Abstract
Receptor activator of NF-κB ligand (RANKL) is a TNFα-like cytokine that is produced by a diverse set of lineage-specific cells and is involved in a wide variety of physiological processes that include skeletal remodeling, lymph node organogenesis, mammary gland development, and thermal regulation. Consistent with these diverse functions, control of RANKL expression is accomplished in a cell-specific fashion via a set of at least 10 regulatory enhancers that are located up to 170 kb upstream of the gene’s transcriptional start site. Here we examined the in vivo consequence of introducing a contiguous DNA segment containing these components into a genetically deleted RANKL null mouse strain. In contrast to RANKL null littermates, null mice containing the transgene exhibited normalized body size, skeletal development, and bone mass as well as normal bone marrow cavities, normalized spleen weights, and the presence of developed lymph nodes. These mice also manifested normalized reproductive capacity, including the ability to lactate and to produce normal healthy litters. Consistent with this, the transgene restored endogenous-like RANKL transcript levels in several RANKL-expressing tissues. Most importantly, restoration of RANKL expression from this segment of DNA was fully capable of rescuing the complex aberrant skeletal and immune phenotype of the RANKL null mouse. RANKL also restored appropriate levels of B220+IgM+ and B220+IgD+ B cells in spleen. Finally, we found that RANKL expression from this transgene was regulated by exogenously administered 1,25(OH)2D3, parathyroid hormone (PTH), and lipopolysaccharide (LPS), thus recapitulating the ability of these same factors to regulate the endogenous gene. These findings fully highlight the properties of the Tnfsf11 gene locus predicted through previous in vitro dissection. We conclude that the mouse Tnfsf11 gene locus identified originally through unbiased chromatin immunoprecipitation with DNA microarray (ChIP-chip) analysis contains the necessary genetic information to direct appropriate tissue-specific and factor-regulated RANKL expression in vivo. Research.
Keywords: RANKL; PTH; 1,25D3; LPS; TRANSCRIPTIONAL REGULATION
Introduction
Receptor activator of NF-κB ligand (RANKL) is a TNFα-like cytokine that is expressed in cells of various lineages and participates in the regulation of a number of diverse biological processes in higher vertebrates. These processes include a primary remodeling function orchestrated by RANKL at various sites in the skeleton(1) as well as activities associated with lymph node organogenesis,(2–4) the development of B and T cells(2) and immune cell response during chronic inflammation.(5–7) They also include RANKL participation in the development of the mammary gland,(8) development of Paneth M cells of the intestine,(9) and in thermoregulation in the central nervous system (CNS).(10) RANKL is expressed largely as a membrane-associated protein,(1,11,12) supporting its central role as a paracrine factor affecting primarily adjacent target cells. In addition, the membrane-associated form can be cleaved to produce a soluble form that can found in the circulation, suggesting its potential role as a factor active at more distant sites as well.(1,13) Importantly, RANKL is part of a regulatory axis that includes osteoprotegerin (OPG), a soluble decoy receptor that diverts the activity of RANKL away from bone fide target cells by preventing RANKL interaction with RANK, the ligand’s cognate receptor.(1) The relevance of this axis in human disease is highlighted in multiple ways, but perhaps most distinctly through the identification of mutations within the TNFSF11 and TNFRSF11A genes that encode RANKL and RANK, respectively. Mutations of this type can lead to either loss or gain of function activities that are exemplified by osteopetrosis or osteoporosis.(14,15) These and other activities have been recapitulated in mice through genetic deletion of either RANKL(2,16) or RANK,(17) and through transgenic overexpression of OPG.(18) In genome-wide association studies (GWASs), less dramatic yet highly correlated effects have been noted between specific single nucleotide polymorphisms (SNPs) located within these gene loci and altered bone mineral density (BMD), although the underlying molecular basis for these relationships is unknown.(19)
Many of the molecular events associated with RANKL action in the skeleton have been identified. RANKL interacts directly with RANK on the surface of specific hematopoietic precursors, thereby promoting their differentiation and subsequent fusion into bone-resorbing osteoclasts.(1) Recent studies suggest that control of RANKL expression specifically from the osteocyte may represent a fundamental mechanism through which normal bone remodeling is achieved in adult mice.(20) Interestingly, RANKL expression from T and B lineage cells does not appear to contribute to the regulation of skeletal bone turnover during normal physiology,(21) although its contribution during chronic inflammation is less certain.(22)
Central to the actions of RANKL is its regulation by an impressive array of systemic as well as local hormones and cytokines that act on both mesenchymal and hematopoietic lineage cells. First and foremost, these include the primary calcemic regulatory hormones 1,25-dihydroxyvitamin D3 (1,25 (OH)2D3)(23) and parathyroid hormone (PTH),(24) which function to control mineral homeostasis, in part through their direct activities in the skeleton. In fact, it was this ability of 1,25(OH)2D3 to promote osteoclast differentiation in co-cultures of osteoblasts and spleen cells that led to the concept of the presence of an osteoclastogenic factor that was ultimately identified as RANKL.(23,25,26) Regulatory factors also include a number of inflammatory cytokines such as TNFα(27) and interleukin 1 (IL-1),(27) the IL-6–type cytokines exemplified by IL-6 and oncostatin M (OSM),(28,29) and prostaglandins such as prostaglandin E2 (PGE2).(29) These regulate RANKL expression from cells of both mesenchymal and hematopoietic origin. Many additional factors that regulate RANKL have been also identified, including progesterone(30) and prolactin,(31) which act as primary regulators of RANKL expression in epithelial cells during mammary gland development and may play a role in the development of breast cancer.(32,33)
Interestingly, initial studies aimed at understanding the molecular mechanisms through which many of these factors control RANKL expression using traditional transcription promoter-reporter methodologies were either unsuccessful or inconclusive, suggesting that a more complex mechanism of regulation was operative. The fundamentals of this mechanism were resolved, however, when unbiased chromatin immunoprecipitation (ChIP) methodologies revealed that 1,25(OH)2D3 and PTH-mediated activation of RANKL occurs via multiple binding sites for their transcriptional mediators vitamin D receptor (VDR) and cAMP response element-binding protein (CREB), respectively.(34,35) Importantly, these regulatory sites were not located near the gene’s promoter, as anticipated, but rather as far upstream of the Tnfsf11 gene’s transcriptional start site (TSS) as 75 to 76 kb.(34,35) In the case of PTH, dissection of the RANKL upstream region during a similar time frame using modified bacterial artificial chromosome (BAC) clones also revealed the presence of this distal region.(36) Of considerable importance, the biological contribution of this element, termed D5, to RANKL expression has been validated in vivo through its genomic deletion from the mouse genome.(36–38) However, further examination has now shown that this intergenic region, which extends upstream from the TSS almost 200 kb, contains at least 10 domains that control RANKL expression, serving to mediate the activity of the inflammatory cytokines,(39,40) additional steroid and polypeptide hormones,(41) PGE2,(30) and signaling pathways induced by T-cell receptor activation.(42) Furthermore, although several of these regulatory regions trigger RANKL expression uniquely in osteoblast lineage cells, others mediate expression in vitro only in T cells and perhaps B cells.(42) Finally, the boundaries of this broad regulatory domain, together with the RANKL transcription unit itself, have been defined by active transcriptional repressor CTCF binding sites that serve to limit the epigenetic changes provoked during activation by hormones.(43) Thus, this series of studies has revealed the breadth of the genetic loci that comprises the mouse(34,39,40,43,44) and human(45,46) TNFSF11 transcription units as well as their associated intergenic regulatory control segments. Interestingly, the application of unbiased ChIP methodologies that now routinely employ DNA sequencing approaches to annotate the genome has revealed that this type of regulatory complexity is common for many genes that exhibit important cellular control.
To determine whether the mouse Tnfsf11 gene locus, is sufficient for appropriate expression and regulation of the RANKL gene, we constructed a contiguous DNA segment spanning all of the elements of the Tnfsf11 gene locus and containing a recombinantly introduced reporter and used this construct to generate transgenic mice. We then examined the capacity of this construct to recapitulate expression of wild-type mouse tissue levels of RANKL mRNA, to mediate upregulation by 1,25(OH)2D3, PTH, and lipopolysaccharides (LPS), and to rescue the debilitating skeletal and immune cell phenotypes associated with RANKL deletion by crossing the transgenic mice with RANKL null mice. We find that this exogenously introduced DNA segment was sufficient to restore each of these aspects of RANKL biology in vivo.
Materials and Methods
Generation of the Tnfsf11 BAC transgene
BAC clones RP23–68C13 and RP23–52A3 were obtained from The BACPAC Resource Center. A recombineering approach was used to insert an appropriate segment of BAC clone RP23–52A3 into the BAC RP23–68C13 to obtain a seamless “full-length” mouse Tnfsf11 BAC clone. Briefly, a 22 kb segment of the BAC RP23–52A3 corresponding to −156 to −178 kb upstream of the RANKL TSS was recombineered to the 3′ end of the BAC RP23–68C13 to produce a RANKL transgene spanning +40 kb to –178 kb relative to the RANKL TSS. Insertion of a cassette containing an internal ribosome entry site-luciferase (IRES-LUC) reporter gene and a thymidine kinase–neomycin (TK-neo) selectable marker was achieved using the GalK system to produce the Tnfsf11 BAC clone construct.(47) The expanded BAC clone was linearized using Not1-mediated digestion and the resulting transgene isolated from the parent vector using pulsed field gene electrophoresis and gel filtration.(48)
Mouse strains
RANKL null (RANKL−/−) transgenic mice were kindly provided by Choi and colleagues.(16) RANKL null mice were generated by deletion of nucleotides 699 to 1089 of exon 5 and 1123 bp of the 3′ untranslated region (UTR) by homologous recombination. RANKL+/+;Tg719 and RANKL+/+;Tg721 transgenic mouse strains were created through pronuclear injection of fertilized C57BL/6 eggs using the recombinantly modified Tnfsf11 BAC clone prepared as above and documented in Fig. 1. A two-step breeding strategy was used, in which reproductively active male RANKL−/− mice were first crossed with either RANKL+/+;Tg719 or RANKL+/+;Tg721 female mice to create either RANKL+/−;Tg719 or RANKL+/−;Tg721 mice. Heterozygous RANKL+/−;Tg719 or RANKL+/−;Tg721 mice were then crossed with RANKL+/− mice to produce the RANKL+/+, RANKL−/−, RANKL−/−;Tg719, RANKL−/−;Tg721 strains as well as additional breeding genotypes RANKL+/−, RANKL+/−;Tg719, or RANKL+/−;Tg721 necessary for study.
