Scholar]
- 37.Wittenhagen, L. M. & Kelley, S. O. (2002) Nat. Struct. Biol. 9, 586–590. [DOI] [PubMed] [Google Scholar]
- 38.Schilling-Bartetzko, S., Franceschi, F., Sternbach, H. & Nierhaus, K. H. (1992) J. Biol. Chem. 267, 4693–4702. [PubMed] [Google Scholar]
- 39.Kumazawa, Y., Schwartzbach, C. J., Liao, H. X., Mizumoto, K., Kaziro, Y., Miura, K., Watanabe, K. & Spremulli, L. L. (1991) Biochim. Biophys. Acta 1090, 167–172. [DOI] [PubMed] [Google Scholar]
- 40.Schwartzbach, C. J. & Spremulli, L. L. (1989) J. Biol. Chem. 264, 19125–19131. [PubMed] [Google Scholar]
- 41.Takai, K. & Yokoyama, S. (2003) Nucleic Acids Res. 31, 6383–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agris, P. F. (2004) Nucleic Acids Res. 32, 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moras, D., Comarmond, M. B., Fischer, J., Weiss, R., Thierry, J. C., Ebel, J. P. & Giege, R. (1980) Nature 288, 669–674. [DOI] [PubMed] [Google Scholar]
- 44.Goddard, J. P. (1977) Prog. Biophys. Mol. Biol. 32, 233–308. [PubMed] [Google Scholar]
- 45.Rich, A. & RajBhandary, U. L. (1976) Annu. Rev. Biochem. 45, 805–860. [DOI] [PubMed] [Google Scholar]
- 46.Wittenhagen, L. M., Roy, M. D. & Kelley, S. O. (2003) Nucleic Acids Res. 31, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravn, K., Wibrand, F., Hansen, F. J., Horn, N., Rosenberg, T. & Schwartz, M. (2001) Eur. J. Hum. Genet. 9, 805–809. [DOI] [PubMed] [Google Scholar]
- 48.Goto, Y., Horai, S., Matsuoka, T., Koga, Y., Nihei, K., Kobayashi, M. & Nonaka, I. (1992) Neurology 42, 545–550. [DOI] [PubMed] [Google Scholar]
- 49.Koga, Y., Nonaka, I., Kobayashi, M., Tojyo, M. & Nihei, K. (1988) Ann. Neurol. 24, 749–756. [DOI] [PubMed] [Google Scholar]
- 50.Yoneda, M., Miyatake, T. & Attardi, G. (1994) Mol. Cell. Biol. 14, 2699–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Enriquez, J. A., Chomyn, A. & Attardi, G. (1995) Nat. Genet. 10, 47–55. [DOI] [PubMed] [Google Scholar]
- 52.Andrews, R. M., Kubacka, I., Chinnery, P. F., Lightowlers, R. N., Turnbull, D. M. & Howell, N. (1999) Nat. Genet. 23, 147. [DOI] [PubMed] [Google Scholar]