Abstract
Background
NKX2-1 encodes a transcription factor with large impact on the development of brain, lung and thyroid. Germline mutations of NKX2-1 can lead to dysfunction and malformations of these organs. Starting from the largest coherent collection of patients with a suspected phenotype to date, we systematically evaluated frequency, quality and spectrum of phenotypic consequences of NKX2-1 mutations.
Methods
After identifying mutations by Sanger sequencing and array CGH, we comprehensively reanalysed the phenotype of affected patients and their relatives. We employed electrophoretic mobility shift assay (EMSA) to detect alterations of NKX2-1 DNA binding. Gene expression was monitored by means of in situ hybridisation and compared with the expression level of MBIP, a candidate gene presumably involved in the disorders and closely located in close genomic proximity to NKX2-1.
Results
Within 101 index patients, we detected 17 point mutations and 10 deletions. Neurological symptoms were the most consistent finding (100%), followed by lung affection (78%) and thyroidal dysfunction (75%). Novel symptoms associated with NKX2-1 mutations comprise abnormal height, bouts of fever and cardiac septum defects. In contrast to previous reports, our data suggest that missense mutations in the homeodomain of NKX2-1 not necessarily modify its DNA binding capacity and that this specific type of mutations may be associated with mild pulmonary phenotypes such as asthma. Two deletions did not include NKX2-1, but MBIP, whose expression spatially and temporarily coincides with NKX2-1 in early murine development.
Conclusions
The high incidence of NKX2-1 mutations strongly recommends the routine screen for mutations in patients with corresponding symptoms. However, this analysis should not be confined to the exonic sequence alone, but should take advantage of affordable NGS technology to expand the target to adjacent regulatory sequences and the NKX2-1 interactome in order to maximise the yield of this diagnostic effort.
INTRODUCTION
NKX2-1 (OMIM *600635), also known as TITF-1, TTF-1 or T/ebp, is a member of the homeodomain-containing NK-2 gene family.1 It encodes a transcription factor, which is expressed during early development of thyroid, lung and forebrain regions, particularly the basal ganglia and hypothalamus.2,3 In thyrocytes, NKX2-1 regulates the expression of thyroglobulin, thyreotropin receptor, thyroid peroxidase (TPO) and pendrin, and is therefore required for both development and maintenance of the thyroid gland. In the lung, NKX2-1 is implicated in the branching of the lobar bronchi, and at later stages of embryonic development, it regulates the expression of surfactant proteins in type II pneumocytes. In the brain, it has a central role in proper specification and migration of interneuron subtypes in the medial ganglionic eminence and telencephalon.4,5 However, little is known about the direct and indirect transcriptional targets of NKX2-1 in the central nervous system.
In mice, the disruption of the Nkx2-1 gene resulted in a severe, lethal phenotype in homozygous animals, characterised by pulmonary hypoplasia, absence of functional thyroid tissue and complex malformations of brain structures, including the basal ganglia, hypothalamus and pituitary. Heterozygous littermates remained unaffected.4 Against this background and in the light of the multifaceted role of NKX2-1 in normal lung, thyroid and brain development and function, a corresponding phenotype was also expected for germline mutations of NKX2-1 in humans. Indeed, patients with large chromosomal deletions of chromosome 14 encompassing NKX2-1 and several other genes showed malfunctioning of the thyroid, the lung and the central nervous system (CNS).6,7 Yet, it was not before the identification of the first intragenic point mutations that these symptoms could be unequivocally assigned to a dysfunction of NKX2-1.8–10 Notably, in contrast to mice, where only inactivation of both alleles resulted in a disease phenotype, all patients in these studies were heterozygous for the mutation, therefore suggesting a dominant mode of inheritance in humans.
Considerable phenotypic variability in terms of organ involvement and expressivity was already evident in these first described patients and their affected relatives. All individuals suffered from a neurological disorder with profound muscular hypotonia and delay in motor development in their first months of life, followed by the onset of a movement disorder, mainly characterised by uncontrolled, jerky movements by means of a truncal chorea and peripheral athetosis. However, a functional defect of the thyroid was present in only half of the patients, variably manifesting as either severe congenital hypothyroidism (CH) or merely elevated thyroid-stimulating hormone (TSH) levels. Even fewer patients presented with additional pulmonary affection ranging from severe neonatal distress to the occurrence of frequent and severe pneumonias in the first two to three years of life.8–10
Since then, 20 case reports and 7 series of patients with a total of 161 patients and 77 mutations have been reported (see online supplementary table 1),6–33 but altogether they have failed to reveal a correlation between type, size and position of mutations and a specific phenotype. The only exceptions were reports on oligodontia in individuals harbouring chromosomal deletions simultaneously encompassing NKX2-1 and PAX9, reflecting the role of PAX9 in odontogenesis.34–36 This overall lack of genotype–phenotype correlation was surprising given the fact that the two activating domains of NKX2-1 are supposed to be differentially involved in thyroid and pituitary development, implying that at least different missense mutations might cause different phenotypes.37
One reason for this unexpected finding could be a biased composition of patient cohorts in all studies published to date. This bias may be attributed to differences in the main emphasis of these studies, either on Benign hereditary chorea (BHC),15,22,38 CH13,14 or chromosomal aberrations,16,39 respectively, or may have originated from particularities in disease naming,17 which could have led to exclusion of patients that do not display the full spectrum of possible symptoms. Therefore, a comprehensive description of mutation frequency and quality within a larger cohort of non-biased patients is still missing. We have taken advantage of our 10-year experience in diagnosing NKX2-1 deficiency8 to collect a cohort of 101 patients with BHC or the combination of thyroid, lung or neurological symptoms. Here we report the results of the most comprehensive screen for point mutations and DNA copy number variants (CNVs) affecting NKX2-1 in patients with an indicative phenotype to date. Starting from the subset of index patients with confirmed NKX2-1 mutations as well as their mutation-positive relatives, we have re-evaluated the spectrum of symptoms associated with this genotype. Moreover, we provide evidence for DNA copy number changes in next proximity to NKX2-1 that do result in a phenotype comparable to that of intragenic mutations.
MATERIALS AND METHODS
Patients and DNA isolation
Over a period of 10 years, samples of 101 patients (57 male, 44 female, age 0–18 years) were referred for diagnosis characterised by clinical symptoms as follows: (i) triad of thyroid dysfunction, neurological deficit and pulmonary disease in 26 patients (25.5%); (ii) thyroid dysfunction and neurological deficit in 48 patients (47.1%); (iii) isolated neurological deficit in 16 patients (15.7%); (iv) neurological deficit and pulmonary malfunction in 8 patients (7.8%) and (v) thyroid dysfunction and pulmonary disease in 4 patients (3.9%). All patients have been classified by experienced clinicians from 35 medical centres in 18 countries. Genomic DNA of all subjects as well as of healthy controls was isolated from peripheral blood leucocytes using the Qiagen DNA blood mini kit (Qiagen, Hilden, Germany). Informed consent for the genetic diagnosis was obtained from the parents according to the local regulations (ethical approval no. EA2-089-06).
DNA sequencing analysis
The entire coding region of NKX2-1 was amplified in two fragments from genomic DNA (forward primer: 5′-CGTCCCCAGACTCGCTCGCTC-3′/5′-CCTCCTCTTCCTTCCTCC-3′; reverse primer: 5′-CTCAGGAGGTGCGGCGAGAGCCGTC-3′/5′-GAGGGAAGCGGTGAGGCAGAG-3′). Exons 1–3 were sequenced using V3 kit and ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA) applying appropriate primers. DNA of patients 16 and 24 were additionally cloned into a TOPO cloning vector (Invitrogen, Groningen, The Netherlands) as described previously to delineate allele distribution. If DNA was available, the inheritance of identified sequence variants was analysed in family members.
NKX2-1 structural homology model
For model design of the human NKX2-1, the already published NMR structure of the homologous rat thyroid transcription factor homeodomain (Protein-Database entry code 1FTT40) served as a structural template. The interacting DNA fragment was arranged according to a nearly identical and superimposed X-ray structure of the NKX2-5 homeodomain (gray backbone) bound with DNA (PDB entry code 3RKQ). The homeobox is constituted by three helices, which are connected by loops (figure 1C). For model preparation, Sybyl 8.1 was used (Tripos Inc., St. Louis, Missouri, 63144, USA) and the image was produced with the PyMOL software (The PyMOL Molecular Graphics System, V.1.3 Schrödinger, LLC).
Figure 1.
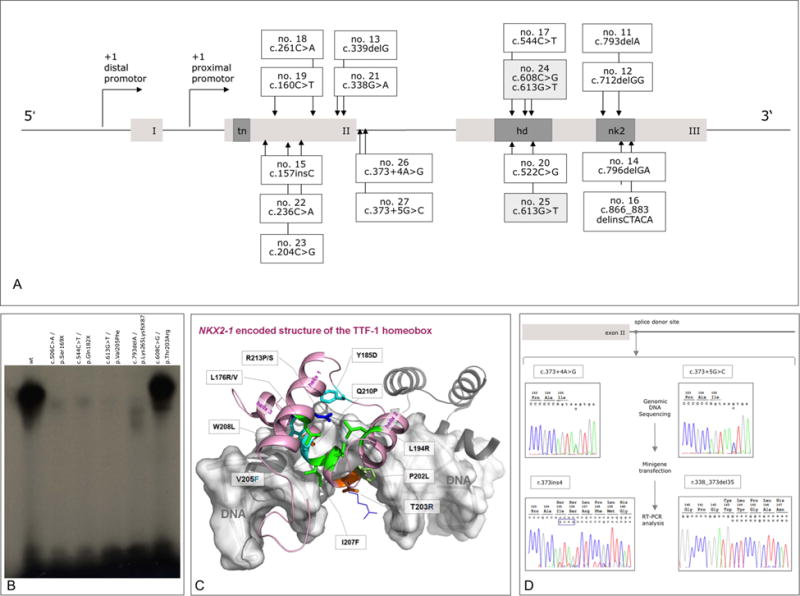
Position and functional consequences of NKX2-1 mutations. (A) Distribution of 17 novel point mutations with respect to the functional domains of NKX2-1 isoform 2 (REFSEQ NM_003317.3). Distal and proximal promoters86,87 are indicated by arrows; functional domains are shown in dark grey (tn, tin domain; hd, homeodomain; nk2, NK2 domain). (B) DNA binding characteristics of mutant NKX2-1. Electromobility shift assay testing wild-type and mutant NKX2-1 binding to a labelled probe corresponding to the NKX2-1 binding site of the TPO promoter. wt (lane 1); c.506C>A/p.Ser169X (8 lane 2); c.544C>T/p.Gln182X (no. 17, lane 3); c.613G>T/p.Val205Phe (nos. 24/1 and 25, lane 4); c.793delA/p. Lys265LysfsX87 (no. 11, lane 5); c.608C>G/p.Thr203Arg (no. 24/2, lane 6); see text for details. (C) Structural homology model. Visualisation of the putative mode of binding between a human NKX2-1 homeobox (light-magenta backbone) and a fragment of DNA (white, translucent surface). The wild-type positions of known naturally occurring mutations are shown as sticks (colour code: green, hydrophobic side chain properties; cyan, aromatic; orange, hydrophilic; blue, positively charged; light green, proline). The side chains and potential orientations of two mutations described in this study—at positions 203 (p. T203R) and 205 (p. V205F)—are additionally visualised as lines. (D) Splicing defects caused by intronic point mutations. Mutant nos. 26 and 27 were analysed by mini gene reporter assays. Sequencing chromatograms of genomic DNA (c.373+4A>G/c.373 +5G>C variants) and RNA after CHO cell transfection and reverse transcription (r.373ins4/r.338_373del35) are depicted in the upper and lower two boxes, respectively.