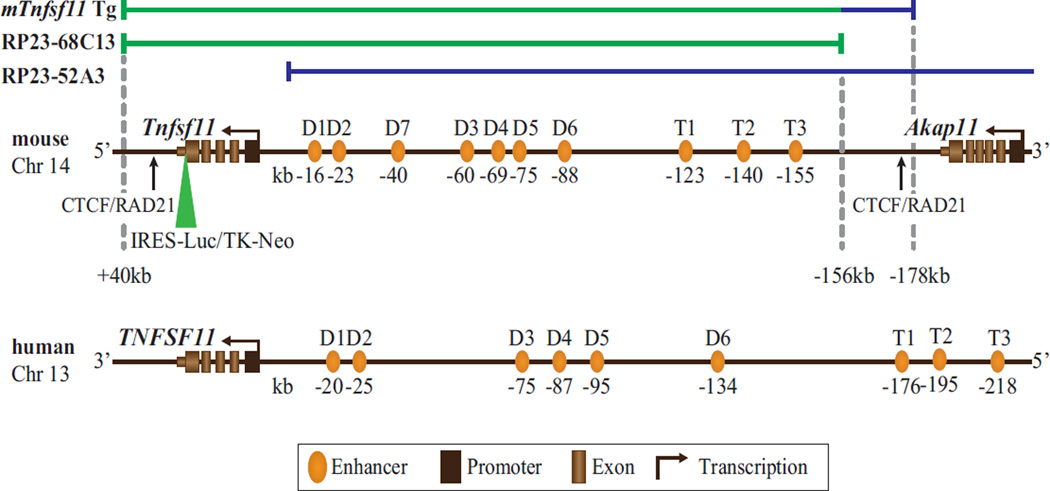
Fig. 1.
Schematic structure of the murine (m) Tnfsf11 and human (h) TNFSF11 gene loci and the mTnfsf11 transgene. The mTnfsf11 gene locus (center), h Tnfsf11 gene locus (bottom), and the mTnfsf11 transgene comprised of its two BAC components RP23–68C13 (green line) and a 22-kb 3′ segment of RP23–52A3 (blue line) spanning the mouse Tnfsf11 gene locus from +40 kb to −178 kb (top) are shown. Across the mTnfsf11 gene locus, exons comprising the transcription units for both Tnfsf11 and Akap11 and previously characterized enhancers with their positions relative to the Tnfsf11 TSS are indicated in kilobases (kb) below the colored ovals. CTCF/RAD21 boundary elements are marked by arrows. The mTnfsf11 BAC transgene also contains an indicated cassette containing an IRES-driven LUC reporter and a TK promoter (TK)-driven neomycin resistance gene (Neo) selection mechanism that was inserted within the 3′-UTR of the gene. Similar features are indicated for the h TNFSF11 gene locus on Chr 13. TSS = transcriptional start site; BAC = bacterial artificial chromosome; LUC = luciferase; IRES = internal ribosome entry site; TK = thymidine kinase; UTR = untranslated region; Chr = chromosome.
Animal studies
The animals were housed in the virus-free Animal Research Facility of University of Wisconsin–Madison. The housing was four animals per cage in high-density ventilated caging with automatic water. The husbandry rooms were monitored to have 12-hour light/dark cycles, 22.2°C temperature, and 45% humidity. RANKL+/+, RANKL−/−;Tg719, and RANKL−/−;Tg721 mice were fed a chow diet, whereas RANKL−/− mice were fed a powdered form of the chow diet (Harlan Teklad, Madison, WI, USA). To induce RANKL expression in vivo, experimental mice were injected with a single intraperitoneal (IP) dose of 1,25 (OH)2D3 (10 ng/g of body weight [bw]) in propylene glycol, PTH (1–84) (230 ng/g bw), or LPS (10 µg/g bw) in phosphate buffered saline (PBS) or a similar volume of appropriate vehicle as control. Animals were stratified to vehicle or treatment groups according to their body weight. Animals were euthanized and tissues collected 6 hours after 1,2(OH)2D3 (SAFC Global, Madison, WI, USA) or LPS (Sigma Aldrich, St. Louis, MO, USA) injection and 1 hour after PTH (Bachem California Inc., Torrance, CA, USA) injection. All injections and tissue collections were performed in the procedure rooms in the Research Animal Facility of the University of Wisconsin–Madison. All animal studies were reviewed and approved by the Research Animal Care and Use Committee of the University of Wisconsin–Madison.
BMD and micro–computed tomography analysis
Animals were sedated using isoflurane prior to and during the BMD measurements. BMDs were measured by dual X-ray absorptiometry (DXA) with a PIXImus densitometer (GE-Lunar Corp, Madison, WI, USA) in a blinded fashion. The total body BMD was measured as whole body minus calvarium, shoulder blades, and first few thoracic vertebrae. Vertebral BMD was measured in a rectangular region of interest (ROI) containing five to six of the lumbar vertebrae. BMD of the femur was measured with an ROI containing the right femur. Quality control of the PIXImus was performed daily using a proprietary skeletal phantom. Lumbar vertebrae 4 (L4) and femurs were fixed in 10% Millonig’s Modified Buffered Formalin (Leica Biosystems, Richmond, IL, USA) for 24 hours and gradually dehydrated through a series of ethanol solutions into 100% ethanol. Bones were scanned using a micro–computed tomography (µCT) instrument (Model µCT40; Scanco Medical, Wayne, PA, USA). Trabecular and cortical measurements were taken by µCT as described.(49,50)
Histology
Femurs were fixed in 10% Millonig’s Modified Buffered Formalin for 24 hours and then decalcified in 14% EDTA for 2 weeks. Dehydrated samples were then embedded in paraffin and cut into 5-µm longitudinal sections. Paraffin was removed and sections were rehydrated prior to staining. The sections were stained with hematoxylin and eosin stain (H&E) or tartrate-resistant acid phosphatase (TRAP), and counterstained with methyl green or 0.3% (wt/vol) toluidine blue. In order to determine osteoclast surface and number, quantitative histomorphometry was performed on the TRAP-stained femoral sections using a computer and digitizer tablet (OsteoMetrics, Decatur, GA, USA) interfaced to a Zeiss Axioscope (Carl Zeiss, Thornwood, NY, USA) with an attached drawing tube. The number of TRAP-positive cells and the surface occupied by TRAP-positive cells on the cancellous perimeter (osteoclast number and surface) were measured directly. Terminology recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research was used in this study.(51)
Gene expression
Tissues were dissected, frozen immediately in liquid nitrogen, and stored at −80°C. Frozen tissues were homogenized in Trizol Reagent (Life Technologies, Grand Island, NY, USA) and RNA was isolated according to the manufacturer’s instructions. Lymphocytes were isolated from bone marrow and spleen using Dynabeads Mouse pan B (B220) and Dynabeads FlowComp Mouse Pan T kits (CD90.2) (Life Technologies) according to the manufacturer’s directions. RNA from the isolated lymphocytes was prepared with Trizol Reagent (Life Technologies). RNA (1 µg) was treated with DNAse I (Life Technologies) and used as a template to synthesize cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). RNA isolation and cDNA production was performed in a blinded fashion. Relative mRNA levels were determined via multiplex TaqMan quantitative reverse transcription–PCR (RTPCR) using VIC-labeled Mouse ACTB and FAM-labeled TaqMan gene expression assays (Applied Biosystems). The following TaqMan Gene Expression probes (Applied Biosystems) were used for RT-PCR: TRAP (Mm00475698_m1), OPG (Mm01205928_m1), IL-6 (Mm0044619_m1), CD69 (Mm01183378_m1), mouse ACTB (4352341E), and RANKL (forward 5′-GGTCTAACCCCTGGACATGTG-3′; reverse 5′-CTTTGCAATGACATGGCATCCT-3′; probe FAM-5′-CTGAGAACCTTGAAATTAAG-3′-NFQ) (nonfluorescent quencher). Relative mRNA levels were calculated using the delta threshold cycle (ΔCt) method.(52)
Blood chemistry
Cardiac blood was collected at the time of euthanasia. Collected blood was maintained at room temperature for 30 minutes followed by centrifugation at 3,400 g for 12 minutes to obtain serum. Soluble RANKL (sRANKL) content in the serum was measured using an R&D Systems (Minneapolis, MN, USA) Quantikine Mouse RANKL (Cat. No. MTR00) kit according to the manual provided by the manufacturer.
Transgene copy number
Mouse tails (0.5 cm or less) were clipped and frozen; genomic DNA (gDNA) was isolated from the samples with proteinase K digestion and phenol/chloroform extraction. We obtained four custom TaqMan primers from Applied Biosystems for quantifying the copy number of the RANKL transgene: one targeted to the deleted region of the Tnfsf11 gene and therefore was absent in the RANKL−/− mice (forward 5′-GGTCTAACCCCTGGACATGTG-3′; reverse 5′-CTTTGCAATGACATGGCATCCT-3′; probe FAM-5′-CTGAGAACCTTGAAATTAAG-3′-NFQ), three targeted to the RANKL enhancer regions D2 (forward 5′-CATCTTCCAGGCCAGGCT-3′; reverse 5′-CAGAATTTGGTTTGCTGGTCTACAA-3′; probe FAM-5′-CAGGCTTCCTATGCAAATAA-3′-NFQ), D5 (forward 5′-ACTTCAACCAGAACCCATTCG-3′; reverse 5′-CTCGCCTCAT TGTGCAATTG-3′; probe FAM-5′-AACCACGGGCTTCACTGTCCTC-3′-NFQ) and T1 (forward 5′-TTTCCACATACCATCACATTTTCTG-3′; reverse 5′-CGGTTAATGCAGCCAACTG-3′; probe FAM-5′-AAACCACTT CCTCTCCCTGCTCC-3′-NFQ) (see Fig. 1). PCR was assembled as a multiplex reaction with FAM-labeled target probes and VIC-labeled TaqMan Copy Number Reference probes, mouse Tfrc (Life Technologies) according to the manufacturer’s directions. Copy number was calculated using Copy Caller v2.0 software (Life Technologies).
Flow cytometry
Femoral marrow and spleen cells were isolated and stained in BD Staining Buffer (BD Biosciences, San Jose, CA, USA). Following red blood cell lysis with ACK Lysing Buffer (Life Technologies), samples were blocked with CD16/CD32 for 5 minutes and then stained for 30 minutes using B-cell panel antibodies (CD45R/ B220-APC-Cy7, CD43-APC, CD24-BV421, IgM-FITC, IgD-BV510, and BP-1-PE) or T-cell panel antibodies (CD3-V450, CD8-Alexa Fluor 700, and CD4-APC). All antibodies were purchased from BD Biosciences. After the removal of unbound antibodies, samples were fixed with 2% formaldehyde for 24 hours and then analyzed by flow cytometry using a BD FACS Aria (BD Biosciences). Analysis of the samples was done using Flowjo Software (FlowJo, LLC, Ashland, OR, USA). Appropriate gates for the cell populations were drawn with guidance of Fluorescence Minus One (FMO) controls; the percentages of cell populations versus single cells were indicated.
