Abstract
Testicular orphan nuclear receptor 4 (TR4) is a member of the nuclear receptor superfamily for which a ligand has not yet been found. In vitro data obtained from various cell lines suggest that TR4 functions as a master regulator to modulate many signaling pathways, yet the in vivo physiological roles of TR4 remain unclear. Here, we report the generation of mice lacking TR4 by means of targeted gene disruption (TR4–/–). The number of TR4–/– pups generated by the mating of TR4+/– mice is well under that predicted by the normal Mendelian ratio, and TR4–/– mice demonstrate high rates of early postnatal mortality, as well as significant growth retardation. Additionally, TR4–/– females show defects in reproduction and maternal behavior, with pups of TR4–/– dams dying soon after birth with no indication of milk intake. These results provide in vivo evidence that TR4 plays important roles in growth, embryonic and early postnatal pup survival, female reproductive function, and maternal behavior.
Keywords: gene expression regulation, knockout mice, reproduction
Members of the nuclear receptor superfamily are known to play important roles in differentiation, development, homeostasis, and metabolism, as well as in disease development and progression (1). Testicular orphan nuclear receptor 4 (TR4) (also known as TAK1 and Nr2c2; www.informatics.jax.org) is closely related to the retinoid X receptor (RXR), chicken ovalbumin upstream promoter-transcription factor (COUP-TF), and hepatocyte nuclear factor 4 (HNF4) in sequence and structure (2), and binds to AGGTCA DNA sequence motifs in direct repeat orientation, with variable spacing, in the promoters of its target genes. As an orphan nuclear receptor with several known regulatory targets (3–11), TR4 may affect many signaling pathways and thus have a major impact on physiological function. A substantial difficulty in determining the function of many orphan receptors, including TR4, has been the inability to identify ligands through which the receptors are activated. In response to this dilemma, creation of mammalian gene knockout models has become a successful strategy through which to study the physiological roles of orphan receptors in vivo (12–15). TR4 remains an orphan receptor with no identified ligand and a relative mystery with respect to physiological function.
TR4 is widely expressed in both embryonic and adult tissues, again suggesting that the receptor affects various developmental and physiological pathways. For example, TR4 is expressed in both neural and nonneural tissues during embryonic development (4, 5, 7). In situ hybridization experiments using probes specific for TR4 have shown transcripts present in actively proliferating cell populations of the brain and peripheral organs at various times throughout embryogenesis, with more restricted expression in adult animals (16–18). Peripheral tissue expression of TR4 has been shown to be nearly ubiquitous, yet it is often cell-specific within particular tissues (19). Northern blot analysis of tissue from the adult rat demonstrated significant TR4 expression in the prostate, adrenal gland, spleen, thyroid gland, and pituitary gland (16), whereas expression in the mouse was found to be highest in the testis, kidney, and skeletal muscle (19).
Upon generation of TR4–/– mice, a complex set of phenotypic abnormalities was found to exist. TR4–/– mice are produced at lower than Mendelian ratios, with a significantly lower proportion of female knockouts produced than male knockouts. Evidence of a growth defect in affected animals is apparent early in postnatal life, and fertility in the knockout mice is greatly reduced compared with wild-type mice. Additionally, female mice lacking TR4 exhibit behavioral abnormalities, including defects in maternal behavior.
Experimental Procedures
TR4–/– Targeting Construct Design and Knockout Mouse Generation. The λ knockout shuttle (KOS) system (20) was used to derive a TR4-targeting vector. Three independent genomic clones spanning exons 4–10 were isolated. The targeting vector was derived from one clone and contained a 2,173-bp deletion that included most of exon 4 and all of exon 5. The genomic sequence encoding the DNA-binding domain of TR4 was replaced by a Lac-Z/Neo selection cassette. The NotI linearized vector was electroporated into strain 129SvEvbrd (LEX1) embryonic stem (ES) cells, and G418/FIAU-resistant ES cell clones were isolated and screened for homologous recombination of the mutant DNA by Southern blot. One targeted ES cell clone was injected into blastocysts of strain C57BL/6 (albino), which were then inserted into pseudopregnant female mice for continuation of fetal development. Resulting chimeric male mice were then mated to C57BL/6 (albino) females to generate animals heterozygous for the mutation. Three sexually mature breeding pairs, with each mouse heterozygous for the disrupted TR4 gene (TR4+/–), were provided by Lexicon Genetics Incorporated).