Electrophoretic mobility shift assay (EMSA)
Wild-type and mutant NKX2-1 proteins were in vitro translated from the pCMX expression vector using the TNT reticulocyte system (Promega, Southampton, UK). Synthetic oligonucleotide probes corresponding to the 3′ NKX2-1 binding site on the thyroglobulin promoter were labelled with [32P]dCTP by Kleenow polymerase and EMSA were performed as described previously.41,42 5 μg of in vitro translated human wild-type NKX2-1 or mutant proteins (NKX2-1 p.Ser169X, NKX2-1 p. Gln182X, NKX2-1 p.Val205Phe, NKX2-1 p.Lys265LysfsX87, NKX2-1 p.Thr203Arg) were incubated with 1 μg BSA in the binding buffer (10 mmol/L TrisHCl pH 7.5, 1 mmol/L EDTA, 4% Ficoll 1 mmol/L dithiothreital and 2 mmol/L PMSF). After incubation for 20 min at 30°C, 100 fmol of the end-radiolabeled oligonucleotide was added to the reaction and incubated for additional 45 min at 30°C. The oligonucleotide 5′-CACTGCCCAGTCAAGTGTTCTTGA-3′ contains the rat NKX2-1 binding site, which is located at nucleotides −72 to −66 relative to the transcription start site of the thyroglobin gene (NCBI accession no. X06162).43–45 Electrophoresis was performed in a 5% native polyacrylamide gel in 0.5×x TBE buffer pH 8.3 with 34 mA at 4°C for approximately 2.5 h. Protein complexes were visualised by autoradiography.
Mini gene reporter assay and cell transfection
The entire genomic region of NKX2-1 (Isoform 2 NM_003317.3) of a wild-type control DNA and two patients’ (26 and 27) DNA was cloned into the eukaryotic expression vector pCS2+. CHO-K1 cells (106/10 cm dish) were transfected with 5 μg plasmid DNA using metafectene (Biontex, Munich, Germany) according to the manufacturer’s protocol. Three days after transfection, cells were harvested, total RNA was extracted using TRIzol (Sigma, Deisenhausen, Germany) and mRNA was isolated with the oligo-Tex mRNA kit (Qiagen, Hilden, Germany). Reverse transcription was performed using oligo-dT primer and M-MLV Reverse Transcriptase (Promega, Mannheim, Germany). Subsequent PCR was performed, and PCR products were directly sequenced.
Customised array CGH analysis
Sixty individuals negative for NKX2-1 point mutations were selected for DNA copy number analysis by means of high-resolution array CGH employing a custom 44k oligonucleotide array (Agilent, Santa Clara, California, USA) representing genomic region chr14: 32055352-41215621 (HG18). As the 5′ breakpoint was excluded in the identified deletion in case 5, results were verified by array CGH on a 244k oligonucleotide array from Agilent following the manufacturer’s instructions (protocol no. G4410-90010). Labelling and hybridisation were performed according to the manufacturer’s recommendations (protocol no. G4410-90010). Arrays were scanned with the G2565BA Agilent Microarray Scanner System (resolution 5 μm; PMT 100% for Cy3/Cy5, respectively) (Agilent Inc. Santa Clara, California, USA). Image processing was performed using Feature Extraction Software v9.1, and data were further analysed in CGH Analytics Software V.3.4.27 (Agilent) using the default settings. All genomic coordinates given in this paper refer to NCBI Build 36 (HG18). Confirmatory experiments were performed with breakpoint spanning PCR and direct sequencing of amplicons. The sequence variant was analysed in family members if DNA specimens were available.
In situ hybridization
For in situ hybridisation, E11.5 NMRI wild-type embryos were used. Mice were mated, and noon of the day of the vaginal plug was considered E0.5. Embryos were dissected in ice-cold 1× PBS, fixed in 4% PFA, dehydrated and embedded in paraffin. 6 μm tissue sections were prepared on a microtome, mounted on silanized slides and dried at 37°C. For hybridisation, probes were labelled with digoxigenin (DIG). The Nkx2-1 probe contained nucleotides c.494-992 (NM_009385.2), the Mbip probe c.574-145 (NM_145442.2), matching the probe used in Ref. 46 Probes were amplified from cDNA by PCR, cloned into the pCRII-vector (Invitrogen, Carlsbad, USA), linearised and transcribed with T7 or Sp6 polymerase. The in situ hybridisation protocol was kindly provided by the Research Group Mundlos (Max Planck Institute for Molecular Genetics, Berlin, Germany) and has been adapted from Ref. 47. Briefly, tissue sections were dewaxed, rehydrated, digested with Proteinase K and acetylated. Hybridisation with DIG-labelled probes was carried out at 68° C. Following posthybridisation washes, sections were incubated with an AP-conjugated a-DIG-antibody (Roche Applied Science, Mannheim, Germany) and signal was detected with NBT/BCIP (Roche Applied Science, Mannheim, Germany).
Detailed phenotyping
For detailed phenotyping of mutation-positive individuals, a total of 28 patients (17 male, 11 female, age 1.5–44 years/18 index patients, 8 relatives) was reanalysed (ethical approval no. EA2-089-06). A questionnaire containing 187 items was used for obtaining clinical data and information on patient history. Video documentation of the patient’s movement disorder was carried out according to standardised protocols, followed by independent classification by two experienced neurologists (using the Burke–Fahn–Marsden Dystonia Rating Scale).
For graphical representation of phenotype data, a heatmap was generated using ‘R’48 (http://www.R-project.org/) and ‘ggplot2’ (http://ggplot2.org/).
RESULTS
NKX2-1 intragenic mutations
Sequencing of 101 patients with a phenotype suggestive for a dysfunction of NKX2-1 revealed 17 intragenic mutations in 17 patients, of which 15 are novel, whereas the remaining two have already been described in unrelated individuals.8 All mutations with respect to their location in the protein are depicted in figure 1A. As listed in more detail in table 1, we have identified six frameshift mutations due to insertion/deletion mutations (indels), one insertion and one deletion–insertion resulting in premature stop codons or elongated protein sequences, seven nonsense and one missense mutation. Furthermore, one compound-heterozygous missense mutation was identified, of which one allelic variant [c.703G>T] occurred de novo, whereas the other [c.698C>G] could also be identified in two siblings and the mother. We carried out EMSA to elucidate the effect of the two missense mutations on NKX2-1 DNA binding capacity in comparison to two nonsense-elongation and one frameshift-elongation mutations. These experiments verified loss of DNA binding capacity for the three nonsense and frameshift mutations as well as for variant Val235Phe, whereas p. Thr233Arg appeared to have a DNA binding capacity equivalent to the wild-type protein (figure 1B). So far, all missense mutations within the homeobox domain were shown to interfere with DNA binding. We developed a homology structure model to delineate the binding capacity of p.Thr233Arg in more detail. The homeobox is constituted of three helices, which are connected by loops. The model demonstrates that the helix 3 is most important for DNA binding, whereby the exact adjustment between the helices 1 and 3 relative to each other is of general significance for NKX2-1 functions. In consequence, mutations at any amino acid in these helices either interfere with the correct fold of the box (intramolecular interactions) or they modulate intermolecular interactions. The wild-type positions of previously described mutations were introduced in the model as well as the side chains and potential orientations of two mutations described in this study—at positions 203 (p.-Thr203Arg) and 205 (p.Val205Phe) (figure 1C).
Table 1.