Statistical evaluation
Data were analyzed using GraphPad Prism 4 software (GraphPad Software, Inc., La Jolla, CA, USA). All values are reported as the mean ± SD and differences between group means were evaluated using one-way ANOVA, two-way ANOVA, or Student’s t test as indicated in the figure legends.
Results
Constructing mRANKLTg/RANKL−/− mouse strains
The mouse Tnfsf11 gene locus on chromosome 14 is depicted in Fig. 1. As can be seen, an intergenic region extending upstream over 200 kb to the neighboring gene Akap11 contains a series of at least 10 regulatory control regions (enhancers, D1 to D7; T1 to T3)(34,35,39,40,42,43) that we have previously shown contribute to the regulated expression of RANKL in osteoblast lineage cells and/or immune cells. Active CTCF/RAD21 sites, which define the apparent functional boundaries of the mouse Tnfsf11 gene, are also indicated.(43) As seen in Fig. 1, this organization is highly conserved within the human TNFSF11 gene as well, although the gene is transcribed from the opposite strand and the regulatory domain spans a much larger segment of DNA. With this information as a guide, we prepared a mouse Tnfsf11 construct that spanned the region from +40 kb to −178 kb (relative to the Tnfsf11 TSS) containing the boundary CTCF/RAD21 sites as well as all other known regulatory sites by coupling intact BAC clone RP23–68C13 to a small segment of mouse BAC RP23–52A3 as illustrated in Fig. 1 using recombineering methodology. After insertion of a selectable IRES-linked luciferase reporter cassette into the 3′ noncoding portion of the final Tnfsf11 exon within the construct (as indicated in the cartoon in Fig. 1), the transgene was introduced by pronuclear injection into C57BL/6 mice. Luciferase-positive offspring were identified and used to create two separate murine RANKL transgenic (RANKLTg) strains termed RANKLTg719 and RANKLTg721, which, as documented in Supporting Fig. 1, contained 8.7 ± 1.4 and 5.3 ± 1.2 copies of the transgene, respectively. Both strains were crossed into the RANKL null (RANKL−/−) background via sequential breeding to create the two “rescue” strains RANKL−/−;Tg719 and RANKL−/−;Tg721. By virtue of luciferase activity, the RANKL−/−;Tg719 strain was predicted to display higher RANKL expression than that of RANKL−/−;Tg721 (data not shown). Individual lines were genotyped, expanded, and then used to assess the phenotype of the rescue strains relative to nontransgenic RANKL +/+ and RANKL−/− littermate controls.
General features of RANKL−/−;Tg719 and RANKL−/−;Tg721 rescue strains
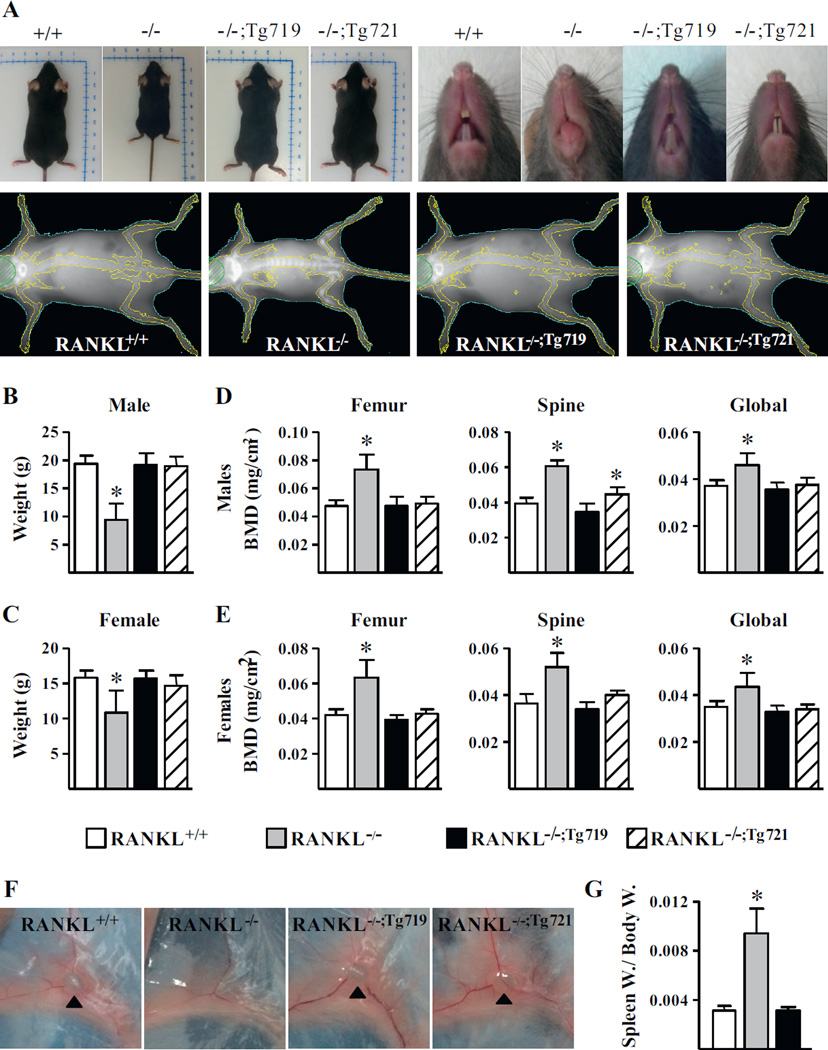
As documented in Fig. 2, although RANKL−/− mice exhibited a series of biological defects including: (1) growth retardation (Fig. 2 A); (2) decreased body weight (Fig. 2 B, C); (3) reduction in skeletal size (Fig. 2 A); (4) increased bone mass, as indicated visually (Fig. 2 A) and via radiographic analysis of BMD in both males and females (Fig. 2 D, E); (5) the formation of club-shaped long bones (Fig. 2 A); (6) the absence of incisor tooth eruption (Fig. 2 A); and (7) limited marrow cavities within long bones (not shown), all characterized previously,(2,16) these features were fully normalized in both the RANKL−/−;Tg721 and RANKL−/−;Tg719 strains of mice. In addition, although RANKL−/− mice failed to produce lymph nodes (Fig. 2 F) and displayed splenomegaly characteristic of extramedullary hematopoiesis as indicated by enlarged spleens seen in Fig. 2 G, the presence of the Tnfsf11 transgene in the RANKL−/−;Tg719 and RANKL−/−;Tg721 rescue strain led to normal lymph node development and spleen size. Importantly, and unlike the RANKL−/− strain, female transgenic mice were reproductively active, giving birth to litters of normal size that grew normally. Based upon this overt reproductive success, we conclude that mammary gland development, subsequent lactation, and the capacity of these strains of mice to mobilize sufficient calcium and phosphorus from their skeleton for the offspring proceeded normally as well. These observations suggested that the Tnfsf11 transgene was indeed active in producing RANKL in a number of potential target tissues.
Fig. 2.
The mTnfsf11 transgene rescues specific features of RANKL−/− mice. Two strains of mice that lack the endogenous RANKL but contain the mTnfsf11 BAC transgene, namely RANKL−/−;719Tg and RANKL−/−;721Tg,were produced. The general phenotype of these rescue strains were compared to the wild-type (RANKL+/+) and RANKL null (RANKL−/−)mice. (A)Whole-body, incisor tooth, and DXA images of male mice from all strains were taken at 5 weeks of age. (B–E)Bodyweight (B, C) and DXABMDs (D, E) of male and female mice were measured at 5weeks of age (n = 3to18mice/genotype). (F) The presence of inguinal lymph nodes in 5-weekold male mice are indicated with black triangles. (G) Spleen weights normalized to body weights of 5-week-old male mice (n = 3 to 4 mice/group). The values shown represent the mean ± SD. All statistical comparisons were derived using one-way ANOVA analysis (B–E) and Student’s t test (G). * p <0.05 compared to the wild-type group. BAC = bacterial artificial chromosome; DXA = dual-energy X-ray absorptiometry; Spleen W. = spleen weight; Body W. = body weight.
Relative expression of RANKL transcripts in skeletal tissues of RANKL−/−;Tg719 and RANKL−/−;Tg721 rescue strains
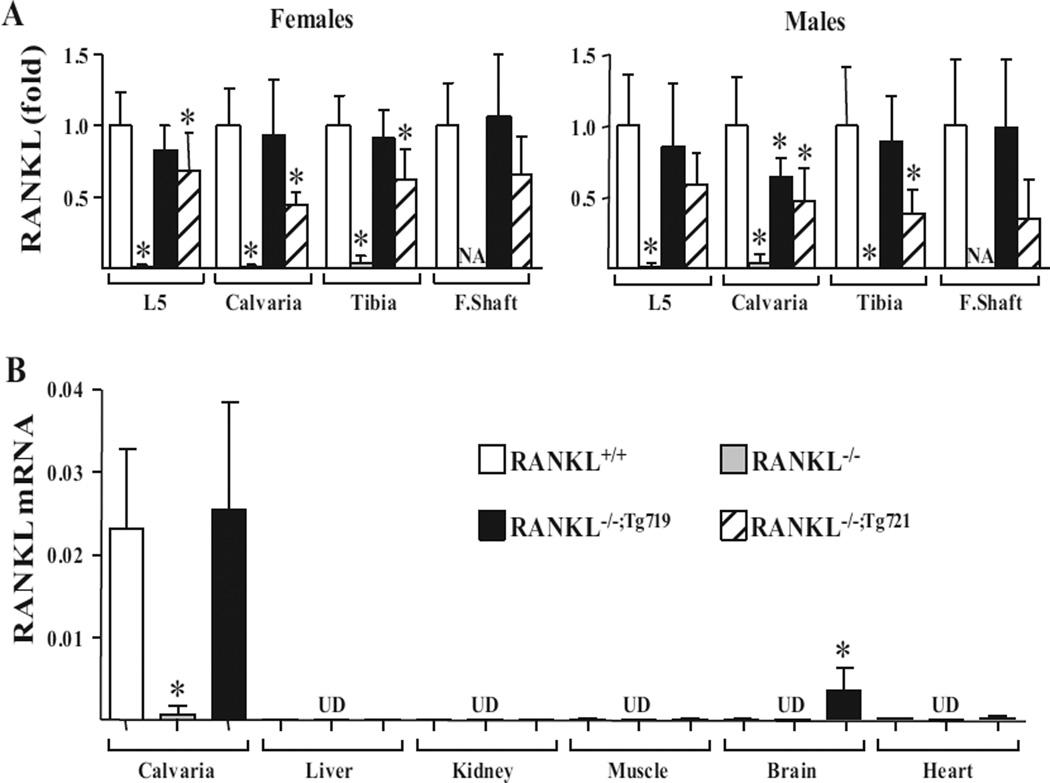
To explore the level of RANKL expression from the Tnfsf11 transgene in the two RANKL−/−;Tg719 and RANKL−/−;Tg721 mouse strains, we first isolated RNA from four independent sites in the skeleton and examined the level of RANKL mRNA relative to that measured in RANKL+/+ and RANKL−/− littermate controls. As shown in Fig. 2 B–E and reported by others,(53) skeletal phenotypes and osteoclast numbers of male and female mice differ even at 1 month of age. Because of these significant gender-specific differences in skeletal phenotype, osteoclast numbers, and the potential role of RANKL in these differences, RANKL RNA levels of the skeletal tissues were quantitated separately in male and female mice. As can be seen in Fig. 3 A, RANKL mRNA was completely absent in samples obtained from the skeletal sites of both sexes of RANKL−/− mice, but its levels in the RANKL−/−;Tg719 strain were equivalent to those observed at the identical skeletal sites in RANKL+/+ mice, although RANKL expression in mice of the RANKL−/−;Tg721 strain was somewhat reduced. Importantly, the results in Fig. 3 B indicate that RANKL transcripts in the RANKL−/−;Tg719 strain were at low to undetectable levels in the kidney, liver, and lung, consistent with of levels of endogenous RANKL in these tissues. The endogenous RANKL gene was expressed at very low levels in brain; however, RANKL transcripts in brain tissue in the RANKL−/−;Tg719 strain were higher than levels of the endogenous gene, likely reflecting a unique ectopic expression of the transgene in this tissue.