Experimental Animals, Genotyping, and Growth Analysis. TR4+/– founders were intercrossed, as well as backcrossed to mice of the C57BL/6 strain. Mice were housed in the vivarium facility of the University of Rochester Medical Center. The animals were provided a standard diet with constant access to food and water, and exposed to a 12-h light/dark cycle. Genomic DNA was isolated from tail or embryo samples, after proteinase K digestion, by means of phenol/chloroform extraction (21). Extracted DNA was used as template for PCR-based genotyping. Primers for amplification of the wild-type and targeted alleles were designed based on sequences within intron 4 (deleted in the TR4–/–), or within the selection cassette and external to the deleted region, respectively. The PCR primer sequences are TR4-107 (wild-type, forward, 5′-GGAGACACACTGCACATGTTCGAATAC-3′; TR4-111, wild-type, reverse, 5′-CACAGCTCATTTCT CTGCTCACTTACTC-3′; Neo-3a, targeted allele, forward, 5′-GCAGCGCATCGCCTTCTATC-3′; and TR4-34, targeted allele, reverse, 5′-TGCAAGCATACTTCTTGTTCC-3′. DNA in PCR-genotyping reactions was amplified in 35 cycles with melting, annealing, and extension temperatures of 94°C, 61°C, and 72°C, respectively. For weight measurements, littermates from TR4+/– matings were weighed every other day, starting at postnatal day 2, until day 30. Subsequently, pups were weighed once per week for 8 additional weeks. Serum levels of insulin-like growth factor-1 (IGF-1) were determined by using a commercially available RIA kit employing the competitive binding immunoassay format (Diagnostic Systems Laboratories, Webster, TX). All experimental protocols were approved by the University Committee on Animal Resources and the office of Environmental Health and Safety before implementation.
RT-PCR Analysis of Gene Expression. For RT-PCR analysis of gene expression, total RNA was isolated from the cerebella of 7-month-old TR4+/+ and TR4–/– mice, by using TRIzol Reagent (Invitrogen). First-strand cDNA synthesis was achieved by using the SuperScript II RNase H Reverse Transcriptase kit (Invitrogen). The sense strand (S) and antisense strand (AS) PCR primer sequences are (m, mouse): mTR4(S), 5′-CATATTCACCACCTCGGACAAC-3′; mTR4(AS), 5′-TGACGCCACAGACCACAC-3′; mTR2(S), 5′-CCGCATCTAATCGCTGGA GAG-3′; mTR2(AS), 5′-GCATAGGAGAAGGCATGGTGAG-3′; β-actin(S), 5′-TGTGCCCA TCTACGAGGGGTATGC-3′; and β-actin(AS), 5′-GGTACATGGTGGTGCCGCCAGACA-3′.
Tissue Preparation, Histology, and Immunostaining. Mice were anesthetized with an overdose of pentobarbital and perfused through the left ventricle with 20 ml of saline (pH 7.3), followed by 20 ml of 10% neutral buffered formalin. Tissues were removed and postfixed by submersion in 10% formalin. Alternatively, fresh tissues were fixed by direct submersion in 10% neutral buffered formalin, before processing. Tissue was processed for embedding and embedded in paraffin. Liver tissues were cut at a thickness of 7 μm, deparaffinized, and stained with a mouse monoclonal anti-human IGF-1 primary antibody (Upstate Biotechnology, Lake Placid, NY), followed by use of a biotinylated anti-mouse secondary antibody (Vector Laboratories). Staining was visualized by using the Vector ABC staining kit followed by DAB substrate (Vector Laboratories). Hematoxylin was used as a nuclear counterstain after immunostaining.
Tests of Female Reproductive Capacity and Maternal Behavior Observations. Age-matched adult TR4+/+ and TR4–/– female mice were each paired with a sexually mature TR4+/+ male for 2.5 weeks and then separated. The number of pups born to each female was recorded. Dams and pups were observed in their home cage after parturition.
Data Analysis. The body weight data were analyzed by ANOVA. IGF-1 RIA data and mortality rates were analyzed by using independent sample one-tailed t tests, assuming equal variance. Ratios of genotypes generated from heterozygous pairings and differences from expected Mendelian ratios, overall and within sex, were analyzed by using the χ2 test. Sex ratios within each genotype were also analyzed by using the χ2 test.