Overview of identified NKX2-1 deletions and intragenic mutations
| No. | DNA sequence | Size | Protein localisation |
Protein level | Inheritance
|
Phenotype
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Affected family members* |
Thyroid dysfunction |
Choreoathetosis/ muscular hypotonia |
Pulmonary malfunction |
Sex | ||||||
| 1 | chr14:g (36042742_36142540)del |
99 798 bp | Entire protein | p.0 | AD | M 2 | Yes | Yes | Yes | M |
| 2 | chr14:g (35138127_37502915)del |
2 364 788 bp | Entire protein | p.0 | AD | M 1 | Yes | Yes | No | M |
| 3 | chr14:g (35221080_36553158)del |
1 332 078 bp | Entire protein | p.0 | n/a | n/a | No | Yes | No | F |
| 4 | chr14:g (36058432_36825446)del |
767 014 bp | Entire protein | p.0 | n/a | n/a | Yes | Yes | No | M |
| 5 | chr14:g (34071433_36246414)del |
2 174 981 bp | Entire protein | p.0 | n/a | n/a | Yes | Yes | No | F |
| 6 | chr14:g (31766511_46944545)del |
15 178 034 bp | Entire protein | p.0 | n/a | n/a | Yes | Yes | No | M |
| 7 | chr14:g (35883857_38226260)del |
2 342 403 bp | Entire protein | p.0 | AD | F 2 | Yes | Yes | Yes | M |
| 8 | chr14:g (35989750_36754641)del |
764 891 bp | Entire protein | p.0 | De novo | Nil | Yes | Yes | Yes | F |
| 9 | chr14:g (35257445_35257464 + 35257487_35984627)del |
727 181 bp | Del excluding NKX2-1 |
p.? | De novo | Nil | Yes | Yes | No | F |
| 10 | chr14:g (35801941_35878529)del |
76 588 bp | Del excluding NKX2-1 |
p.? | AD | M 2/F 2 | No | Yes | No | M |
| 11 | c.793delA | NK2 | p.Lys265LysfsX87 | De novo | Nil | Yes | Yes | No | M | |
| 12 | c.712delGG | NK2 | p.Gly238ArgfsX170 | AD | F 2 | Yes | Yes | Yes | M | |
| 13 | c.339delG | TN-HD | p.Trp113CysfsX16 | De novo | Nil | No | Yes | No | M | |
| 14 | c.796delGA | NK2 | p.Asp266Argfs142X | De novo | Nil | Yes | Yes | No | F | |
| 15 | c.157insC | TN-HD | p.Met53HisfsX355 | De novo | Nil | Yes | Yes | Yes | F | |
| 16 | c.866_883delinsCTACA | NK2-X | p.Gln289ProfsX59 | De novo | Nil | Yes | Yes | Yes | M | |
| 17 | c.544C>T | HD | p.Gln182X | De novo | Nil | Yes | Yes | Yes | F | |
| 18 | c.261C>A | TN-HD | p.Cys87X | AD | F 2 | Yes | Yes | Yes | M | |
| 19 | c.160C>T | TN-HD | p.Gln55X | Father n/a; mother WT |
n/a | Yes | Yes | No | F | |
| 20 | c.522C>G | HD | p.Tyr174X | n/a | n/a | Yes | Yes | Yes | M | |
| 21 | c.338G>A | TN-HD | p.Trp113X | n/a | n/a | Yes | Yes | No | M | |
| 22 | c.236C>A | TN-HD | p.Ser79X | AD | F 2 | Yes | Yes | No | F | |
| 23 | c.204C>G | TN-HD | p.Tyr68X | AD | F 1 | Yes | Yes | Yes | M | |
| 24 | [c.608C>G]+[c.613G>T] | HD | [p.Thr203Arg]+[p. Val205Phe] | AD/de | F 3 | Yes | Yes | No | M | |
| 25 | c.613G>T | HD | p.Val205Phe | De novo | Nil | Yes | Yes | No | F | |
| 26 | c.373+4A>G/r.373ins4 | TN-HD (splice donor site) | p.Ile155SerfsX126 | De novo | Nil | Yes | Yes | Yes | M | |
| 27 | c.373+5G>C/r.338_373del35 | TN-HD (splice donor site) | p.Trp143CysfsX138 | n/a | n/a | Yes | Yes | No | M | |
| 28 |
chr14:g (32446471_41666818)del |
9.2 Mbp | Entire protein | p.0 | De novo | Nil | Yes | Yes | Yes | M |
| 29 | c.585nsGG | HD | p.Leu195GlyfsX3 | De novo | Nil | Yes | Yes | Yes | M | |
| 30 | c.506C>A | HD | p.Ser169X | n/a | n/a | Yes | Yes | No | M | |
| 31 | c.261C>A | TN-HD | p.Cys87X | AD | F 1 | Yes | Yes | Yes | M | |
| 32 | c.613G>T | HD | p.Val205Phe | De novo | Nil | Yes | Yes | Yes | M | |
Nomenclature for mutations according to recommendations of the Human Genome Variation Society (http://www.hgvs.org/mutnomen/); reference sequence REFSEQ Isoform 2 NM_003317.3; P, entire protein; HD, homeodomain; NK2, NK2-specific domain; AD, autosomal-dominant; n/a, information not available; TN-HD, sequence between tin- and homeodomain; M, male; F, female;
excluding index patient; mutations 29–32 as reported previously by authors.8
The italicised values refer to cases that have not been identified in the present genotyping study, but earlier in the Krude et al 2002 study8.
In addition, we performed mini gene reporter assays for the two intronic mutations that potentially affect splice site mutations (nos. 26 and 27). Both mutations result in splice variants that are each truncating the functional domains in exon 3 by a frame shift (figure 1D).
NKX2-1 CNV
DNA of 60 patients negative for NKX2-1 point mutations was further investigated for the presence of copy number changes in the NKX2-1 genomic region by high-resolution array CGH. By using a 44k customised oligonucleotide array, we identified 10 heterozygous deletions affecting the chromosomal region 14q13 in 10 patients. All 10 CNVs are differing in size and were verified by breakpoint spanning PCR and sequencing (table 1, figure 2B and online supplementary figure S2). The largest deletion is spanning 15.17 Mb (no. 6, chr14: 31766412-46945775) and comprises 45 RefSeq genes, including NKX2-1. The NKX2-1 gene comprises two exon and one intron covering 3306 bp. Incomplete loss of NKX2-1 has exclusively been observed in no. 4, where the deletion was limited to exon 1 and the distal NKX2-1 promoter. Figure 2A displays the extent of all CNVs in their genomic context, according to the UCSC Genome Browser (HG18).
Figure 2.
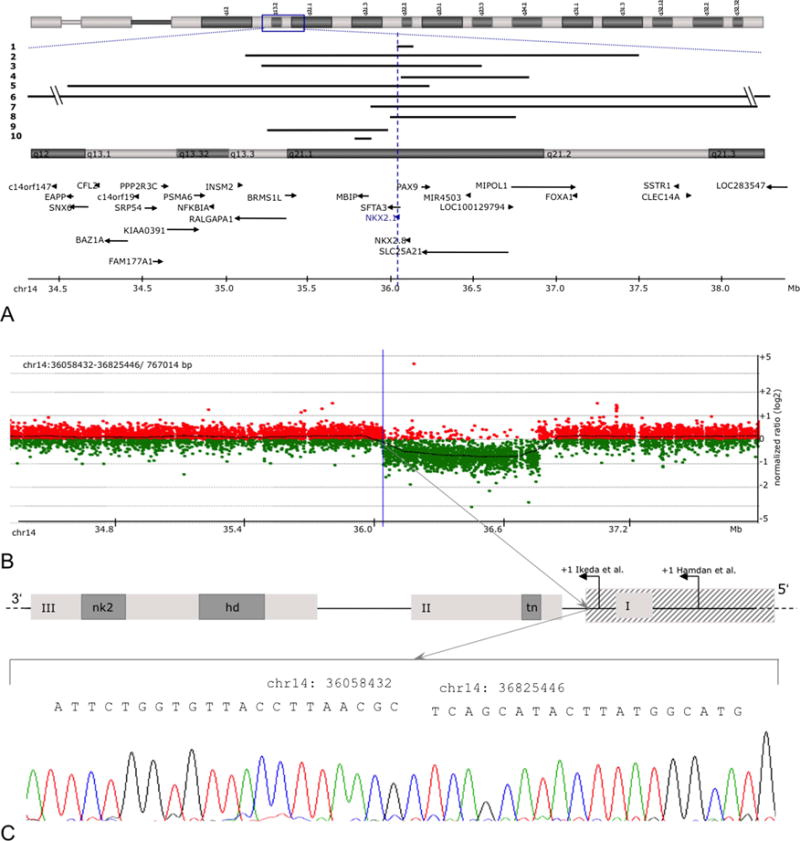
Disease-associated copy number variants (CNVs). (A) Localisation of all CNVs reported in this study. The vertical line highlights the position of NKX2-1 on chromosome 14q13.3. (B) Array CGH profile of deletion no. 4 encompassing chr14: 35989750-36754641 (array CGH results of all other cases are summarised in online supplementary figure 2). (C) CNV verification by breakpoint spanning PCR and amplicon sequencing on the example of the proximal border of deletion no. 4 (hatched box).
CNV affecting MBIP
Two CNVs, among them the smallest aberration encompassing 76.67 kb (no. 10, chr14: 35802212-35878882), did not include NKX2-1, but led to a heterozygous loss of the 5′-neighbouring gene MBIP, which encodes for the MAP3K12 binding inhibitory protein (figure 3A, table 1). Deletion no. 9 occurred de novo in a patient with chorea and mild hypothyroidism (3B.1) while deletion no. 10, which exclusively affects MBIP, was transmitted in a large kindred with chorea of dominant inheritance (figure 3B.2). We performed Mbip in situ hybridisation of early mice embryos and detected an identical pattern of expression in comparison to NKX2-1, including the basal ganglia, forebrain, thyroid and lung (figure 3C). We sequenced the coding region of MBIP in 74 patients negative for NKX2-1 mutation, but did not detect any alteration (data not shown).
Figure 3.