Fig. 3.
The mTnfsf11 transgene is expressed in a tissue-specific manner with mRNA levels comparable to those of endogenous Tnfsf11. (A) RANKL mRNA levels in (L5), calvaria, tibia, and F.Shaft of 5-week-old male and female mice were measured by quantitative RT-PCR (n = 4 to 16 mice/ genotype). RANKL mRNA levels are indicated for each tissues as fold-change normalized to RANKL+/+ levels. Due to lack of marrow space and therefore F.Shafts in RANKL−/− mice, RANKL mRNA measurements could not be obtained (NA). (B) RANKL mRNA levels in calvaria, liver, kidney, muscle, brain, and heart of 4-week-old male RANKL+/+, RANKL−/−, and RANKL−/−;Tg719 mice measured by quantitative RT-PCR (n = 4 mice/genotype). All quantitative RT-PCR values were normalized to the housekeeping gene beta-actin and indicated as mean ± SD. All statistical comparisons were derived using a one-way ANOVA analysis. * p <0.05 compared to the wild-type group. NA = not available; F.Shaft = femur shaft; UD = level undetectable by RT-PCR; L5 = lumbar vertebra 5.
Despite considerable effort; we were unable to detect either endogenous or transgene-derived RANKL protein by Western blot analysis, a result consistent with difficulties associated with detecting low levels of RANKL protein using currently available antibodies. However, we were able to measure sRANKL levels in the circulation. Although circulating sRANKL levels were undetectable in RANKL−/− mice, both RANKL−/−;Tg719 and RANKL−/−;Tg721 rescue strains produced significant levels of sRANKL (Supporting Fig. 2). Importantly, the levels of sRANKL in the RANKL−/−;Tg719 strain were similar to that of RANKL+/+ mice, whereas sRANKL levels of the RANKL−/−;Tg721 strain were significantly lower than wild-type controls (Supporting Fig. 2). These results indicate that tissue-specific RANKL transcripts are indeed being produced by the Tnfsf11 transgene and the levels of the transcripts at individual sites, as well as the circulating sRANKL levels in the RANKL−/−;Tg719 strain are similar to that seen in RANKL+/+ mice. In addition, as predicted from the initial luciferase measurements that were used to identify the original transgenic strains (data not shown), production of RANKL mRNA and circulating sRANKL in RANKL−/−;Tg721 mice was somewhat lower.
RANKL production from the Tnfsf11 transgene in the RANKL−/−;Tg719 and RANKL−/−;Tg721 strains of mice rescues the growth plate phenotype of the RANKL−/− mouse and restores osteoclast formation
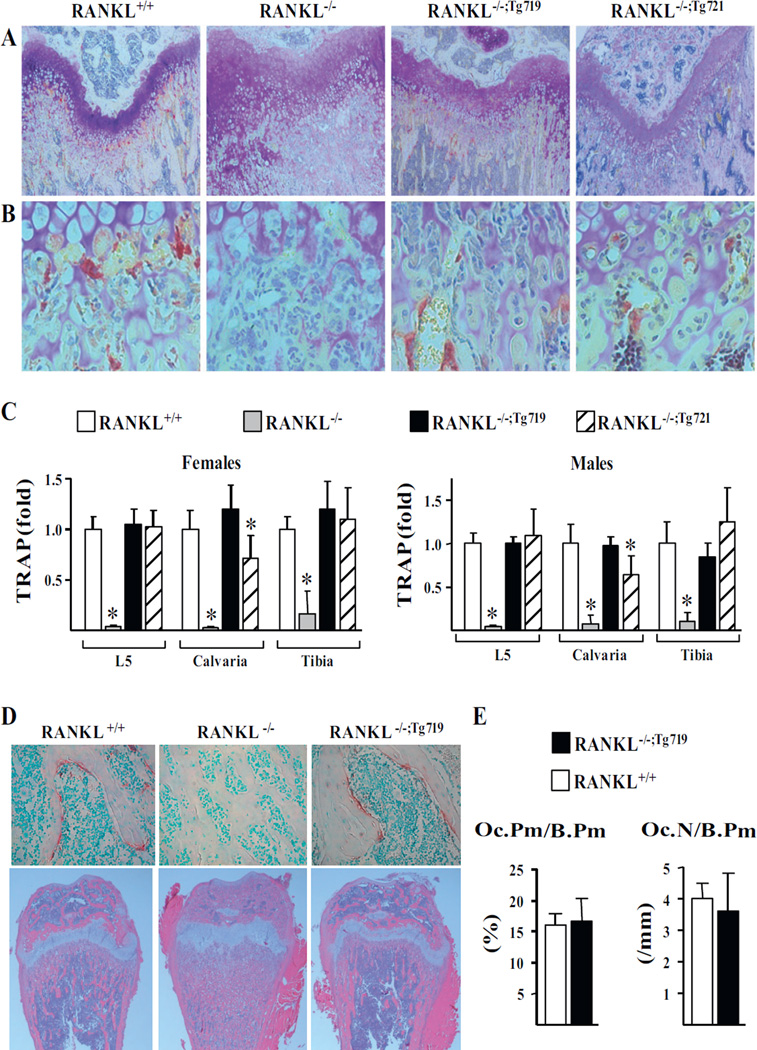
Based upon the above observations, we next examined whether the RANKL produced in the RANKL−/−;Tg719 and RANKL−/−;Tg721 mouse strains was capable of fully restoring the skeletal deficiencies apparent in the RANKL−/− mouse. As can be seen in the histological section documented in Fig. 4 A, D, although the growth plate in the RANKL−/− strain was disorganized as a consequence of the loss of RANKL expression, growth plate sections isolated from the bones of both transgenic rescue strains were normal and appeared to be similar to those obtained from RANKL+/+ mice. In addition, as seen in the cross-sections depicted in Fig. 4 B, D, although TRAP stained osteoclasts were undetectable in RANKL−/− mice, they were clearly present in both the RANKL+/+, RANKL−/−;Tg721, and RANKL−/−;Tg719 strains of mice. Indeed, as can be seen in Fig. 4 E, both osteoclast number and the percentage of cancellous bone surface covered by osteoclasts were equivalent in both the RANKL+/+ and RANKL−/−;Tg719 strains of mice, indicating that the Tnfsf11 transgene was capable of fully restoring the process of osteoclast formation. These results were confirmed through the quantitation of TRAP mRNA in tissue samples from L5 vertebrae, tibias, and calvaria from the skeletons of mice from both rescue strains whose levels, as seen in Fig. 4 C, were equivalent to those quantitated in the tissues of RANKL+/+ mice. These results indicate that the expression of RANKL from the Tnfsf11 transgene fully restored the deficiency in osteoclast formation observed in the RANKL−/− mouse, likely leading to the recovery of normal bone remodeling (Fig. 4 D).
Fig. 4.
The mTnfsf11 transgene is capable of rescuing growth plate organization and supporting osteoclastogenesis. (A, B) Femoral sections of 5-week-old male mice of the indicated genotype were stained with TRAP and toluidine blue to examine growth plate organization (A) and osteoclast formation (B). (C) TRAP mRNA levels in L5, calvaria, and tibia of 5-week-old female and male mice of the indicated genotype were measured using quantitative RT-PCR. All values were normalized to beta-actin and represent the mean of 3 to 16 animals per group. (D) TRAP and methyl green–stained (top) and H&E-stained (bottom) femoral sections of 5-week-old male mice. (E) Histomorphometric analyses of Oc.Pm/B.Pm and Oc.N/B.Pm were performed in TRAP-stained and methyl green–stained femoral sections of 5-week-old male mice of the indicated genotype (n = 3 animals/ group). All statistical comparisons of RT-PCR were derived using one-way ANOVA analysis and statistical comparisons of histomorphometric parameters were performed using Student’s t test. * p <0.05 compared to the wild-type group. Oc.Pm/B.Pm = osteoclast perimeter per bone perimeter; Oc.N/B.Pm = osteoclasts number per bone perimeter; H&E = hematoxylin and eosin stain; TRAP = tartrate-resistant acid phosphatase; L5 = lumbar vertebra 5.