Results
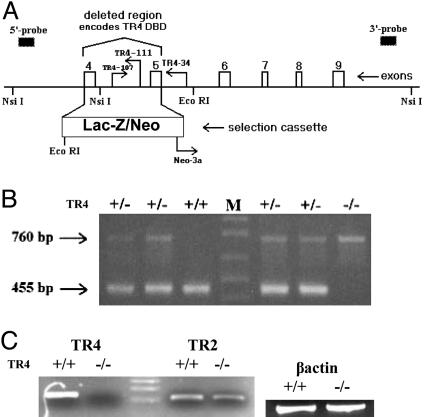
Generation and Confirmation of TR4–/– Mice. A Lac-Z/Neo selection cassette was constructed and inserted into the TR4 gene, replacing exons 4 and 5, as well as the intervening intron 4 (Lexicon Genetics) (Fig. 1A). Elimination of the DNA-binding domain (DBD) of TR4 (exons 4 and 5) renders the receptor functionally inactive, because it can no longer act as a transcription factor to regulate the expression of target genes. PCR amplification using wild-type primers yields a product of 455 bp, and use of TR4–/– primers yields a product of 760 bp (Fig. 1 A and B). RT-PCR amplification of cerebellar RNA, using primers complimentary to the DBD of the TR4 gene, demonstrated that TR4 transcript was present in samples prepared from TR4+/+ mice, but was not found in TR4–/– samples (Fig. 1C). It was hypothesized that the closely related TR2 gene may be upregulated in compensation for the loss of TR4. Fig. 1C demonstrates that TR2 levels are not significantly different in tissue from TR4–/– mice compared with TR4+/+.
Fig. 1.
TR4–/– targeting construct and genotype confirmation. (A) A segment of TR4 genomic DNA is shown. The Lac-Z/Neo selection cassette is inserted between introns 3 and 5. DBD, DNA-binding domain. (B) PCR genotyping yielded 455-bp fragments for wild-type (+/+) and 760-bp fragments for targeted (–/–) alleles. (C) Expression of TR4 and TR2 mRNA in cerebellar tissue from TR4+/+ and TR4–/– mice by means of RT-PCR. β-actin levels were determined as a control for template amount.
TR4–/– Mice Are Generated from Heterozygous Pairings at Less Than Expected Mendelian Ratios. From 110 TR4+/– breeding pairs, 751 pups were produced and genotyped. The ratios of genotypes produced were significantly different from expected Mendelian ratios (Table 1). TR4–/– mice were found to make up only 13.6% of the total pups produced, when TR4+/+ and TR4–/– pups would each be expected to account for 25% of the total number. Interestingly, of the entire population of TR4–/– mice produced, significantly fewer were female (30.4%), compared with the equal sex ratio that was expected.
Table 1. TR4 genotype ratios produced by heterozygous breeding pairs.
| +/+(%) | +/-(%) | -/-(%) | |
|---|---|---|---|
| Female | 114 (46.5) | 212 (52.5) | 31** (30.4) |
| Male | 131 (53.5) | 192 (47.5) | 71 (69.6) |
| Total | 245 (32.6) | 404 (53.8) | 102*** (13.6) |
| Expected Mendelian ratios | 187.75 (25.0) | 375.5 (50.0) | 187.75 (25.0) |
| Mortality between 3 and 5 weeks | 1 | 2 | 12*** |
TR4-/- female mice are generated at a significantly lower rate than are male TR4-/- mice (**, P < 0.005). TR4-/- mice are generated at less than expected Mendelian ratios, and TR4-/- mortality rates are significantly higher than those of TR4+/+ or TR4+/- mice (***, P < 0.001).
Loss of TR4–/– Embryos Begins at Embryonic Day (E) 13–E14.5. Pregnant female TR4+/– mice that had been mated to male TR4+/– mice were killed at time points ranging from E7.5–E22. Resultant genotype ratios of the embryos present suggest a loss of TR4–/– embryos beginning at E13–E14.5 (Table 2). This embryological stage corresponds to late fetal organogenesis and the beginning stages of fetal growth and development (22). Embryological expression of TR4 has demonstrated particularly abundant TR4 transcript in the ventricular zones of the brain, as well as in the striatum and cerebellar primordium at E14–E16, with expression extending to the spinal cord by E16 (4), suggesting a role for TR4 in neuronal proliferation and development of particular brain regions that may be lethal when disrupted.
Table 2. Genotype ratios of embryos produced from heterozygous pairings.
| Embryonic day | No. of embryos/no. of litters | Expected Mendelian ratio +/+: +/-: -/- | Actual ratio +/+: +/-: -/- |
|---|---|---|---|
| 7.5 | 8/1 | 2:4:2 | 2:4:2 |
| 11.5-12.5 | 39/4 | 9.75:19.5:9.75 | 15:15:9 |
| 14.5-15.5 | 35/5 | 8.75:17.5:8.75 | 13:16:6 |
| 16.5-17.5 | 46/5 | 11.5:23:11.5 | 18:22:6 |
| 18.5-P0/E22 | 101/12 | 25.25:50.5:25.25 | 47:38:16 |
| Total | 229/27 | 57.25:114.5:57.25 | 95:95:39 |
Dams were killed within the indicated range of days after conception. The time of conception was estimated to be 0.5 days prior to the observance of a vaginal plug. P0/E22, postnatal day 0/embryonic day 22.