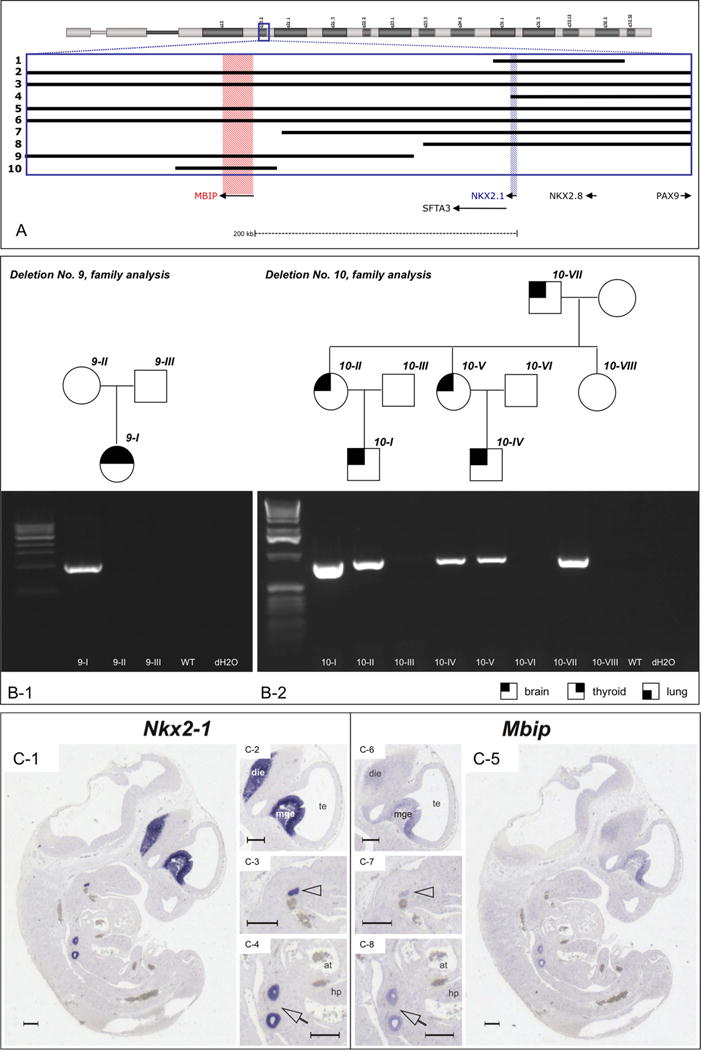
Copy number variants (CNVs) not including NKX2-1. (A) Close-up of chr14: 35,739,446-36,221,024 (Hg18). The positions of NKX2-1 and MBIP are highlighted in blue and red, respectively. Note that CNVs nos. 9 and 10 do not affect NKX2-1. (B) Breakpoint spanning PCR verifying the deletion in index patients nos. 9 and 10 and their relatives; see legend at the bottom for phenotype symbols. All genomic coordinates are given for Hg18. (C) Nkx2-1 and Mbip co-expression in mouse embryonic tissues. Nkx2-1 and Mbip in situ hybridisations on adjoining sagittal sections of an E11.5 wild-type embryo. Nkx2-1 and Mbip have common expression domains in the diencephalon, the median ganglionic eminence, the lung anlage and the thyroid primordium. C1–C4, Nkx2.1; C5–C8, Mbip in situ hybridisation. Panels C2–C4 and C6–C8 represent enlargements of C1 and C5, respectively. Arrowheads, thyroid primordium; arrows, lung anlage; die, diencephalon; mge, median ganglionic eminence; te, telencephalic vesicle; at, atrium; hp, hepatic primordium. Scale bars represent 250 μm.
Phenotype spectrum in index patients
Altogether, we have identified 25 new cases of NKX2-1 mutations and two additional patients with small deletions close to NKX2-1 affecting the adjacent MBIP gene. Among them, 11 patients displayed the complete triad of movement, thyroid and lung symptoms, 13 patients the dual combination of movement defects and thyroid dysfunction, and 3 patients showed isolated movement disorder (figure 4A). Phenotype of cases with NKX2-1 mutation did not differ from the two cases of MBIP mutations, as the latter were both affected by chorea associated with mild thyroid dysfunction in one of them. In all patients, neurological impairment was present, characterised by benign choreoathetosis preceded by muscular hypotonia in early infancy. Within the group of mutation-negative cases (figure 4B), 15 patients were affected by the triad of movement, thyroid and pulmonary symptoms, putatively caused either by variants in the NKX2-1 enhancer and/or promoter region or by mutations in another yet unknown candidate gene.
Figure 4.
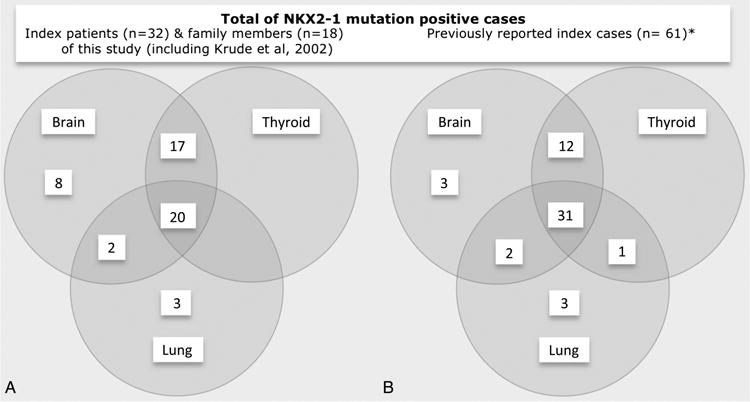
Brain, thyroid and lung involvement in patients with and without NKX2-1 mutations. Venn diagrams demonstrating that the triad of symptoms is enriched in mutation-positive patients. (A) Symptoms in patients with NKX2-1 deficiency (27/101). (B) Symptoms in patients without NKX2-1 deficiency (74/101). Two further Venn diagrams, one focusing on the phenotype of 18 family members and another one including also the 5 index patients of our previous study,8 are given in online supplementary figure S3. Two further Venn diagrams demonstrating that symptom involvement is identical in index cases (n=32, including also the five index patients of our previous study8 to that of family members (n=18), except three patients with isolated asthma phenotype) are given in online supplementary figure S3.
Overall, the phenotypic spectrum of all 32 index mutation-positive patients that we identified in this report as well in our initial publication from 2002 (see online supplementary figure S3A) is dominated by the occurrence of a movement disorder in all patients. However, accompanying thyroid dysfunction was the most consistent combination of symptoms, followed by the triad of thyroid, lung and movement affection, whereas only three patients showed an isolated movement disorder. Moreover, comparison of the index cases who were initially examined for their presumably more severe phenotype with other affected family members who represent thereby an unbiased cohort of mutation-positive patients revealed an identical spectrum of symptom association (see online supplementary figure S3).
Extended phenotyping
In order to define a more detailed phenotype of NKX2-1 deficiency in a larger cohort of patients, we performed a clinical study in 28 mutation-positive patients (7 deletions/21 point mutations) by recollecting all available clinical and history data as well as video documentation of individual movement statuses (figure 5A,B).
Figure 5.
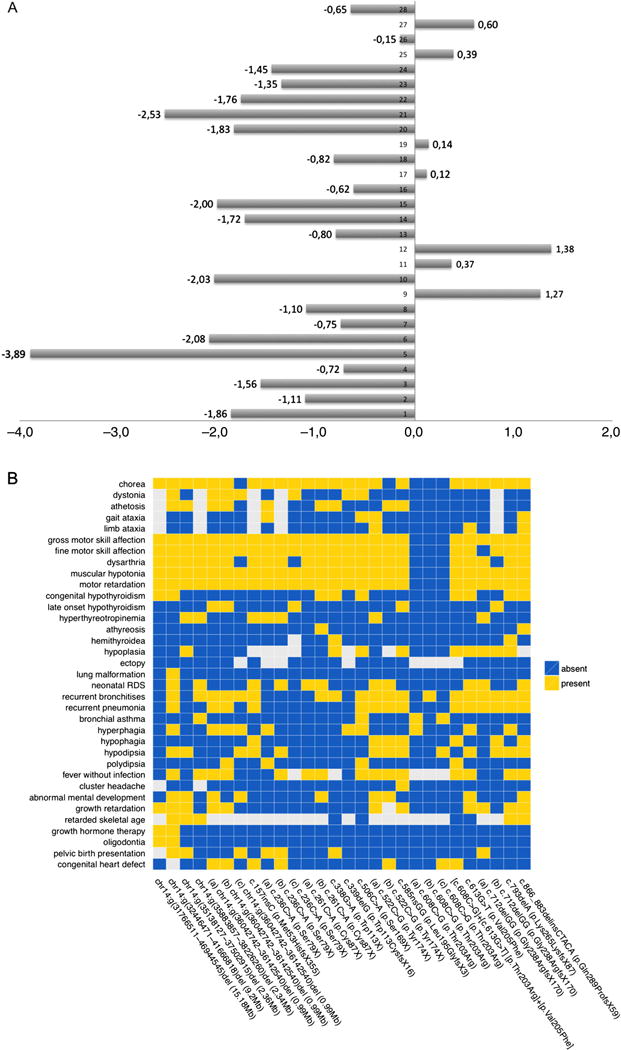
Additional symptoms in patients with NKX2-1 mutations. (A) Body height standard deviation score (SDS) values of detailed phenotype cohort. Median value −0.84 SDS; patients 1–18 carrying NKX2-1 mutations: 1, chr14:g (36042742_36142540)del; 2, chr14:g (36042742_36142540)del; 3, chr14:g (36042742_36142540)del; 4, chr14:g (35138127_37502915)del; 5, chr14:g (32446471_41666818)del; 6, chr14:g (31766511_46944545)del; 7, chr14:g (35883857_38226260)del; 8, c.522C>G_p.Tyr174X; 9, c.522C>G_p. Tyr174X; 10, c.585nsGG_p. Leu195GlyfsX3; 11, c.613G>T_p. Val205Phe; 12, [c.608C>G] +[c.613G>T]_[p.Thr203Arg]+[p. Val205Phe]; 13, c.608C>G_p. Val205Phe; 14, c.608C>G_p. Val205Phe; 15, c.608C>G_p. Val205Phe; 16, c.338G>A_p.Trp113X; 17, c.261C>A_p.Cys87X; 18, c.261C>A_p.Cys87X; 19, c.506C>A_p. Ser169X; 20, c.157insC_p. Met53HisfsX355; 21, c.236C>A_p. Ser79X; 22, c.236C>A_p.Ser79X; 23, c.236C>A_p.Ser79X; 24, c.866_883delinsCTACA_p. Gln289ProfsX59; 25, c.793delA_p. Lys265LysfsX87;26, c.712delGG_p. Gly238ArgfsX170; 27, c.712delGG_p. Gly238ArgfsX170; 28, c.339delG_p. Trp113CysfsX16. (B) Heatmap summarising the results of detailed phenotyping in 18 index patients and 10 relatives with NKX2-1 deficiency. The presence and absence of symptoms is indicated by yellow and blue boxes, respectively. Missing values are shown in grey. The first seven columns to the left depict the phenotypes of the patients affected by a copy number variant. Point mutations are sorted according to their genomic position. The letters (a), (b) and (c) heading the description of the mutation indicate family members sharing the same mutation.