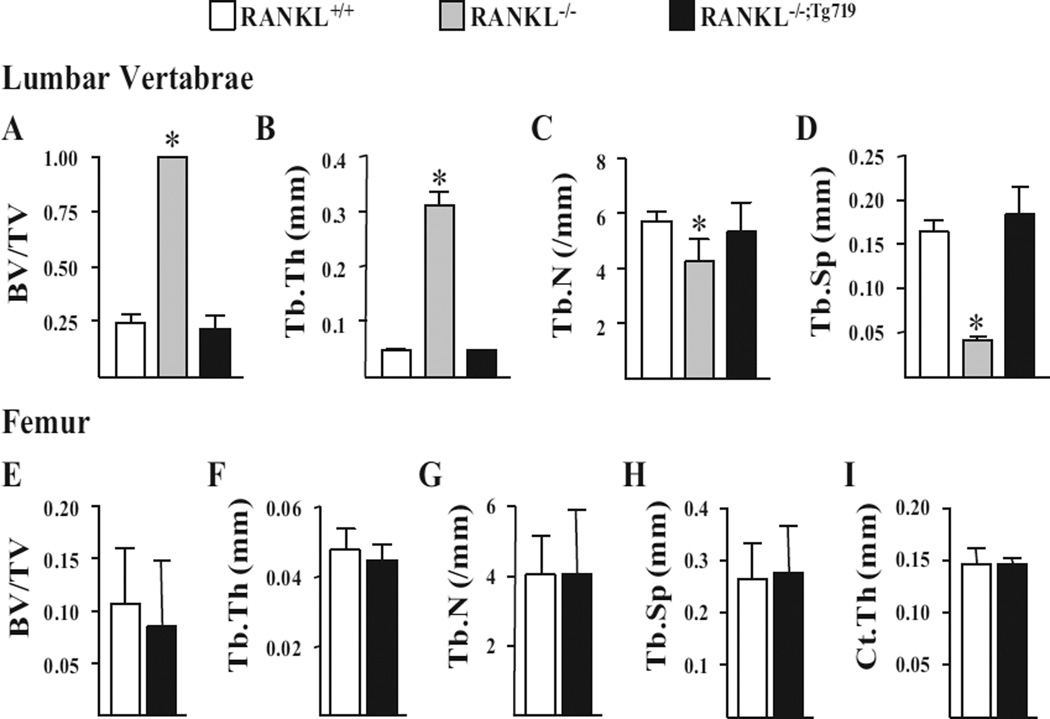
RANKL production from the Tnfsf11 transgene in the RANKL−/−;Tg719 and RANKL−/−;Tg721 strains of mice rescues the skeletal microarchitecture of the RANKL−/− mouse
To assess the quality of bone that resulted from Tnfsf11 transgene expression, we next measured bone architectural parameters in the vertebrae and femurs of RANKL+/+, RANKL−/−, and RANKL−/−;Tg719 mice by µCT analysis. Accordingly, although analysis of the RANKL−/− strain exhibited higher vertebral and femoral cancellous bone volume (Fig. 5 A, E) with thicker trabeculae (Fig. 5 B, F) that were reduced in number (Fig. 5 C, G) and more closely spaced (Fig. 5 D, H), each of these parameters in the RANKL−/−;Tg719 strain was normalized relative to RANKL+/+ mice. Moreover, and consistent with the equivalency in osteoclast numbers and surface (Fig. 4 C–E), all cancellous bone parameters in spine and femur (Fig. 5 A–H), as well as the cortical thickness in femoral midshafts (Fig. 5 I), were indistinguishable between RANKL+/+ and RANKL−/−;Tg719 rescue mice. These results indicate that when expressed at levels observed in the RANKL−/−;Tg719 strain of mice, the Tnfsf11 transgene is capable of fully rescuing all skeletal parameters measured that were aberrant in the RANKL−/− mouse.
Fig. 5.
The mTnfsf11 transgene is capable of rescuing the osteopetrotic phenotype of RANKL−/− mice. L4 vertebrae and femur of 5-week-old mice of the indicated genotypes were used for theµCT analysis. (A–H) Vertebral (A–D) and femoral (E–H) cancellous bone architecture was determined by measuring BV/ TV (A, E), Tb.Th (B, F), Tb.N(C, G), and Tb.Sp (D, H) viaµCT analysis. (I) Ct.Th was measured at the femoral midshaft. All values represent the mean and SD with 4 to 6 animals per group. Statistical comparisons of the vertebral µCT analysis (A–D)were accomplished using one-way ANOVA and femoral µCt analysis (E–I) were accomplished using Student’s t test. * p <0.05 compared to the wild-type group. µCT = micro–computed tomography; BV/TV = bone volume over tissue volume; Tb.Th = trabecular thickness; Tb.N = trabecular number; Tb.Sp = trabecular spacing; Ct.Th = cortical thickness; L4 = lumbar vertebra 4.
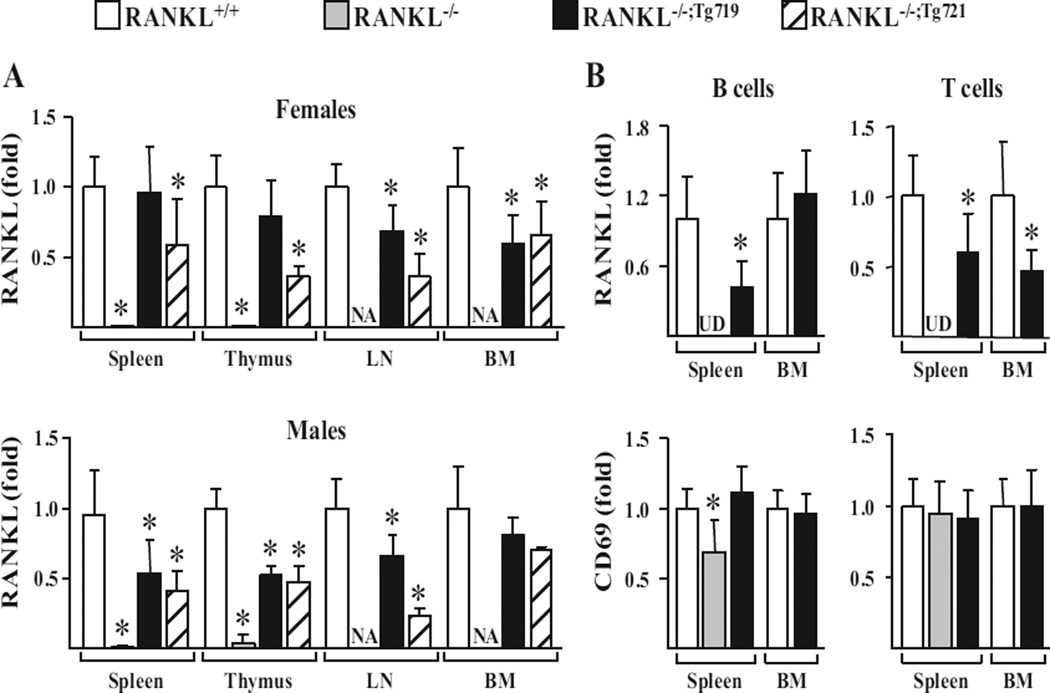
Relative expression of RANKL transcripts in lymphoid tissues of RANKL−/−;Tg719 and RANKL−/−;Tg721 rescue strains
Initial observations established that the Tnfsf11 transgene was capable of both restoring lymph node organogenesis and preventing extramedullary hematopoiesis, as evidenced by the absence of splenomegaly observed in RANKL−/− mice, suggesting that RANKL expression had been restored in immune tissues. To examine this directly, we isolated RNA from spleen, thymus, lymph nodes, and bone marrow of RANKL+/+ and RANKL−/− mice, and the two RANKL−/−;Tg rescue strains, and quantitated the levels of RANKL mRNA in these samples. As can be seen in Fig. 6 A, RANKL mRNA was completely absent in all samples obtained from the tissues isolated from both sexes of RANKL−/− mice, but the levels identified in the tissues of the two rescue strains were strikingly increased, although in some cases not fully restored to levels seen in the RANKL+/+ mouse. Thus, although RANKL transcript levels were fully normalized in the spleen and thymus of females derived from the RANKL−/−;Tg719 strain, they were statistically lower in the lymph nodes and bonemarrow of those same females as well as in all of the immune tissues obtained from male mice (Fig. 6 A). In addition, as seen in skeletal tissues, the overall expression of RANKL mRNA in all immune tissues derived from the RANKL−/−;Tg721 strain was generally lower than that measured in the RANKL−/−;Tg719 strain. These data indicate that although RANKL mRNA expression levels are strongly elevated in immune tissues as a consequence of Tnfsf11 transgene expression in the RANKL−/−;Tg719 strain, their levels appear to be fully restored only in spleen and thymus of female mice.
Fig. 6.
The mTnfsf11 transgene is expressed in lymphoid tissues and in both T cells and B cells. (A) RANKL mRNA expression in the spleen, thymus, LN, and BM of 5-week-old female and male mice of the indicated genotypes were measured via quantitative RT-PCR (n = 3 to 16 mice/genotype). (B) RNA was obtained from B cells and T cells isolated from bone marrow and spleen of 5- to 6-week-old male mice of the indicated genotypes and used to measure RANKL and CD69 mRNA levels via quantitative RT-PCR (n = 8 to 9 mice/genotype). All values were normalized to beta-actin. Statistical comparisons were accomplished using one-way ANOVA when more than two groups were being compared (A) and Student’s t test was used when two groups were being compared (B). * p <0.05 compared to the wild-type group. LN = lymph node; BM = bone marrow; UD = level undetectable by RT-PCR.
Relative expression of RANKL transcripts in splenic-derived and bone marrow–derived B cells and T cells of RANKL−/−;Tg719 and RANKL−/−;Tg721 rescue strains
To explore the above phenomenon further, we isolated B and T cells from splenic and bone marrow tissues obtained from RANKL+/+, RANKL−/−, and RANKL−/−;Tg719 mice and measured the expression of RANKL mRNA from the isolated cells. As can be seen in Fig. 6 B, mRNA for CD69, a control gene whose expression increases upon lymphocyte activation, was generally unaffected, suggesting that isolation of lymphocytes were comparable between all genotypes. However, the levels of RANKL mRNA in B and T cells were highly elevated in the RANKL−/−;Tg719 strain compared to the levels that were undetectable in the RANKL−/−mouse, although this elevation did not reach levels of RANKL mRNA observed in RANKL+/+–derived B and T cells from both tissue sources (Fig. 6 B). The absence of bone marrow prevented the isolation of B and T cells from this source in the RANKL−/− mouse strain.
Effects of RANKL expression on B-cell and T-cell development
RANKL is known to participate in B-cell and T-cell lineage maturation. As a consequence, we obtained bone marrow and splenic tissue from RANKL+/+, RANKL−/−, and RANKL−/−;Tg719 mice, selectively identified subpopulations of these cells by flow cytometry and quantitated their numbers. As documented in Table 1, the percentage of B cells obtained from RANKL+/+ and RANKL−/−;Tg719 bone marrow in each maturation stage was not statistically different, although because of the absence of marrow in the RANKL−/− strain, a selective differential could not be evaluated among the B-cell stages, as has been explored in detail by others.(2,21) An examination of maturation and lineage stages of B cells and T cells derived from the spleens of these mice, however, indicated that the RANKL−/− mouse was deficient in B220+IgM+ and B220+IgD+ B cells and that the presence of the Tnfsf11 transgene restored these maturation states to levels seen in RANKL+/+ mice. CD3+, CD4+, and CD8+ T-cell levels were not different across the three strains of mice, suggesting that, as reported,(2,21) RANKL expression did not influence their developmental progression.