Reduced Postnatal Growth of TR4–/– Mice. At the time of birth, TR4–/– mice were visibly indistinguishable from TR4+/+ or TR4+/– littermates, with no obvious defects in suckling ability. However, it was found that both male and female TR4–/– mice were smaller than TR4+/+ and TR4+/– mice at similar ages (Fig. 2B). TR4–/– pups displayed a range of significant weight reduction, between 20% and 56%, compared with their TR4+/+ and TR4+/– counterparts, up to 12 weeks of age (Fig. 2B). These data show that weight differences in TR4–/– mice compared with their TR4+/+ and TR4+/– littermates are apparent during the first postnatal week, and that a reduction in growth rate is obvious up to 1–2 weeks after the normal time of weaning (3 weeks). Thereafter, the growth curves are nearly parallel, with a 20–30% reduction in weight maintained into adulthood (Fig. 2).
Fig. 2.
Growth retardation in TR4–/– mice. (A) Male mice at 2 months of age. (B) Mouse growth curves. TR4–/– pups display a range of significant weight reduction (P < 0.05), except on day 4 in males (16% reduction, P < 0.1). For both sexes: TR4+/+, n = 15; TR4+/–, n = 15; male TR4–/–, n = 6; female TR4–/–, n = 5 up to day 22, and n = 3 from day 24 to day 86.
Analysis of Growth-Related Hormone Levels. Levels of growth hormone (GH) and IGF-1, a downstream target of GH, were measured in serum from age-matched TR4+/+ and TR4–/– mice or analyzed by means of immunostaining. Overall pituitary structure, as well as somatotroph numbers and levels of GH produced in the anterior pituitary, are similar in sections from TR4+/+ and TR4–/– mice (data not shown). However, serum levels of IGF-1 were found to be reduced by 34% in TR4–/– mice compared with TR4+/+ controls (Fig. 3A), and immunostaining for IGF-1 in liver sections demonstrated reduced IGF-1 immunoreactivity in TR4–/– hepatocytes compared with TR4+/+ (Fig. 3B).
Fig. 3.
Reduced levels of IGF-1 in TR4–/– mice. (A) Serum levels of IGF-1 in 7-month-old male mice by means of RIA. A significantly lower serum IGF-1 level was observed in TR4–/– mice compared with TR4+/+ (P = 0.05, n = 3 for each genotype). (B) Immunostaining for IGF-1 in liver tissue from 4-month-old TR4+/+ and TR4–/– male mice. Light brown color designates positive staining. The tissues were counterstained with hematoxylin, ×400 magnification.
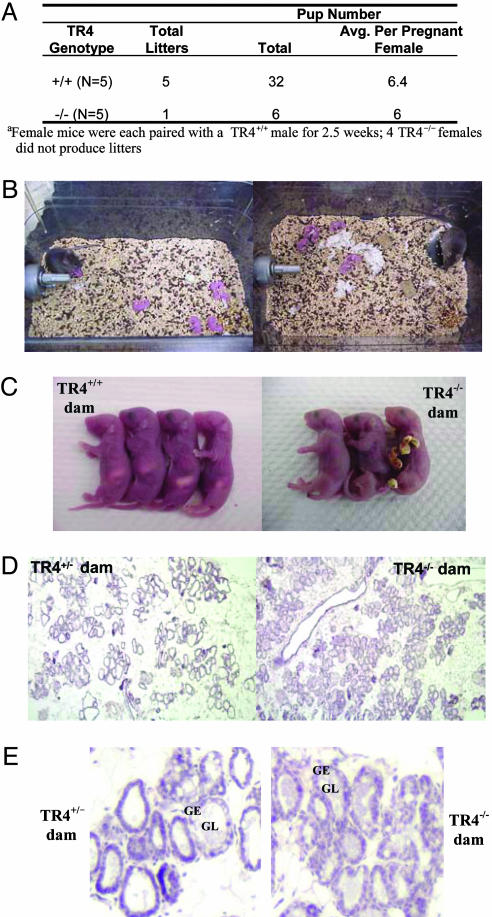
Reduced Fertility in Female TR4–/– Mice. Short-term mating experiments to test TR4–/– female reproductive capacity demonstrated impaired TR4–/– fertility. Only one of five TR4–/– females produced a litter after a 2.5-week mating period, whereas all TR4+/+ females produced litters (Fig. 4A).
Fig. 4.
Reproductive deficiencies and lack of maternal behavior among TR4–/– dams. (A) Female reproductive results after short-term pairing (2.5 weeks). (B) Observations of TR4–/– dams with pups at postpartum days 1 (Left) and2(Right). TR4–/– dams do not build nests, collect pups to a single location, crouch over pups, or nurse their offspring. (C) Pups of TR4–/– dams die within 24–36 h after birth with no milk in their stomachs. (D) Histology of mammary gland tissue from a TR4+/– female and from a TR4–/– female, on postpartum day 1. The presence of luminal secretions (pink staining) suggests normal milk production in TR4–/– mice. The images were obtained at ×1,000 magnification, with subsequent digital reduction of image size. (E) A portion of the mammary gland sections stained in D at full ×1,000 magnification. GL, glandular lumen; GE, glandular epithelium.