To date, a wide range of neurological descriptions has been used for the movement disorders in the available reports on NKX2-1 deficiency, which is namely ‘choreoathetosis’ in the majority of cases. We aimed to gain an objective neurological classification through independent judgement. In 17 of 22 videos, movement disorder was classified as chorea, athetosis was stated in 9, ataxia in 7 and dystonia in 6. As either chorea or athetosis was present in all examined patients, our data are suggesting that choreoathetosis is in fact the most frequent form of movement disorder in NKX2-1 deficiency.
In addition to the gross motor disorder, 23 patients were affected by dysarthria, while speech comprehension was normal in all. An attentiveness disorder was mentioned in 22 patients. Mental development was quantified by IQ testing in 12 patients, showing low normal levels in most cases (exact values were available in three individuals with 80, 116 and 119 IQ points, respectively (K-ABC test and CPM-test)). EEG recording was available in 16 patients, with 3 to have suspicion of focal lesions, while no seizures were reported in any of them. MRI of the CNS was available in nine patients, with no major pathological findings (Rathkes pouches n=2 and hypoplastic pallidum n=1).
In 23 patients, thyroid defects were present as CH (n=11), hyperthyreotropinemia (n=12) and thyroid dysgenesis (n=14, among them athyreosis n=2, hemithyroid n=2, thyroid hypoplasia n=10).
Careful case history revealed a high incidence of lung affection, that is, in 22 of the 28 patients. In 13 patients, a respiratory distress syndrome with need for oxygen treatment during the neonatal period was reported. Fifteen patients were affected by recurrent pneumonias requiring antibiotic treatment (incidence ranging from 3 per annum to 12 per annum) in the first 6 years of life. In one patient, the pneumonic episodes persisted beyond that age, prior to diagnosis of a lung sequester. Spontaneous pneumothoraces (four relapses) occurred in another patient.
According to the murine phenotype in knockout animals with severe hypothalamic defects, we extended our exploration to various other symptoms, herein focusing on hypothalamic function. In 19 patients, bouts of fever episodes without evidence for infectious causes were recorded; 11 patients had increased appetite with reduced satiety, although BMI values were normal in all patients; in 14 patients, thirst was abnormally diminished, and in 10 patients a disturbed rhythm of sleep was reported.
Notably, patients’ height standard deviation score (SDS) values revealed a trend for lower body length (median value −0.84 SDS, see figure 5A), and two patients even received growth hormone therapy (SDS values −3.89 and −2.08, respectively).
In seven individuals, a congenital heart defect (ventricular or atrial septum defect, respectively) was present, of which none needed surgical intervention and three patients were seen with oligodontia. Furthermore, six pregnancies had breech presentation at birth, hypothetically giving evidence for intrauterine hypokinesia due to muscular hypotonia.
DISCUSSION
Despite the key role of NKX2-1 in thyroid development,49 several comprehensive screening approaches failed to detect NKX2-1 mutations in patients with CH.50–52 Only when the presumptive NKX2-1 deficiency phenotype had been extended so that it also took into account the impact of NKX2-1 on development and function of other organs such as lung and brain, the first mutations were identified.8,9
Core phenotype associated with NKX2-1 deficiency
Neurological impairment, mainly in the form of choreoathetosis, was present in all index patients identified in this study. This is in accordance with all reports in literature so far, apart from those neonatal cases with severe lung affection, who died before the onset of neurological symptoms.20,28 The triad of symptoms (including defects of thyroid and lung) was significantly enriched in mutation-positive individuals, although non-neurological features were less frequent and their presence varied to a much higher extent. In line with literature, our analysis revealed a high rate of thyroid defects (75%) with mild TSH elevation in the majority of patients. We detected a higher frequency of pulmonary disorder (78%) than previously reported, with neonatal respiratory distress (46%) and recurrent pneumonia with need for antibiotic treatment (53%) as the prevailing features. The reasons for the organ-specific differences in response to NKX2-1 deficiency remain elusive, but some facts suggest that the lower tolerance towards NKX2-1 deregulation in brain might be connected to the postnatal function of NKX2-1 in this organ. NKX2-1 is expressed in brain throughout life, especially in cholinergic and GABAergic neurons of the basal ganglia, both in mice and human.53–55 Moreover, structural brain defects have not been described in NKX2-1 mutation-positive patients so far, and they are oligosymptomatic in the first years of their life. Importantly, NKX2-1-deficient patients in this cohort were predominantly not affected by epilepsy or severe mental retardation. This was unexpected given the phenotype of Nkx2-1-deficient mice with respect to cortical interneurons5 and recent bioinformatic analysis predicting NKX2-1 as a candidate gene for epilepsy in humans.56
New symptoms associated with NKX2-1 deficiency: abnormal height, cardiac septum defects and asthma
Our comprehensive phenotypic characterisation revealed novel NKX2-1-associated hypothalamic symptoms, including frequent occurrence of fever without infection, dysrhythmic sleep and a trend towards lower body height SDS values. Altogether, this suggests an influence of NKX2-1 on the hypothalamus–pituitary–growth axis in accordance with the severe disturbance of the hypothalamic function in the knockout mice.4 Furthermore, we confirmed the occurrence of attention deficit disorder as described by Gras and colleagues.15
Atrial and ventricular septum defects (VSD, ASD) were present in 7 out of 28 index patients with NKX2-1 mutations. Congenital heart defects are a known comorbidity of thyroid malformations, with ASD as the most frequent manifestation.57 Thus—at first sight—the cardiac findings observed in this study may reflect that proper heart development depends on normal thyroid hormone activity, for example, the postnatal surge of T3 levels, which normally triggers the switch from cardiac myosin heavy chain isoform MYH7 to MYH6.58 Mutations in MYH6 have already been identified in familial cases of atrial septum defects59). However, cardiac defect frequency observed in NKX2-1 mutation-positive patients of this study is much higher than the reported prevalence in patients with thyroid malfunction without genotype-based stratification (7/28 vs 13.8/100057,60). Moreover, one patient included in this study presented with congenital heart defect in the absence of any thyroidal defect. Both observations point at a direct role of NKX2-1 in heart development and the aetiology of cardiac septum defects, which is independent of thyroid hormone activity. This idea is supported by the fact that mutations in NKX2-5 have already been identified as a cause of ASD, and that both members of the NKX family share several direct and indirect downstream targets, including key regulators of heart development such as GATA4, GATA6 and HOP.61–63
There is accumulating evidence for a role of NKX2-1 in the development and progression of leukaemia, thyroid and bronchial carcinoma.64–69 Furthermore, an association of germline mutations and the development of neoplasiae, for example, multinodular goitre and papillary thyroid carcinoma,60 has been discussed. This may impact the use of congenital NKX2-1 mutations as a prognostic marker in future. In principle, the identification of congenital NKX2-1 mutations is a favourable result as it predicts that disease will hardly progress and it holds out the prospect that symptoms will improve at later stages of life.70,71 This positive aspect may be tarnished in future by uncertainties about an increased cancer risk that might be associated with congenital NKX2-1 mutations. Unfortunately, the young average age of patients in this study did not allow any conclusive assessment of cancer risk associated with congenital NKX2-1 mutations. However, one patient died of bronchial carcinoma at the age of 46 without having any history of smoking.
Quality of mutations and their correlation to phenotype
In line with previous studies, all intragenic point mutations identified in this study differ in position and quality. Thus, the exclusive clustering of missense mutations within the DNA binding homeodomain is unlikely to indicate a mutational hot spot, but rather reflects a lower tolerance towards single amino acid exchanges within the homeodomain compared with the surrounding transactivation domains. There, only nonsense or frame-shift mutations in the N-terminal part, and potentially also in the C-terminal part,37 can compromise protein functionality.
We have performed accurate phenotyping in index patients and their affected relatives. This enabled us to monitor the symptoms caused by the very same NKX2-1 mutation in different individuals. In general, intrafamiliar phenotypic variability was smaller than the variability between unrelated patients carrying different mutations (figure 5B). Neurological symptoms turned out to be the most consistent feature within families, while the intrafamiliar spectrum of thyroid and lung symptoms could range from no impairment at all to severe hypothyroidism with agenesis of the gland and severe neonatal respiratory distress, respectively.
Our family studies also highlighted the possible consequences of NKX2-1 mutations not altering its ability to bind to DNA. So far all mutations located within the homeodomain have shown loss of function due to interference with DNA binding.8,13,15,21–25,27,28 Here we report the first variant within the homeodomain (p.Thr203Arg) that does not affect DNA binding capacity. The mutation was present on the maternal allele of an index patient, who also carried a second missense mutation (p.Arg205Phe) mapping to the homeodomain encoded on his paternal allele. As can be inferred from the structural homology model, which depicts the intercalation of homeodomain helices and DNA binding groove (figure 1C), the p.Thr203Arg variant neither seems to play a role in the stabilisation of the helix conformation nor to interact directly with DNA. In line with this assumption, and in contrast to the p.-Arg205Phe mutation on the paternal allele, experimental validation by EMSA showed no deterioration of DNA binding capacity associated with the p.Thr203Arg variant.