Table 1.
RANKL Transgene is Capable of Supporting Normal B-Cell and T-Cell Development
| RANKL+/+ | RANKL−/− | RANKL−/−;Tg719 | |
|---|---|---|---|
| Bone marrow | |||
| PreProB cells (B220+CD24lowBP1−) (%) | 3.6 ± 0.6 | × | 3.2 ± 0.7 |
| ProB cells (B220+CD24+BP1−) (%) | 3.8 ± 1.0 | × | 3.4 ± 0.9 |
| PreB cells (B220+CD24+BP1+) (%) | 9.4 ± 1.3 | × | 8.6 ± 3.0 |
| Immature B cells (B220+CD24+IgM+IgD−) (%) | 14.7 ± 2.9 | × | 12.6 ± 3.0 |
| Transitional B cells (B220+CD24+IgMhighIgDlow) (%) | 3.3 ± 0.6 | × | 2.7 ± 1.0 |
| Early mature B cells (B220+CD24+IgMhighIgDhigh) (%) | 1.0 ± 0.8 | × | 1.1 ± 0.6 |
| Late mature B cells (B220+CD24+IgMlowIgDhigh) (%) | 1.7 ± 1.4 | × | 1.9 ± 0.9 |
| CD3+ T cells (%) | 1.9 ± 0.4 | × | 1.7 ± 0.6 |
| Spleen | |||
| B220+IgM+ B cells (%) | 26.6 ± 3.2 | 15.0 ± 4.8* | 28.6 ± 3.5 |
| B220+IgD+ B cells (%) | 40.6 ± 3.8 | 16.2 ± 5.2* | 43.3 ± 4.7 |
| CD3+ T cells (%) | 35.6 ± 5.8 | 37.4 ± 3.3 | 32.1 ± 3.1 |
| CD4+ T cells (%) | 21.9 ± 4.7 | 22.9 ± 3.3 | 19.4 ± 1.1 |
| CD8+ T cells (%) | 14.2 ± 1.4 | 15.0 ± 1.4 | 13.2 ± 1.9 |
Values are mean ± SD of 3 to 4 animals per genotype. All animals used in the experiments were 6-week-old female littermates. Flow cytometry analysis of the B and T cells in bone marrow and spleen were done after red blood cell lysis.
p<0.05 compared to wild-type as evaluated by one-way ANOVA.
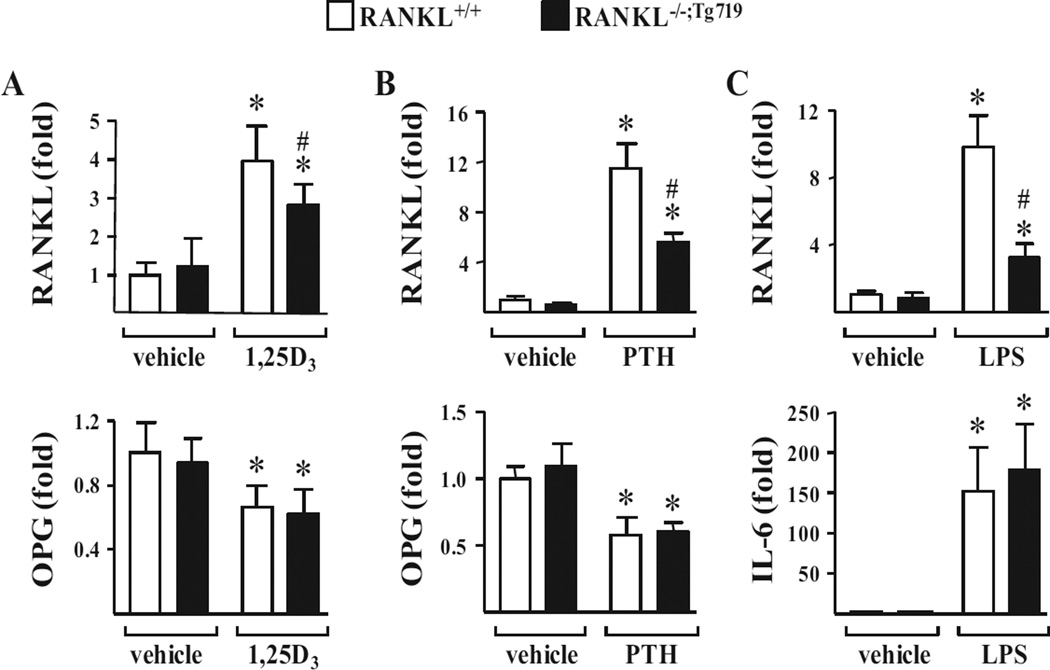
The Tnfsf11 transgene recapitulates specific regulatory capabilities inherent to its endogenous counterpart
As discussed earlier, the Tnfsf11 gene is regulated by numerous systemic hormones and local cytokines whose actions have been localized to a series of upstream enhancers that are contained within the Tnfsf11 transgene. In a final series of experiments, we tested whether these upstream enhancers retained the capacity to regulate the expression of RANKL from the RANKL−/−;Tg719 strain of mice. To this end, we administered a single dose of 1,25(OH)2D3 (10 ng/g bw), PTH (100 ng/g bw), or the cytokine inducer LPS (100 µg/g bw) to cohorts of RANKL+/+ and RANKL−/−;Tg719 mice and measured the levels of RANKL and control OPG or IL-6 mRNAs isolated from L5 vertebrae at 6 hours, 1 hour, or 6 hours later, respectively. As can be seen in Fig. 7 A, 1,25(OH)2D3 strongly induced RANKL mRNA expression in both strains, an induction that was also observed for PTH and documented in Fig. 7 B. However, the induction of RANKL mRNA in the RANKL−/−;Tg719 strain did not fully reach that observed in the RANKL+/+ mouse. Likewise, as can be seen in Fig. 7 C, LPS also induced RANKL mRNA in the RANKL−/−;Tg719 strain, although again, this level of induction was reduced relative to that seen in the RANKL+/+ mouse. Control mRNAs (OPG or IL-6 mRNAs) in both strains were similarly regulated by all three modulators, and comparable observations were made in calvaria, femur shafts, and bone marrow as documented in Supporting Fig. 3. We conclude that although the transgene retains the capacity to regulate expression of its cognate RANKL mRNA by exogenously applied factors, the level of induction from the transgene in the RANKL−/−;Tg719 strain is slightly diminished. The reasons for this are unknown, although they contrast with the ability of the transgene to fully rescue the organ, skeletal, and immune tissue phenotypes of the RANKL−/− mouse.
Fig. 7.
mTnfsf11 transgene expression is inducible with 1,25(OH)2D3, PTH, and LPS. All animals used in the following experiments were littermates. (A) Seven-week-old to 8-week-old female RANKL+/+ or RANKL−/−;Tg719 mice were injected i.p. with 1,25(OH)2D3 (1,25D3) (10 ng/g of bw in propylene glycol) or vehicle (propylene glycol). The animals were euthanized and the tissues were collected 6 hours after injection. RANKL and OPG mRNA expression in the L5 was measured via quantitative RT-PCR (n = 4 to 6 mice/group). (B) Eight-week-old male RANKL+/+ or RANKL−/−;Tg719 mice were injected with PTH (1–84) (230 ng/g bw in PBS) or vehicle (PBS) and the same tissues as in A collected 1 hour after injection. RANKL and OPG mRNA expression in L5 was measured via quantitative RT-PCR (n = 4 mice/group). (C) Five-week-old female RANKL+/+ or RANKL−/−;Tg719 mice were injected with LPS (10 µg/g bw in PBS) or vehicle (PBS) and tissues were collected 6 hours after injection. RANKL and IL-6 mRNA expression in L5 was measured via quantitative RT-PCR (n = 4 to 5 mice/group). All values indicated represent the mean ± SD and statistical comparisons were accomplished using two-way ANOVA. * p <0.05 effect of treatment within the same genotype, #p <0.05 effect of genotype within the same treatment. bw = body weight; OPG = osteoprotegerin; IL-6 = interleukin-6; L5 = lumbar vertebra 5; LPS = lipopolysaccharide; PTH = parathyroid hormone.
Discussion
In this report, we prepared a 220-kb mouse Tnfsf11 transgene that contained the RANKL transcription unit, together with its extended upstream regulatory domain, and assessed the activity of this transgene in vivo in the context of the RANKL-null mouse. We show that this transgene produced basal RANKL transcripts in RANKL−/− mice that are transgene copy number–dependent, tissue-specific, and synthesized at levels that are generally consistent with those transcribed from the endogenous Tnfsf11 gene. The levels of RANKL mRNA produced by this transgene are capable of (1) restoring a normal growth pattern; (2) restoring lymph node organogenesis; (3) enabling osteoclast-dependent tooth eruption; and (4) preventing the splenomegaly that occurred as a result of the development of extramedullary hematopoiesis due to bone marrow occlusion, all of which are features of the RANKL−/− mouse. RANKL expression from this transgene also restores reproductive capacity, facilitates normal mammary gland development and lactation, and rescues the ability of these mice to mobilize skeletal calcium and phosphorus levels that are essential for the development of healthy offspring. Overall, these results demonstrate that the Tnfsf11 transgene recapitulates the tissue-specific expression of RANKL mRNA transcribed from the endogenous Tnfsf11 gene, and that the mRNA produced resulted in levels of RANKL protein which fully restored the major phenotypic deficits that are apparent in the RANKL−/− mouse.
In addition to the production of appropriate levels of RANKL mRNA and circulating sRANKL, the presence of the transgene in the RANKL−/−;Tg719 strain fully rescued all of the aberrant skeletal and immune features observed in the RANKL−/− mouse.(2,16) At the level of the skeleton, for example, the transgene rescued virtually all of the defects associated with the deficiency in osteoclast formation and activity seen in RANKL−/− mice, leading to a normalization of BMD, growth plate organization, and all parameters examined that were associated with skeletal integrity and intact bone remodeling. Interestingly, although the RANKL−/−;Tg721 strain expressed lower levels of RANKL mRNA and sRANKL, all of the features associated with the growth plate, BMD, and recovery of lymph node organogenesis appeared normal. These results suggest that the levels of RANKL protein necessary for diverse functions in the mouse are subject to variation, a hypothesis that is consistent with the fact that although RANKL−/− mice are profoundly compromised, mice heterozygous for the mutant Tnfsf11 allele exhibit minimal phenotypic deficiencies. Additional studies, particularly those that directly measure the level of RANKL protein in specific tissues, will be necessary to assess this hypothesis. Finally, the transgene also normalized the pattern of B-cell maturation in the spleen, as identified by the recovery of B220+IgM+ and B220+IgD+ B-cell populations and restored the extensive maturation states present in the rescued bone marrow.