Loss of Maternal Behavior in TR4–/– Dams. Observations of TR4–/– mothers suggest defects in maternal behavior. TR4–/– dams do not build nests, collect pups to a single location, crouch over pups, or nurse their offspring, and pups of TR4–/– dams die within 24–36 h after birth with no milk in their stomachs (Fig. 4 B and C). Histology of mammary gland tissue from a TR4+/– female (TR4+/– females show normal reproductive capacity and maternal behavior) and from a TR4–/– female, on postpartum day 1, demonstrates no obvious defect in milk production in the TR4–/– mouse (Fig. 4D). The magnified mammary gland structures (Fig. 4D) show milk (pink staining) within the glandular lumen (GL) (Fig. 4E). However, the mammary glands of the TR4–/– dam are abnormally full of milk, which is consistent with the lack of maternal behavior and thus lack of pup nursing observed in these animals. In contrast, the TR4+/– mother shows mammary glands with both full and empty luminal regions (Fig. 4 D and E), suggesting that milk is drained by pup suckling and restored through continuing milk production. Although histological analysis of mammary gland structures supports the observed lack of maternal behavior among TR4–/– dams, few samples have been analyzed thus far. It is important to note that pups may develop preferences for particular nipples, and with small litters not making use of all available nipples, mammary gland structures may appear histologically similar to those of the TR4–/– mothers, even with normal displays of maternal behavior and pup suckling activity. Additionally, preliminary analysis of oxytocin levels in the mouse hypothalamus suggests a reduction in expression in TR4–/– mice compared with TR4+/+ mice (data not shown). Due to the reduced expression of oxytocin in TR4–/– mice, it is possible that these mothers have abnormal milk ejection capacity, as is the case in the oxytocin-deficient oxytocin knockout and Mf3 knockout mice (23, 24). However, with the defect in maternal behavior displayed by TR4–/– mothers, it will be difficult to test the milk ejection reflex.
Eye Pathology Among TR4–/– Mice. An additional phenotype observed among TR4–/– mice is the accumulation of fluid surrounding the eye (Fig. 5). The eyelids of TR4–/– mice often appeared swollen with excess moisture or a dried, crusty area surrounding the eyes. In this condition, the mice either refrained from, or were prevented from, opening their eyes widely. The increased release and/or buildup of eye secretions indicate possible abnormalities of lacrimal gland and/or immune function due to the loss of TR4 expression.
Fig. 5.
Eye pathology in TR4–/– mice. Male mice at 10 weeks of age are pictured. In the TR4–/– mouse, secretions have accumulated around the eye, and the eyelids appear inflamed.
Discussion
The number of homozygous TR4–/– pups generated from heterozygous pairings was just over 50% of the expected number based on normal Mendelian ratios of genotypes (Table 1). Without evidence that significant numbers of pups are dying perinatally, the data obtained suggest increased embryologic mortality of TR4–/– mice. It is known that TR4 is expressed in the mouse embryo as early as E9, with particularly high expression in various regions of the developing nervous system during subsequent prenatal development (4). Further analysis of embryonic genotype ratios indicated a loss of TR4–/– embryos starting at E13–E14.5, with a more pronounced reduction of TR4–/– embryos by the late stages of embryogenesis (E16.5 to birth) (Table 2). Another closely related orphan nuclear receptor, COUP-TFI, has a significantly overlapping spatial and temporal expression pattern with TR4 (18). Interestingly, COUP-TFI null mutants display defects in formation of the ninth cranial ganglion, and disrupted nerve projection and arborization in additional regions of the developing mouse brain (12). Unlike the TR4–/– mice, however, COUP-TFI null mutants are found at Mendelian ratios throughout embryogenesis and at birth, but nearly all die perinatally (12). Further exploration of fetal development with regard to neurogenesis, angiogenesis, cardiac development, and hematopoiesis may help to explain the significant loss of TR4–/– embryos observed.
The growth defect observed among TR4–/– mice (Fig. 2) is similar in rate of growth reduction observed among other small mouse mutants (25–28), yet relatively unique in the timing of the onset of growth reduction. Unlike the 2-week postnatal latency to growth reduction in the Snell, Ames, and little dwarf mutants (28), TR4–/– animals display significant growth reduction as early as postnatal day 2 (the first time point at which measurements were collected, Fig. 2B). Interestingly, a mutant mouse deficient in the winged helix gene Mf3, which encodes a transcription factor highly expressed in the central nervous system of the developing embryo, shows growth reduction as early as 2–3 days after birth (24). Also similar to TR4, Mf3 is expressed in the neural tube at E9.5, as well as in the diencephalon and midbrain. Furthermore, the Mf3 mutant animals show variations in phenotype with considerable embryonic lethality, 30% preweaning mortality among those surviving to birth, and surviving adult mutants that, although small in size, can live to over 1 year of age (24).