Only the index patient who carried both mutations presented with the characteristic phenotype that is typically associated with NKX2-1 deficiency, including neurological symptoms. His three relatives, who solely carried the mutation that had no effect on DNA binding (p.Thr203Arg), lacked most of these symptoms, but not all. Congenital heart defect was diagnosed in one of the three relatives, asthma or recurrent bronchial infection was present in all of them. This observation can be interpreted in two ways: (i) only the second mutation (p.Arg205Phe) in the index patient is causative, while ASD and pulmonary problems in his relatives are not related to mutations in NKX2-1; or (ii) the mild pulmonary and cardiac symptoms are caused by impaired functionality of NKX2-1 that is not mediated by its DNA binding capacity. Although the observed haploinsufficiency of NKX2-1 argue in favour of the first hypothesis, there is also evidence in support of the second one. Minoo and colleagues recently discovered that the homeodomain of NKX2-1 physically interacts with FOXA1 in a DNA-independent manner in order to control the expression of surfactant protein C (SP-C) and Clara cell protein (Ccsp).72 Both SpC and Ccsp are essential for proper function of the lung, and their deregulation has already been implicated in the aetiology of asthma and recurrent pulmonary infections.73,74 In light of these facts, it is tempting to speculate that the characteristic NKX2-1 phenotype and its variability are not only attributed to impaired DNA binding, but are also a reflection of altered protein–protein interaction characteristics of NKX2-1, or more precisely, one of its isoforms. Thus, preservation of DNA binding capacity of mutated NKX2-1 alone may not be an eligible argument to rule out disease association. This may especially apply to those rare cases that lack neurological symptoms, including the recently described missense mutation p.Phe198Leu, where, unfortunately, its functional implications have not been investigated.75 Noteworthy, HOP, one of the shared downstream targets of NKX2-1 and NKX2-5 (see above), is a homeodomain protein that lacks any DNA binding potential.76
CNVs outside of NKX2-1 result in a phenotype similar to that of intragenic mutations
Given the impact of DNA CNVs on the aetiology of congenital disorders, we have complemented our sequence analysis by testing patients negative for NKX2-1 point mutations by means of array CGH employing a high-resolution custom array. We have identified 10 novel heterozygous deletions. In line with previous reports,14,16 they all differed in size and position. This makes it unlikely that recurrence of NKX2-1 deletions is the consequence of a specific genomic architecture that renders this region particularly prone to rearrangements. Our data also corroborate the reported lack of correlation between CNV size, which ranges from 76 kb to 15.17 Mb in our study, and severity of disease.14
Strikingly, two of the deletions did not encompass NKX2-1 at all. Similar observations have been made recently and have fuelled speculations that removal of regulatory elements downstream of NKX2-1 might produce a phenotype similar to intragenic loss.39,77 At least two prospective enhancer elements related to forebrain and thyroid function seem to exist with or in close proximity to the genomic interval encompassed by the two deletions identified in this study.78,79 For one of these conserved sequences, a forebrain expression has already been shown (http://enhancer.lbl.gov/cgi-bin/imagedb3.pl?form=presentation&show=1&experiment_id=1538&organism_id=1) and the other one has been highlighted as potential enhancer by a recent meta-analysis of GWAS data related to thyroid function and TSH levels.79 Unfortunately, re-evaluation of public data on higher-order chromatin organisation and CTCF binding, a protein involved in DNA looping, did not reveal any particular chromosome conformation that conclusively indicates any connections of NKX2-1 with a specific regulatory element in this region80–82 (see online supplementary figure S5). Nevertheless, there seems to exist selective pressure to keep NKX2-1 in next proximity to the interval deleted in the two patients as this region, together with NKX2-1, is part of one of the largest blocks of synteny in humans83 and also represents the core region of the recurrent amplicon reported in thyroid and bronchial carcinomas.64–66 However, synteny and co-amplification could also signal the presence of genes, whose co-regulation with NKX2-1 is essential for proper development and function of brain, thyroid and lung. Proceeding on this assumption, distortion of the fine-tuned stoichiometry of their products could mimic an NKX2-1 deficiency like phenotype. Two genes recently discussed in the context of CNVs outside NKX2-1 are NPAS3 and RALGAPA1.77 Malfunction of these two genes can lead to respiratory distress and cognitive disorders, respectively. Yet, both genes are not mapping within the commonly deleted region of the two cases from our study. The only gene mapping to this interval is MBIP (MAP3K12 binding inhibitory protein 1), which encodes a component of the ATAC (Ada Two-A containing) complex. This histone acetyltransferase complex is supposed to translate extracellular cues into chromatin modifications and consequently altered gene expression.84 Interrogation of protein interaction databases by means of String9.0585 and literature mining did not reveal apparent commonalities between MBIP and NKX2-1, apart from the fact that both proteins are—directly or indirectly—involved in the regulation of protein acetylation. In order to elucidate more information on possible functional connections between the two proteins, we have evaluated expression patterns of Mbip in mouse embryos at embryonic day 11.5. In situ hybridisation revealed that the Mbip expression pattern spatially and temporally coincides with that of Nkx2-1, for example, in the thyroid anlage, lung and forebrain (figure 3C). This fits to the recent description of Mbip expression in progenitor cells of the murine lateral ganglionic eminence,46 which is also known as the major expression domain of Nkx2-1 in the forebrain at the same stage of embryonic development.4 53 This coincidence renders deregulation of MBIP a plausible cause of a NKX2-1 deficiency like phenotype and has prompted us to sequence MBIP in those 74 patients of our cohort, who were negative for NKX2-1 mutations. In contrast to what would have been expected under the assumption of a causative role of MBIP for this phenotype, no single mutation was identified in this gene.
In summary, our study expands the phenotypic spectrum recurrently associated with NKX2-1 deficiency, with some of the symptoms maybe not exclusively linked to alterations of DNA binding capacity of this transcription factor. In our view, the high incidence of NKX2-1 mutations justifies the regular molecular screening in corresponding patients in order to distinguish them from others with resembling phenotypes at early stages of disease, but considerable different outcomes. However, these diagnostic efforts should not be confined to the gene itself, but should also comprise the analysis of regulatory elements and interaction partners of NKX2-1.
Supplementary Material
Acknowledgments
We thank all the patients and their families for their cooperation and support.
Footnotes
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/jmedgenet-2013-102248).
Correction notice This article has been corrected since it was published Online First. The second corresponding author is now included and Markus Schuelke name has been updated.
Contributors Genotyping experiments and analysis (Sanger sequencing and array CGH) as well as preparation of figures were done by AT. Extended phenotype data collection and analysis was carried out by SS-H. Further experimental data were generated by PS (mouse embryonic hybridisation experiments). IM and SJ (technical assistance on array CGH and Sanger sequencing), CD (EMSA experiments), HB (Mini Gene Reporter Assay & Cell Transfection experiments) and GK (NKX2-1 structural homology model). Furthermore, JK and MS assisted on analysis of extended phenotype data. GE, AS and H-HR contributed to interpretation of data and AG to study design, respectively. CB, KB, H-JC, PC, FZ, MG, JH, SI, CH, KK, BP, DR and IS did patients’ phenotyping and sample collection. AT, RU and HK designed the study, interpreted data and wrote the manuscript, and are therefore the guarantors of the work.
Competing interests None.
Patient consent Obtained.
Ethics approval Institutional Review Board of Charité, University Medicine Berlin.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Kim Y, Nirenberg M. Drosophila NK-homeobox genes. Proc Natl Acad Sci USA. 1989;86:7716–20. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guazzi S, Price M, De Felice M, Damante G, Mattei MG, Di Lauro R. Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 1990;9:3631–9. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuno K, Gonzalez FJ, Kimura S. Thyroid-specific enhancer-binding protein (T/EBP): cDNA cloning, functional characterization, and structural identity with thyroid transcription factor TTF-1. Mol Cell Biol. 1991;11:4927–33. doi: 10.1128/mcb.11.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–9. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 5.Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–70. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 6.Devriendt K, Vanhole C, Matthijs G, de Zegher F. Deletion of thyroid transcription factor-1 gene in an infant with neonatal thyroid dysfunction and respiratory failure. N Engl J Med. 1998;338:1317–18. doi: 10.1056/NEJM199804303381817. [DOI] [PubMed] [Google Scholar]