Both the pattern of RANKL mRNA expression from the Tnfsf11 transgene and the relative levels of expression across several tissues compared to that seen in the RANKL+/+ mouse suggest that this transgene carries all the genetic information necessary for the appropriate expression of RANKL mRNA across multiple tissues. Perhaps most interesting; however, is the ability of the transgene to direct the regulation of RANKL mRNA by 1,25 (OH)2D3, PTH, and LPS in a fashion similar to that observed for the endogenous gene. This feature provides further validation that the Tnfsf11 transgene we have constructed contains key elements that mediate functional regulation in cis. It is important to note, however, that although these activities of the transgene provide support for the presence of important regulatory domains, they do not provide additional evidence for the locations of these regulatory elements that are indicated in Fig. 1. Indeed, this confirmation is derived, in part, from the skeletal loss of function phenotype that emerged as a result of the genomic deletion of the D5 enhancer in the Tnfsf11 gene in vivo that was characterized by the gene’s significantly reduced transcriptional response to both 1,25(OH)2D3 and PTH and by the low osteoclast number and elevated bone mass in D5 null mice.(36–38) Several additional genomic enhancer deletions have been created recently within the mouse Tnfsf11 regulatory domain in vivo, and the phenotypes resulting from these individual deletions are providing additional confirmation of the precise regulatory functions we have previously ascribed for each of these specific regions (Onal et al., unpublished data). A question does emerge as to why the RANKL mRNA response to 1,25(OH)2D3, PTH, and LPS from the transgene is not as robust as that seen from the endogenous gene in RANKL+/+ mice. One possibility is that the integration site of the transgene exerts a specific impact on regulatory expression. Alternative explanations include the possibility that insertion of the luciferase cassette into the 3′-UTR may have affected appropriate control of RANKL mRNA stability, that copy number may cause an unexpected dispersal of limited endogenous regulatory factors, or that additional cis regulatory components located outside the boundaries of the transgene may contribute. Although the latter is possible, it seems highly unlikely given the co-regulatory role of these elements in directing tissue-specific expression of the transgene and the ability of the transgene to fully rescue the phenotype of the RANKL−/− mouse. However, none of these issues detract from the conclusion that the Tnfsf11 transgene appears to retain the genetic information that is sufficient for appropriate tissue-specific and regulated expression of RANKL mRNA.
The complex regulation of the Tnfsf11 gene through multiple elements located many kilobases from the gene’s TSS was originally thought to represent a paradigm shift in the mechanism through which a gene could be regulated.(34) More recently, however, extensive application of unbiased genomewide methods in defining transcription factor binding sites now suggests that the regulatory features of this gene may represent the norm rather than the exception, particularly in genes whose products represent important regulatory components in their own right.(54) Indeed, our recent studies of not only the VDR, but transcription factors such as RUNX2, C/EBPβ, and CREB, as well as epigenetic enhancer signatures exemplified by specific histone modifications, support this conclusion for a number of genes of regulatory importance.(55–58) It is important to note that, due to the distal nature of many of these regulatory elements, genes immediately adjacent to active enhancers may not represent their regulable targets.(47,59,60) Thus, additional evidence is required to establish a linkage between regulatory elements and nearby genes. To this end, we have used modified BAC clones such as the transgene described here to explore enhancer-gene relationships for a number of genes in vitro and in vivo, including those for Tnfsf11, Vdr, and Cyp24a1.(36,47,48,59,61) The creation of these recombinant BACs have been particularly useful, because they can be used directly as transgenes in mice, analogous to the Tnfsf11 gene described herein. Interestingly, the development of gene-editing methods, as most recently exemplified by clustered regularly interspaced short palindromic repeats (CRISPR) systems,(62) will likely supplant the use of randomly integrated transgenes to explore regulatory function. This approach will not, however, allow the humanization of mice as has been accomplished through the use of human BAC transgenes in specific gene null backgrounds,(48,63) nor will it permit determination of whether a particular regulatory region is sufficient for appropriate expression in vivo.
In recent studies, a human TNFSF11 transgenic mouse has been created that mediates overexpression of human RANKL in wild-type mice.(64) This transgene, containing 74 kb of upstream sequence relative to the TSS, produces exceptionally large amounts of RANKL mRNA in numerous tissues that results in severe osteoporosis. As is suggested by our established boundaries of the human TNFSF11 gene depicted in Fig. 1, it seems unlikely that the human transgene used in the Rinotas and colleagues(64) report contains sufficient distal genetic information to direct either appropriate tissue-specific or hormonally-regulated RANKL expression. In this report, human RANKL (hRANKL) mRNA from this transgene is produced in a wide variety of tissues in mice that do not normally express the gene,(64) and no information was provided to indicate that the transgene was regulated by such factors such as 1,25(OH)2D3 or PTH. Overexpression of the construct in the background of a mouse carrying a Tnfsf11 allele that produces an inactive RANKL protein was able to rescue some aspects of the skeletal phenotype.(64) However, it is unclear whether this was due to expression of the transgene in the appropriate cells or, alternatively, to incorporation of transgenic RANKL into trimers with the inactive protein, and thereby functionally complement the RANKL mutant. Further analysis of this transgene will be required to understand the nature of its expression in the mouse, but at this point it appears that this transgene is comprised of a segment of DNA that simply overexpresses human RANKL in a nonspecific and unregulated manner. We predict that a segment of human DNA with boundaries as identified in Fig. 1 will be necessary to achieve authentic tissue-specific expression and full regulatory capabilities in vivo.
In summary, we have prepared a mouse Tnfsf11 transgene whose boundaries were predetermined through an extensive series of studies using unbiased ChIP techniques, BAC clone mutagenesis, and specific enhancer dissection. This segment of DNA retains the ability to direct appropriate RANKL mRNA expression in both a tissue-specific and regulated fashion in mice and is capable of rescuing the major aberrant developmental, skeletal, hematopoietic, and immune cell deficiencies inherent to the RANKL null mouse. These studies demonstrate that we have accurately defined the boundaries of RANKL gene regulatory elements. Such information will be essential for understanding the role of previously identified SNPs contributing to human bone mass and for a more complete understanding of RANKL gene regulation.
Supplementary Material
Acknowledgments
This work was supported by NIAMS grant AR-074993 and NIDDK grant DK-072281, both to JWP. We thank members of the Pike laboratory for their contributions to this work. We also thank members of the UW Biochemistry Animal Support Staff, the UAMS Animal Resources Staff, and the UWCCC Flow Cytometry Laboratory for their contributions to this effort.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures
All authors state that they have no conflicts of interest.
Authors’ roles: Study design: JWP, KAB, and MO. Study conduct: MO. Establishment of animal model: KAB and JJG. Data collection: MO, EMR, ALD, HCS, JX, and MP. Data analysis: MO. Data interpretation: MO, JWP, and CAO. Drafting manuscript: JWP and MO. Revising manuscript content: MO, JWP, and CAO. Approving final version of manuscript: JWP. JWP takes responsibility for the integrity of the data analysis.
References
- 1.Lacey DL, Timms E, Tan HL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 2.Kong YY, Yoshida H, Sarosi I, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397(6717):315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 3.Kim D, Mebius RE, MacMicking JD, et al. Regulation of peripheral lymph node genesis by the tumor necrosis factor family member TRANCE. J Exp Med. 2000;192(10):1467–1478. doi: 10.1084/jem.192.10.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hess E, Duheron V, Decossas M, et al. RANKL induces organized lymph node growth by stromal cell proliferation. J Immunol. 2012;188(3):1245–1254. doi: 10.4049/jimmunol.1101513. [DOI] [PubMed] [Google Scholar]
- 5.Kong YY, Feige U, Sarosi I, et al. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature. 1999;402(6759):304–309. doi: 10.1038/46303. [DOI] [PubMed] [Google Scholar]
- 6.Takayanagi H, Iizuka H, Juji T, et al. Involvement of receptor activator of nuclear factor kappaB ligand/osteoclast differentiation factor in osteoclastogenesis from synoviocytes in rheumatoid arthritis. Arthritis Rheum. 2000;43(2):259–269. doi: 10.1002/1529-0131(200002)43:2<259::AID-ANR4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Horwood NJ, Kartsogiannis V, Quinn JM, Romas E, Martin TJ, Gillespie MT. Activated T lymphocytes support osteoclast formation in vitro. Biochem Biophys Res Commun. 1999;265(1):144–150. doi: 10.1006/bbrc.1999.1623. [DOI] [PubMed] [Google Scholar]
- 8.Fata JE, Kong YY, Li J, et al. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell. 2000;103(1):41–50. doi: 10.1016/s0092-8674(00)00103-3. [DOI] [PubMed] [Google Scholar]
- 9.Knoop KA, Kumar N, Butler BR, et al. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J Immunol. 2009;183(9):5738–5747. doi: 10.4049/jimmunol.0901563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanada R, Leibbrandt A, Hanada T, et al. Central control of fever and female body temperature by RANKL/RANK. Nature. 2009;462(7272):505–509. doi: 10.1038/nature08596. [DOI] [PubMed] [Google Scholar]
- 11.Itoh K, Udagawa N, Matsuzaki K, et al. Importance of membrane- or matrix-associated forms of M-CSF and RANKL/ODF in osteoclastogenesis supported by SaOS-4/3 cells expressing recombinant PTH/PTHrP receptors. J Bone Miner Res. 2000;15(9):1766–1775. doi: 10.1359/jbmr.2000.15.9.1766. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi N, Akatsu T, Udagawa N, et al. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 1988;123(5):2600–2602. doi: 10.1210/endo-123-5-2600. [DOI] [PubMed] [Google Scholar]
- 13.Hikita A, Yana I, Wakeyama H, et al. Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF-kappaB ligand. J Biol Chem. 2006;281(48):36846–36855. doi: 10.1074/jbc.M606656200. [DOI] [PubMed] [Google Scholar]
- 14.Sobacchi C, Frattini A, Guerrini MM, et al. Osteoclast-poor human osteopetrosis due to mutations in the gene encoding RANKL. Nat Genet. 2007;39(8):960–962. doi: 10.1038/ng2076. [DOI] [PubMed] [Google Scholar]