Mice with growth retardation often display defects in pituitary structure or function, which may become apparent as an ultimate defect in GH or thyroid-stimulating hormone (TSH) production or secretion (25–27, 29). Despite the lack of structural abnormalities of the pituitaries of TR4–/– mice, it is possible that TR4 affects pituitary hormone-initiated signaling at points further downstream. TR4 has been shown to bind to direct repeat 4 thyroid hormone response elements and compete for binding to, and thus regulation of, thyroid hormone target genes (3, 9). Indeed, mice carrying mutations of the thyroid hormone receptors, individually (TRβ–/–), or in combination (TRα1–/–β–/–), display growth impairments (30, 31). Therefore, a disruption of regulation of the thyroid hormone receptor signaling pathway by means of loss of TR4 may contribute to the growth defect observed in TR4–/– mice. Also, as a downstream mediator of GH action in the liver, IGF-1 is known to be important in postnatal growth based on IGF-1 gene knockout studies (32, 33). TR4 has been shown to be expressed in mouse hepatocytes, as well as to induce expression of apolipoprotein E (apoE) (34), a functional constituent of plasma lipoproteins that promotes the receptor-mediated uptake of such lipoproteins by the liver. Through use of TR4–/– mice, it has been confirmed that loss of TR4 reduces expression of apoE in liver. With confirmation of the presence and functional importance of TR4 in the liver, it is possible that TR4 also affects the GH/IGF-1 signaling pathway. IGF-1 has been shown to be reduced in other knockout mouse models with growth abnormalities, such as the steroid receptor coactivator knockout (SRC-3–/–) mouse, in the absence of corresponding defects in growth hormone secretion (35, 36). The discovery of low serum IGF-1 levels (Fig. 3A) and reduced IGF-1 immunoreactivity in liver sections from TR4–/– mice (Fig. 3B) indicates that an IGF-1 deficiency may contribute to the growth retardation observed in TR4–/– mice. Reduction of IGF-1 levels without corresponding reduction in GH has been suggested to indicate partial GH resistance (37), and correlate with growth retardation in other mouse models (36).
More recently, it has been found that glucose levels are significantly reduced in newborn TR4–/– mice, as well as in young TR4–/– animals up to 4 weeks of age, paralleling the reduced growth rate of TR4–/– mice (N.-C.L., Y.-F.L., and C.C., unpublished results). Based on finding of hypoglycemia in young TR4–/– mice, it is possible that a glucoregulatory defect, resulting in perinatal and early postnatal hypoglycemia, contributes to TR4–/– growth retardation.
Based on the high expression of TR4 in the male reproductive tract, particularly the testes, it would be expected that disruption of the gene in a mouse model would result in reduced male fertility. Indeed, fertility is reduced in both male (38) (unpublished results) and female (Fig. 4A) TR4–/– mice. TR4–/– males show a disruption of spermatogenesis during late meiotic prophase and through subsequent meiotic divisions, and testis sections from TR4–/– mice show degeneration of primary spermatocytes and necrotic tubules (38).
The significant lack of maternal behavior among female TR4–/– mice (Fig. 4 B and C), as well as our preliminary data indicating reduced oxytocin expression in the hypothalamic regions of TR4–/– mice, suggests that defects in regulation of levels of this peptide hormone may result from the disruption of TR4 expression. Oxytocin is produced in neurons of the paraventricular and supraoptic nuclei of the hypothalamus, as well as in specific tissues and cell types peripherally (39). Already known to be a TR4 target gene (40), the physiological functions of oxytocin have been well studied, and this peptide hormone is known to affect both central and peripheral systems, as well as behavior (reviewed in ref. 39). Oxytocin has a pronounced anxiolytic effect (39, 41–44) and is required for the milk ejection reflex to occur in nursing dams (23). A reduction in the anxiety-reducing effects of oxytocin in TR4–/– dams may contribute to their abnormal maternal behavior, whereas it is also possible that the low levels of oxytocin present in the mice prevents milk ejection.
In conclusion, TR4–/– mice display defects in postnatal growth, and female TR4–/– mice show reduced fertility and severe defects in maternal behavior, resulting in neonatal death of their offspring. These in vivo data demonstrate that TR4 plays an essential part in various physiological functions related to growth and reproduction. Future studies uncovering the mechanisms of TR4 action related to these and other physiological pathways may lead to the identification of a ligand(s) for TR4, as well as yet undiscovered roles of the receptor.