- 7.Iwatani N, Mabe H, Devriendt K, Kodama M, Miike T. Deletion of NKX2.1 gene encoding thyroid transcription factor-1 in two siblings with hypothyroidism and respiratory failure. J Pediatr. 2000;137:272–6. doi: 10.1067/mpd.2000.107111. [DOI] [PubMed] [Google Scholar]
- 8.Krude H, Schutz B, Biebermann H, von Moers A, Schnabel D, Neitzel H, Tonnies H, Weise D, Lafferty A, Schwarz S, DeFelice M, von Deimling A, van Landeghem F, DiLauro R, Gruters A. Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2–1 haploinsufficiency. J Clin Invest. 2002;109:475–80. doi: 10.1172/JCI14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pohlenz J, Dumitrescu A, Zundel D, Martine U, Schonberger W, Koo E, Weiss RE, Cohen RN, Kimura S, Refetoff S. Partial deficiency of thyroid transcription factor 1 produces predominantly neurological defects in humans and mice. J Clin Invest. 2002;109:469–73. doi: 10.1172/JCI14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breedveld GJ, van Dongen JW, Danesino C, Guala A, Percy AK, Dure LS, Harper P, Lazarou LP, van der Linde H, Joosse M, Gruters A, MacDonald ME, de Vries BB, Arts WF, Oostra BA, Krude H, Heutink P. Mutations in TITF-1 are associated with benign hereditary chorea. Hum Mol Genet. 2002;11:971–9. doi: 10.1093/hmg/11.8.971. [DOI] [PubMed] [Google Scholar]
- 11.Petek E, Plecko-Startinig B, Windpassinger C, Egger H, Wagner K, Kroisel PM. Molecular characterisation of a 3.5 Mb interstitial 14q deletion in a child with several phenotypic anomalies. J Med Genet A. 2003;40:e47. doi: 10.1136/jmg.40.4.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devos D, Vuillaume I, de Becdelievre A, de Martinville B, Dhaenens CM, Cuvellier JC, Cuisset JM, Vallee L, Lemaitre MP, Bourteel H, Hachulla E, Wallaert B, Destee A, Defebvre L, Sablonniere B. New syndromic form of benign hereditary chorea is associated with a deletion of TITF-1 and PAX-9 contiguous genes. Mov Disord. 2006;21:2237–40. doi: 10.1002/mds.21135. [DOI] [PubMed] [Google Scholar]
- 13.Carre A, Szinnai G, Castanet M, Sura-Trueba S, Tron E, Broutin-L’Hermite I, Barat P, Goizet C, Lacombe D, Moutard ML, Raybaud C, Raynaud-Ravni C, Romana S, Ythier H, Leger J, Polak M. Five new TTF1/NKX2.1 mutations in brain-lung-thyroid syndrome: rescue by PAX8 synergism in one case. Hum Mol Genet. 2009;18:2266–76. doi: 10.1093/hmg/ddp162. [DOI] [PubMed] [Google Scholar]
- 14.Teissier R, Guillot L, Carre A, Morandini M, Stuckens C, Ythier H, Munnich A, Szinnai G, de Blic J, Clement A, Leger J, Castanet M, Epaud R, Polak M. Multiplex Ligation-dependent Probe Amplification improves the detection rate of NKX2.1 mutations in patients affected by brain-lung-thyroid syndrome. Horm Res Paediatr. 2012;77:146–51. doi: 10.1159/000337214. [DOI] [PubMed] [Google Scholar]
- 15.Gras D, Jonard L, Roze E, Chantot-Bastaraud S, Koht J, Motte J, Rodriguez D, Louha M, Caubel I, Kemlin I, Lion-Francois L, Goizet C, Guillot L, Moutard ML, Epaud R, Heron B, Charles P, Tallot M, Camuzat A, Durr A, Polak M, Devos D, Sanlaville D, Vuillaume I, Billette de Villemeur T, Vidailhet M, Doummar D. Benign hereditary chorea: phenotype, prognosis, therapeutic outcome and long term follow-up in a large series with new mutations in the TITF1/NKX2–1 gene. J Neurol Neurosurg Psychiatry. 2012;83:956–62. doi: 10.1136/jnnp-2012-302505. [DOI] [PubMed] [Google Scholar]
- 16.Santen GW, Sun Y, Gijsbers AC, Carre A, Holvoet M, Haeringen A, Lesnik Oberstein SA, Tomoda A, Mabe H, Polak M, Devriendt K, Ruivenkamp CA, Bijlsma EK. Further delineation of the phenotype of chromosome 14q13 deletions: (positional) involvement of FOXG1 appears the main determinant of phenotype severity, with no evidence for a holoprosencephaly locus. J Med Genet A. 2012;49:366–72. doi: 10.1136/jmedgenet-2011-100721. [DOI] [PubMed] [Google Scholar]
- 17.Willemsen MA, Breedveld GJ, Wouda S, Otten BJ, Yntema JL, Lammens M, de Vries BB. Brain-Thyroid-Lung syndrome: a patient with a severe multi-system disorder due to a de novo mutation in the thyroid transcription factor 1 gene. Eur J Pediatr. 2005;164:28–30. doi: 10.1007/s00431-004-1559-x. [DOI] [PubMed] [Google Scholar]
- 18.Moya CM, Perez de Nanclares G, Castano L, Potau N, Bilbao JR, Carrascosa A, Bargada M, Coya R, Martul P, Vicens-Calvet E, Santisteban P. Functional study of a novel single deletion in the TITF1/NKX2.1 homeobox gene that produces congenital hypothyroidism and benign chorea but not pulmonary distress. J Clin Endocrinol Metab. 2006;91:1832–41. doi: 10.1210/jc.2005-1497. [DOI] [PubMed] [Google Scholar]
- 19.Nagasaki K, Narumi S, Asami T, Kikuchi T, Hasegawa T, Uchiyama M. Mutation of a gene for thyroid transcription factor-1 (TITF1) in a patient with clinical features of resistance to thyrotropin. Endocr J. 2008;55:875–8. doi: 10.1507/endocrj.k08e-124. [DOI] [PubMed] [Google Scholar]
- 20.Kleinlein B, Griese M, Liebisch G, Krude H, Lohse P, Aslanidis C, Schmitz G, Peters J, Holzinger A. Fatal neonatal respiratory failure in an infant with congenital hypothyroidism due to haploinsufficiency of the NKX2–1 gene: alteration of pulmonary surfactant homeostasis. Arch Dis Child. 2010 doi: 10.1136/adc.2009.180448. [DOI] [PubMed] [Google Scholar]
- 21.Nettore IC, Ferrara AM, Mirra P, Sibilio A, Pagliara V, Kamoi Kay CS, Lorenzoni PJ, Werneck LC, Bruck I, Coutinho Dos Santos LH, Beguinot F, Salvatore D, Ungaro P, Fenzi G, Scola RH, Macchia PE. Identification and functional characterization of a novel mutation in the NKX2-1 gene: comparison with the data in the literature. Thyroid. 2013;23:675–82. doi: 10.1089/thy.2012.0267. [DOI] [PubMed] [Google Scholar]
- 22.Asmus F, Horber V, Pohlenz J, Schwabe D, Zimprich A, Munz M, Schoning M, Gasser T. A novel TITF-1 mutation causes benign hereditary chorea with response to levodopa. Neurology. 2005;64:1952–4. doi: 10.1212/01.WNL.0000164000.75046.CC. [DOI] [PubMed] [Google Scholar]
- 23.do Carmo Costa M, Costa C, Silva AP, Evangelista P, Santos L, Ferro A, Sequeiros J, Maciel P. Nonsense mutation in TITF1 in a Portuguese family with benign hereditary chorea. Neurogenetics. 2005;6:209–15. doi: 10.1007/s10048-005-0013-1. [DOI] [PubMed] [Google Scholar]
- 24.Provenzano C, Veneziano L, Appleton R, Frontali M, Civitareale D. Functional characterization of a novel mutation in TITF-1 in a patient with benign hereditary chorea. J Neurol Sci. 2008;264:56–62. doi: 10.1016/j.jns.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 25.Glik A, Vuillaume I, Devos D, Inzelberg R. Psychosis, short stature in benign hereditary chorea: a novel thyroid transcription factor-1 mutation. Mov Disord. 2008;23:1744–7. doi: 10.1002/mds.22215. [DOI] [PubMed] [Google Scholar]
- 26.Ferrara AM, De Michele G, Salvatore E, Di Maio L, Zampella E, Capuano S, Del Prete G, Rossi G, Fenzi G, Filla A, Macchia PE. A novel NKX2.1 mutation in a family with hypothyroidism and benign hereditary chorea. Thyroid. 2008;18:1005–9. doi: 10.1089/thy.2008.0085. [DOI] [PubMed] [Google Scholar]
- 27.Maquet E, Costagliola S, Parma J, Christophe-Hobertus C, Oligny LL, Fournet JC, Robitaille Y, Vuissoz JM, Payot A, Laberge S, Vassart G, Van Vliet G, Deladoey J. Lethal respiratory failure and mild primary hypothyroidism in a term girl with a de novo heterozygous mutation in the TITF1/NKX2.1 gene. J Clin Endocrinol Metab. 2009;94:197–203. doi: 10.1210/jc.2008-1402. [DOI] [PubMed] [Google Scholar]
- 28.Gillett ES, Deutsch GH, Bamshad MJ, McAdams RM, Mann PC. Novel NKX2.1 mutation associated with hypothyroidism and lethal respiratory failure in a full-term neonate. J Perinatol. 2013;33:157–60. doi: 10.1038/jp.2012.50. [DOI] [PubMed] [Google Scholar]
- 29.Kleiner-Fisman G, Rogaeva E, Halliday W, Houle S, Kawarai T, Sato C, Medeiros H, St George-Hyslop PH, Lang AE. Benign hereditary chorea: clinical, genetic, and pathological findings. Ann Neurol. 2003;54:244–7. doi: 10.1002/ana.10637. [DOI] [PubMed] [Google Scholar]
- 30.Doyle DA, Gonzalez I, Thomas B, Scavina M. Autosomal dominant transmission of congenital hypothyroidism, neonatal respiratory distress, and ataxia caused by a mutation of NKX2-1. J Pediatr. 2004;145:190–3. doi: 10.1016/j.jpeds.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Asmus F, Devlin A, Munz M, Zimprich A, Gasser T, Chinnery PF. Clinical differentiation of genetically proven benign hereditary chorea and myoclonus-dystonia. Mov Disord. 2007;22:2104–9. doi: 10.1002/mds.21692. [DOI] [PubMed] [Google Scholar]
- 32.Hamvas A, Deterding RR, Wert SE, White FV, Dishop MK, Alfano DN, Halbower AC, Planer B, Stephan MJ, Uchida DA, Williames LD, Rosenfeld JA, Lebel RR, Young LR, Cole FS, Nogee LM. Heterogeneous Pulmonary Phenotypes Associated With Mutations in the Thyroid Transcription Factor Gene NKX2-1. Chest. 2013;144:794–804. doi: 10.1378/chest.12-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salerno T, Peca D, Menchini L, Schiavino A, Petreschi F, Occasi F, Cogo P, Danhaive O, Cutrera R. Respiratory insufficiency in a newborn with congenital hypothyroidism due to a new mutation of TTF-1/NKX2-1 gene. Pediatr Pulmonol. 2013;49:E42–4. doi: 10.1002/ppul.22788. [DOI] [PubMed] [Google Scholar]
- 34.Stockton DW, Das P, Goldenberg M, D’Souza RN, Patel PI. Mutation of PAX9 is associated with oligodontia. Nat Genet. 2000;24:18–19. doi: 10.1038/71634. [DOI] [PubMed] [Google Scholar]
- 35.Devos H, Rodd C, Gagne N, Laframboise R, Van Vliet G. A search for the possible molecular mechanisms of thyroid dysgenesis: sex ratios and associated malformations. J Clin Endocrinol Metab. 1999;84:2502–6. doi: 10.1210/jcem.84.7.5831. [DOI] [PubMed] [Google Scholar]
- 36.Guala A, Falco V, Breedveld G, De Filippi P, Danesino C. Deletion of PAX9 and oligodontia: a third family and review of the literature. Int J Paediatr Dent. 2008;18:441–5. doi: 10.1111/j.1365-263X.2008.00915.x. [DOI] [PubMed] [Google Scholar]