- 15.Guerrini MM, Sobacchi C, Cassani B, et al. Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations. Am J Hum Genet. 2008;83(1):64–76. doi: 10.1016/j.ajhg.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim N, Odgren PR, Kim DK, Marks SC, Choi Y. Diverse roles, of the, tumor necrosis, factor family, member TRANCE, in skeletal, physiology revealed, by TRANCE, deficiency partial, rescue by, a lymphocyte-expressed, transgene. Proc Natl Acad Sci USA. 2000;97(20):10905–10910. doi: 10.1073/pnas.200294797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Sarosi I, Yan XQ, et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis, regulation of bone mass, calcium metabolism. Proc Natl Acad Sci USA. 2000;97(4):1566–1571. doi: 10.1073/pnas.97.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucay N, Sarosi I, Dunstan C, et al. Osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Genes Dev. 1998;12(9):1260–1268. doi: 10.1101/gad.12.9.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivadeneira F, Styrkarsdottir U, Estrada K, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41(11):1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O’Brien CA. Matrix-embedded cells control osteoclast formation. Nat Med. 2011;17(10):1235–1241. doi: 10.1038/nm.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onal M, Xiong J, Chen X, et al. Receptor activator of nuclear factor kB ligand (RANKL) protein expression by B lymphocytes contributes to ovariectomy-induced bone loss. J Biol Chem. 2012;287(35):29851–29860. doi: 10.1074/jbc.M112.377945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danks L, Takayanagi H. Immunology and bone. J Biochem. 2013;154(1):29–39. doi: 10.1093/jb/mvt049. [DOI] [PubMed] [Google Scholar]
- 23.Yasuda H, Shima N, Nakagawa N, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor, is identical to TRANCE/RANKL. Proc Natl Acad Sci USA. 1998;95(7):3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SK, Lorenzo JA. Parathyroid hormone stimulates TRANCE and inhibits osteoprotegerin messenger ribonucleic acid expression in murine bone marrow cultures: correlation with osteoclast-like cell formation. Endocrinology. 1999;140(8):3552–3561. doi: 10.1210/endo.140.8.6887. [DOI] [PubMed] [Google Scholar]
- 25.Takeda S, Yoshizawa T, Nagai Y, et al. Stimulation of osteoclast formation by 1,25-dihydroxyvitamin D requires its binding to vitamin D receptor (VDR) in osteoblastic cells: studies using VDR knockout mice. Endocrinology. 1999;140(2):1005–1008. doi: 10.1210/endo.140.2.6673. [DOI] [PubMed] [Google Scholar]
- 26.Fuller K, Chambers TJ. Generation of osteoclasts in cultures of rabbit bone marrow and spleen cells. J Cell Physiol. 1987;132(3):441–452. doi: 10.1002/jcp.1041320306. [DOI] [PubMed] [Google Scholar]
- 27.Hofbauer LC, Lacey DL, Dunstan CR, Spelsberg TC, Riggs BL, Khosla S. Interleukin-1beta and tumor necrosis factor-alpha, but not interleukin-6, stimulate osteoprotegerin ligand gene expression in human osteoblastic cells. Bone. 1999;25(3):255–259. doi: 10.1016/s8756-3282(99)00162-3. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien CA, Gubrij I, Lin SC, Saylors RL, Manolagas SC. STAT3 activation in stromal/osteoblastic cells is required for induction of the receptor activator of NF-kappaB ligand and stimulation of osteoclastogenesis by gp130-utilizing cytokines or interleukin-1 but not 1,25-dihydroxyvitamin D3 or parathyroid hormone. J Biol Chem. 1999;274(27):19301–19308. doi: 10.1074/jbc.274.27.19301. [DOI] [PubMed] [Google Scholar]
- 29.Liu XH, Kirschenbaum A, Yao S, Levine AC. Interactive effect of interleukin-6 and prostaglandin E2 on osteoclastogenesis via the OPG/RANKL/RANK system. Ann N Y Acad Sci. 2006;1068:225–233. doi: 10.1196/annals.1346.047. [DOI] [PubMed] [Google Scholar]
- 30.Obr AE, Grimm SL, Bishop KA, Pike JW, Lydon JP, Edwards DP. Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells. Mol Endocrinol. 2013;27(11):1808–1824. doi: 10.1210/me.2013-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seriwatanachai D, Thongchote K, Charoenphandhu N, et al. Prolactin directly enhances bone turnover by raising osteoblast-expressed receptor activator of nuclear factor kappaB ligand/osteoprotegerin ratio. Bone. 2008;42(3):535–546. doi: 10.1016/j.bone.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Tanos T, Sflomos G, Echeverria PC, et al. Progesterone/RANKL is a major regulatory axis in the human breast. Sci Transl Med. 2013;5(182):182ra. doi: 10.1126/scitranslmed.3005654. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Gupta A, Hu H, Chatterton RT, Clevenger CV, Khan SA. Comment on “Progesterone/RANKL is a major regulatory axis in the human breast”. Sci Transl Med. 2013;5(215):215le. doi: 10.1126/scitranslmed.3006883. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26(17):6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Yamazaki M, Zella LA, et al. Multiple enhancer regions located at significant distances upstream of the transcriptional start site mediate RANKL gene expression in response to 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2007;103(3–5):430–434. doi: 10.1016/j.jsbmb.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Q, Manolagas SC, O’Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol. 2006;26(17):6453–6468. doi: 10.1128/MCB.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galli C, Zella LA, Fretz JA, et al. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149(1):146–153. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onal M, Galli C, et al. The RANKL distal control region is required for the increase in RANKL expression, but not the bone loss, associated with hyperparathyroidism or lactation in adult mice. Fu Q. 2012;26(2):341–348. doi: 10.1210/me.2011-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop KA, Meyer MB, Pike JW. A novel distal enhancer mediates cytokine induction of mouse RANKL gene expression. Mol Endocrinol. 2009;23(12):2095–2110. doi: 10.1210/me.2009-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S, Yamazaki M, Shevde NK, Pike JW. Transcriptional control of receptor activator of nuclear factor-kappaB ligand by the protein kinase A activator forskolin and the transmembrane glycoprotein 130-activating cytokine, oncostatin M, is exerted through multiple distal enhancers. Mol Endocrinol. 2007;21(1):197–214. doi: 10.1210/me.2006-0315. [DOI] [PubMed] [Google Scholar]
- 41.Leibbrandt A, Penninger JM. RANK/RANKL: regulators of immune responses and bone physiology. Ann N Y Acad Sci. 2008;1143:123–150. doi: 10.1196/annals.1443.016. [DOI] [PubMed] [Google Scholar]
- 42.Bishop KA, Coy HM, Nerenz RD, Meyer MB, Pike JW. Mouse Rankl expression is regulated in T cells by c-Fos through a cluster of distal regulatory enhancers designated the T cell control region. J Biol Chem. 2011;286(23):20880–20891. doi: 10.1074/jbc.M111.231548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martowicz ML, Meyer MB, Pike JW. The mouse RANKL gene locus is defined by a broad pattern of histone H4 acetylation and regulated through distinct distal enhancers. J Cell Biochem. 2011;112(8):2030–2045. doi: 10.1002/jcb.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim MS, Fujiki R, Kitagawa H, Kato S. 1alpha,25(OH) 2D3-induced DNA methylation suppresses the human CYP27B1 gene. Mol Cell Endocrinol. 2007;265–266:168–173. doi: 10.1016/j.mce.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Nerenz RD, Martowicz ML, Pike JW. An enhancer 20 kilobases upstream of the human receptor activator of nuclear factor-kappaB ligand gene mediates dominant activation by 1,25-dihydroxyvitamin D3. Mol Endocrinol. 2008;22(5):1044–1056. doi: 10.1210/me.2007-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bishop KA, Wang X, Coy HM, Meyer MB, Gumperz JE, Pike JW. Transcriptional regulation of the human TNFSF11 gene in t cells via a cell type-selective set of distal enhancers. J Cell Biochem. 2014;116:320–330. doi: 10.1002/jcb.24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2010;285(20):15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SM, Bishop KA, Goellner JJ, O’Brien CA, Pike JW. Mouse and human BAC transgenes recapitulate tissue-specific expression of the vitamin D receptor in mice and rescue the VDR-null phenotype. Endocrinology. 2014;155(6):2064–2076. doi: 10.1210/en.2014-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong J, Piemontese M, Thostenson JD, Weinstein RS, Manolagas SC, O’Brien CA. Osteocyte-derived RANKL is a critical mediator of the increased bone resorption caused by dietary calcium deficiency. Bone. 2014;66:146–154. doi: 10.1016/j.bone.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25(7):1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 51.Dempster DW, Compston JE, Drezner MK, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28(1):2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Glatt V, Canalis E, Stadmeyer L, Bouxsein ML. Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J Bone Miner Res. 2007;22(8):1197–1207. doi: 10.1359/jbmr.070507. [DOI] [PubMed] [Google Scholar]
- 54.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St John HC, Bishop KA, Meyer MB, et al. The osteoblast to osteocyte transition: epigenetic changes and response to the vitamin d3 hormone. Mol Endocrinol. 2014;28(7):1150–1165. doi: 10.1210/me.2014-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer MB, Benkusky NA, Lee CH, Pike JW. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. J Biol Chem. 2014;289(28):19539–19554. doi: 10.1074/jbc.M114.578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pike JW, Meyer MB. Fundamentals of vitamin D hormone-regulated gene expression. J Steroid Biochem Mol Biol. 2014;144PA:5–11. doi: 10.1016/j.jsbmb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyer MB, Benkusky NA, Pike JW. The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. J Biol Chem. 2014;289(23):16016–16031. doi: 10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zella LA, Meyer MB, Nerenz RD, Lee SM, Martowicz ML, Pike JW. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol Endocrinol. 2010;24(1):128–147. doi: 10.1210/me.2009-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fretz J, Zella L, Kim S, Shevde N, Pike J. 1,25-Dihydroxyvitamin D3 regulates the expression of low-density lipoprotein receptor-related protein 5 via deoxyribonucleic acid sequence elements located downstream of the start site of transcription. Mol Endocrinol. 2006;20(9):2215–2230. doi: 10.1210/me.2006-0102. [DOI] [PubMed] [Google Scholar]
- 61.Zella LA, Kim S, Shevde NK, Pike JW. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol Endocrinol. 2006;20(6):1231–1247. doi: 10.1210/me.2006-0015. [DOI] [PubMed] [Google Scholar]
- 62.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SM, Goellner JJ, O’Brien CA, Pike JW. A humanized mouse model of hereditary 1,25-dihydroxyvitamin D-resistant rickets without alopecia. Endocrinology. 2014;155(11):4137–4148. doi: 10.1210/en.2014-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rinotas V, Niti A, Dacquin R, et al. Novel genetic models of osteoporosis by overexpression of human RANKL in transgenic mice. J Bone Miner Res. 2014;29(5):1158–1169. doi: 10.1002/jbmr.2112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.