Acknowledgments
We thank Raymond Baggs for assistance with techniques involving experimental animals and pathological analyses and Karen Wolf for help in manuscript preparation. This work was supported by National Institutes of Health Grants DK63212 and DK56984 (to C.C.), the George Whipple Professorship Endowment (to C.C.), and a Department of Veterans Affairs Merit Review Award (to C.K.M.).
Abbreviations: TR4, testicular orphan nuclear receptor 4; IGF-1, insulin-like growth factor-1; GH, growth hormone; En, embryonic day n.
References
- 1.Evans, R. M. (1988) Science 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laudet, V. (1997) J. Mol. Endocrinol. 19, 207–226. [DOI] [PubMed] [Google Scholar]
- 3.Lee, Y. F., Pan, H. J., Burbach, P. H., Morkin, E. & Chang, C. (1997) J. Biol. Chem. 272, 12215–12220. [DOI] [PubMed] [Google Scholar]
- 4.Young, W. J., Smith, S. M. & Chang, C. (1997) J. Biol. Chem. 272, 3109–3116. [DOI] [PubMed] [Google Scholar]
- 5.Young, W. J., Lee, Y. F., Smith, S. M. & Chang, C. (1998) J. Biol. Chem. 273, 20877–20885. [DOI] [PubMed] [Google Scholar]
- 6.Collins, L. L., Lin, D., Mu, X. & Chang, C. (2000) J. Biol. Chem. 276, 27316–27321. [DOI] [PubMed] [Google Scholar]
- 7.Lee, Y. F., Young, W. J., Burbach, J. P. H. & Chang, C. (1998) J. Biol. Chem. 273, 13437–13443. [DOI] [PubMed] [Google Scholar]
- 8.Lee, H., Lee, Y., Burbach, P. & Chang, C. (1995) J. Biol. Chem. 270, 30129–30133. [DOI] [PubMed] [Google Scholar]
- 9.Lee, Y. F., Young, W. J., Lin, W. J., Shyr, C. R. & Chang, C. (1999) J. Biol. Chem. 274, 16198–16205. [DOI] [PubMed] [Google Scholar]
- 10.Hwang, S., Burbach, P. H. & Chang, C. (1998) Endocrine 8, 169–175. [DOI] [PubMed] [Google Scholar]
- 11.Lee, Y. F., Lee, H. J. & Chang, C. (2002) J. Steroid Biochem. Mol. Biol. 81, 291–308. [DOI] [PubMed] [Google Scholar]
- 12.Qiu, Y., Pereira, F. A., DeMayo, F. J., Lydon, J. P., Tsai, S. Y. & Tsai, M. J. (1997) Genes Dev. 11, 1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooney, A. J., Lee, C. T., Lin, S., Tsai, S. Y. & Tsai, M. (2001) Trends Endocrinol. Metab. 12, 247–251. [DOI] [PubMed] [Google Scholar]
- 14.Steinmayr, M., Andre, E., Conquet, F., Rondi-Reig, L., Delhaye-Bouchaud, N., Auclair, N., Daniel, H., Crepel, F., Mariani, J., Sotelo, C. & Becker-Andre, M. (1998) Proc. Natl. Acad. Sci. USA 95, 3960–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shyr, C., Collins, L. L., Mu, X., Platt, K. A. & Chang, C. (2002) Mol. Cell. Biol. 22, 4661–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang, C., Da Silva, S. L., Ideta, R., Lee, Y. F., Yeh, S. & Burbach, J. P. H. (1994) Proc. Natl. Acad. Sci. USA 1994, 6040–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshikawa, T., DuPont, B. R., Leach, R. J. & Detera-Wadleigh, S. D. (1996) Genomics 35, 361–366. [DOI] [PubMed] [Google Scholar]
- 18.van Schaick, H. S. A., Rosmalen, J. G. M., Lopes da Silva, S., Chang, C. & Burbach, J. P. H. (2000) Mol. Brain Res. 77, 104–110. [DOI] [PubMed] [Google Scholar]
- 19.Hirose, T., Fujimoto, W., Yamaai, T., Kim, H. K., Matsuura, H. & Jetten, A. M. (1994) Mol. Endocrinol. 8, 1667–1680. [DOI] [PubMed] [Google Scholar]
- 20.Wattler, S., Kelly, M. & Nehls, M. (1999) BioTechniques 26, 1150–1156, 1158, 1160. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, B., Beddington, R., Costantini, F. & Lacy, E. (1994) in Manipulating the Mouse Embryo (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 296–298.
- 22.Hogan, B., Beddington, R., Costantini, F. & Lacy, E. (1994) in Manipulating the Mouse Embryo (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 19–113.
- 23.Young, W. S., 3rd, Shepard, E., Amico, J., Hennighausen, L., Wagner, K. U., LaMarca, M. E., McKinney, C. & Ginns, E. I. (1996) J. Neuroendocrinol. 8, 847–853. [DOI] [PubMed] [Google Scholar]
- 24.Labosky, P. A., Winnier, G. E., Jetton, T. L., Hargett, L., Ryan, A. K., Rosenfeld, M. G., Parlow, A. F. & Hogan, B. L. M. (1997) Development (Cambridge, U.K.) 124, 1263–1274. [DOI] [PubMed] [Google Scholar]
- 25.Lin, S., Lin, C. R., Gukovsky, I., Lusis, A. J., Sawchenko, P. E. & Rosenfeld, M. G. (1993) Nature 364, 208–213. [DOI] [PubMed] [Google Scholar]
- 26.Li, S., Crenshaw, E. B. I., Rawson, E. J., Simmons, D. M., Swanson, L. W. & Rosenfeld, M. G. (1990) Nature 347, 528–533. [DOI] [PubMed] [Google Scholar]
- 27.Sornsen, M. W., Wu, W., Dasen, J. S., Flynn, S. E., Norman, D. J., O'Connell, S. M., Gukovsky, I., Carriere, C., Ryan, A. K., Miller, A. P., et al. (1996) Nature 384, 327–333. [DOI] [PubMed] [Google Scholar]
- 28.Voss, J. W. & Rosenfeld, M. G. (1992) Cell 70, 527–530. [DOI] [PubMed] [Google Scholar]
- 29.Kendall, S. K., Samuelson, L. C., Saunders, T. L., Wood, R. I. & Camper, S. A. (1995) Genes Dev. 9, 2007–2019. [DOI] [PubMed] [Google Scholar]
- 30.Gothe, S., Wang, Z., Ng, L., Kindblom, J. M., Barros, A. C., Ohlsson, C., Vennstrom, B. & Forrest, D. (1999) Genes Dev. 13, 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneshige, M., Kaneshige, K., Zhu, X., Dace, A., Garrett, L., Carter, T. A., Kazlauskaite, R., Pankratz, D. G., Wynshaw-Boris, A., Refetoff, S., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 13209–13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, J. P., Baker, J., Perkins, A. S., Robertson, E. J. & Efstratiadis, A. (1993) Cell 75, 59–72. [PubMed] [Google Scholar]
- 33.Baker, J., Liu, J. P., Robertson, E. J. & Efstratiadis, A. (1993) Cell 75, 73–82. [PubMed] [Google Scholar]
- 34.Kim, E., Xie, S., Yeh, S. D., Lee, Y. F., Collins, L. L., Hu, Y. C., Shyr, C. R., Mu, X. M., Liu, N. C., Chen, Y. T., et al. (2003) J. Biol. Chem. 278, 46919–46926. [DOI] [PubMed] [Google Scholar]
- 35.Xu, J., Liao, L., Ning, G., Yoshida-Komiya, H., Deng, C. & O'Malley, B. W. (2000) Proc. Natl. Acad. Sci. USA 97, 6379–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skalhegg, B. S., Huang, Y., Su, T., Idzerda, R. L., McKnight, G. S. & Burton, K. A. (2002) Mol. Endocrinol. 16, 630–639. [DOI] [PubMed] [Google Scholar]
- 37.Wolf, M., Bohm, S., Brand, M. & Kreymann, G. (1996) Eur. J. Endocrinol. 135, 729–737. [DOI] [PubMed] [Google Scholar]
- 38.Mu, X., Lee, Y., Liu, N., Chen, Y., Kim, E., Shyr, C. & Chang, C. (2004) Mol. Cell. Biochem. 24, 5887–5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gimpl, G. & Fahrenholz, F. (2001) Physiol. Rev. 81, 629–683. [DOI] [PubMed] [Google Scholar]
- 40.Burbach, J. P., Van Schaick, H. S., Da Silva, C., Asbreuk, H. J. & Schmidt, M. P. (1998) Adv. Exp. Med. Biol. 449, 29–37. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy, M. M. (1995) Adv. Exp. Med. Biol. 395, 235–245. [PubMed] [Google Scholar]
- 42.McCarthy, M. M., McDonald, C. H., Brooks, P. J. & Goldman, D. (1996) Physiol. Behav. 60, 1209–1215. [DOI] [PubMed] [Google Scholar]
- 43.Windle, R. J., Shanks, N., Lightman, S. L. & Ingram, C. D. (1997) Endocrinology 138, 2829–2834. [DOI] [PubMed] [Google Scholar]
- 44.Uvnas, M. K., Ahlenius, S., Hillegaart, V. & Alster, P. (1994) Pharmacol. Biochem. Behav. 49, 101–106. [DOI] [PubMed] [Google Scholar]