- 37.Silberschmidt D, Rodriguez-Mallon A, Mithboakar P, Cali G, Amendola E, Sanges R, Zannini M, Scarfo M, De Luca P, Nitsch L, Di Lauro R, De Felice M. In vivo role of different domains and of phosphorylation in the transcription factor Nkx2-1. BMC Dev Biol. 2011;11:9. doi: 10.1186/1471-213X-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inzelberg R, Weinberger M, Gak E. Benign hereditary chorea: an update. Parkinsonism Relat Disord. 2011;17:301–7. doi: 10.1016/j.parkreldis.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Dale RC, Grattan-Smith P, Nicholson M, Peters GB. Microdeletions detected using chromosome microarray in children with suspected genetic movement disorders: a single-centre study. Dev Med Child Neurol. 2012;54:618–23. doi: 10.1111/j.1469-8749.2012.04287.x. [DOI] [PubMed] [Google Scholar]
- 40.Esposito G, Fogolari F, Damante G, Formisano S, Tell G, Leonardi A, Di Lauro R, Viglino P. Analysis of the solution structure of the homeodomain of rat thyroid transcription factor 1 by 1H-NMR spectroscopy and restrained molecular mechanics. Eur J Biochem/FEBS. 1996;241:101–13. doi: 10.1111/j.1432-1033.1996.0101t.x. [DOI] [PubMed] [Google Scholar]
- 41.Al Taji E, Biebermann H, Limanova Z, Hnikova O, Zikmund J, Dame C, Gruters A, Lebl J, Krude H. Screening for mutations in transcription factors in a Czech cohort of 170 patients with congenital and early-onset hypothyroidism: identification of a novel PAX8 mutation in dominantly inherited early-onset non-autoimmune hypothyroidism. Eur J Endocrinol. 2007;156:521–9. doi: 10.1530/EJE-06-0709. [DOI] [PubMed] [Google Scholar]
- 42.Dame C, Sola MC, Lim KC, Leach KM, Fandrey J, Ma Y, Knopfle G, Engel JD, Bungert J. Hepatic erythropoietin gene regulation by GATA-4. J Biol Chem. 2004;279:2955–61. doi: 10.1074/jbc.M310404200. [DOI] [PubMed] [Google Scholar]
- 43.Civitareale D, Lonigro R, Sinclair AJ, Di Lauro R. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 1989;8:2537–42. doi: 10.1002/j.1460-2075.1989.tb08391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Sanctis L, Corrias A, Romagnolo D, Di Palma T, Biava A, Borgarello G, Gianino P, Silvestro L, Zannini M, Dianzani I. Familial PAX8 small deletion (c.989_992delACCC) associated with extreme phenotype variability. J Clin Endocrinol Metab. 2004;89:5669–74. doi: 10.1210/jc.2004-0398. [DOI] [PubMed] [Google Scholar]
- 45.Musti AM, Avvedimento EV, Polistina C, Ursini VM, Obici S, Nitsch L, Cocozza S, Di Lauro R. The complete structure of the rat thyroglobulin gene. Proc Natl Acad Sci USA. 1986;83:323–7. doi: 10.1073/pnas.83.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tucker ES, Segall S, Gopalakrishna D, Wu Y, Vernon M, Polleux F, Lamantia AS. Molecular specification and patterning of progenitor cells in the lateral and medial ganglionic eminences. J Neurosci. 2008;28:9504–18. doi: 10.1523/JNEUROSCI.2341-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113:235–48. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- 48.Dessau RB, Pipper CB. “R”–project for statistical computing. Ugeskr Laeger. 2008;170:328–30. [PubMed] [Google Scholar]
- 49.Antonica F, Kasprzyk DF, Opitz R, Iacovino M, Liao XH, Dumitrescu AM, Refetoff S, Peremans K, Manto M, Kyba M, Costagliola S. Generation of functional thyroid from embryonic stem cells. Nature. 2012;491:66–71. doi: 10.1038/nature11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lapi P, Macchia PE, Chiovato L, Biffali E, Moschini L, Larizza D, Baserga M, Pinchera A, Fenzi G, Di Lauro R. Mutations in the gene encoding thyroid transcription factor-1 (TTF-1) are not a frequent cause of congenital hypothyroidism (CH) with thyroid dysgenesis. Thyroid. 1997;7:383–7. doi: 10.1089/thy.1997.7.383. [DOI] [PubMed] [Google Scholar]
- 51.Perna MG, Civitareale D, De Filippis V, Sacco M, Cisternino C, Tassi V. Absence of mutations in the gene encoding thyroid transcription factor-1 (TTF-1) in patients with thyroid dysgenesis. Thyroid. 1997;7:377–81. doi: 10.1089/thy.1997.7.377. [DOI] [PubMed] [Google Scholar]
- 52.Hishinuma A, Kuribayashi T, Kanno Y, Onigata K, Nagashima K, Ieiri T. Sequence analysis of thyroid transcription factor-1 gene reveals absence of mutations in patients with thyroid dysgenesis but presence of polymorphisms in the 5′ flanking region and intron. Endocr J. 1998;45:563–7. doi: 10.1507/endocrj.45.563. [DOI] [PubMed] [Google Scholar]
- 53.Germain ND, Banda EC, Becker S, Naegele JR, Grabel LB. Derivation and isolation of NKX2.1-positive basal forebrain progenitors from human embryonic stem cells. Stem Cells Dev. 2013;22:1477–89. doi: 10.1089/scd.2012.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niquille M, Minocha S, Hornung JP, Rufer N, Valloton D, Kessaris N, Alfonsi F, Vitalis T, Yanagawa Y, Devenoges C, Dayer A, Lebrand C. Two specific populations of GABAergic neurons originating from the medial and the caudal ganglionic eminences aid in proper navigation of callosal axons. Dev Neurobiol. 2013;73:647–72. doi: 10.1002/dneu.22075. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Y, Flandin P, Vogt D, Blood A, Hermesz E, Westphal H, Rubenstein J. Ldb1 is essential for development of Nkx2.1 lineage derived GABAergic and cholinergic neurons in the telencephalon. Dev Biol. 2013;385:94–106. doi: 10.1016/j.ydbio.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woods JO, Singh-Blom UM, Laurent JM, McGary KL, Marcotte EM. Prediction of gene-phenotype associations in humans, mice, and plants using phenologs. BMC Bioinformatics. 2013;14:203. doi: 10.1186/1471-2105-14-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olivieri A, Stazi MA, Mastroiacovo P, Fazzini C, Medda E, Spagnolo A, De Angelis S, Grandolfo ME, Taruscio D, Cordeddu V, Sorcini M. A population-based study on the frequency of additional congenital malformations in infants with congenital hypothyroidism: data from the Italian Registry for Congenital Hypothyroidism (1991–1998) J Clin Endocrinol Metab. 2002;87:557–62. doi: 10.1210/jcem.87.2.8235. [DOI] [PubMed] [Google Scholar]
- 58.van Tuyl M, Blommaart PE, de Boer PA, Wert SE, Ruijter JM, Islam S, Schnitzer J, Ellison AR, Tibboel D, Moorman AF, Lamers WH. Prenatal exposure to thyroid hormone is necessary for normal postnatal development of murine heart and lungs. Dev Biol. 2004;272:104–17. doi: 10.1016/j.ydbio.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 59.Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, Loughna S, Robinson TE, Dearlove AM, Ribas G, Bonser AJ, Thomas NR, Scotter AJ, Caves LS, Tyrrell GP, Newbury-Ecob RA, Munnich A, Bonnet D, Brook JD. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet. 2005;37:423–8. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- 60.Ngan ES, Lang BH, Liu T, Shum CK, So MT, Lau DK, Leon TY, Cherny SS, Tsai SY, Lo CY, Khoo US, Tam PK, Garcia-Barcelo MM. A germline mutation (A339 V) in thyroid transcription factor-1 (TITF-1/NKX2.1) in patients with multinodular goiter and papillary thyroid carcinoma. J Natl Cancer Inst. 2009;101:162–75. doi: 10.1093/jnci/djn471. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Rath N, Hannenhalli S, Wang Z, Cappola T, Kimura S, Atochina-Vasserman E, Lu MM, Beers MF, Morrisey EE. GATA and Nkx factors synergistically regulate tissue-specific gene expression and development in vivo. Development. 2007;134:189–98. doi: 10.1242/dev.02720. [DOI] [PubMed] [Google Scholar]
- 62.Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, Hannenhalli S, Epstein JA. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell. 2010;19:450–9. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brewer AC, Alexandrovich A, Mjaatvedt CH, Shah AM, Patient RK, Pizzey JA. GATA factors lie upstream of Nkx 2.5 in the transcriptional regulatory cascade that effects cardiogenesis. Stem Cells Dev. 2005;14:425–39. doi: 10.1089/scd.2005.14.425. [DOI] [PubMed] [Google Scholar]
- 64.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba II, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–8. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, Vuerhard M, Buijs-Gladdines J, Kooi C, Klous P, van Vlierberghe P, Ferrando AA, Cayuela JM, Verhaaf B, Beverloo HB, Horstmann M, de Haas V, Wiekmeijer AS, Pike-Overzet K, Staal FJ, de Laat W, Soulier J, Sigaux F, Meijerink JP. Integrated Transcript and Genome Analyses Reveal NKX2-1 and MEF2C as Potential Oncogenes in T Cell Acute Lymphoblastic Leukemia. Cancer Cell. 2011;19:484–97. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 67.Fabbro D, Di Loreto C, Beltrami CA, Belfiore A, Di Lauro R, Damante G. Expression of thyroid-specific transcription factors TTF-1 and PAX-8 in human thyroid neoplasms. Cancer Res. 1994;54:4744–9. [PubMed] [Google Scholar]
- 68.Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, Magnusdottir DN, Matthiasdottir S, Stacey SN, Skarphedinsson OB, Helgadottir H, Li W, Nagy R, Aguillo E, Faure E, Prats E, Saez B, Martinez M, Eyjolfsson GI, Bjornsdottir US, Holm H, Kristjansson K, Frigge ML, Kristvinsson H, Gulcher JR, Jonsson T, Rafnar T, Hjartarsson H, Mayordomo JI, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Kong A, Stefansson K. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41:460–4. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsuse M, Mitsutake N, Tanimura S, Ogi T, Nishihara E, Hirokawa M, Fuziwara CS, Saenko VA, Suzuki K, Miyauchi A, Yamashita S. Functional characterization of the novel BRAF complex mutation, BRAF(V600delinsYM), identified in papillary thyroid carcinoma. Int J Cancer. 2013;132:738–43. doi: 10.1002/ijc.27709. [DOI] [PubMed] [Google Scholar]
- 70.Fernandez M, Raskind W, Matsushita M, Wolff J, Lipe H, Bird T. Hereditary benign chorea: clinical and genetic features of a distinct disease. Neurology. 2001;57:106–10. doi: 10.1212/wnl.57.1.106. [DOI] [PubMed] [Google Scholar]
- 71.Kleiner-Fisman G, Lang AE. Benign hereditary chorea revisited: A journey to understanding. Mov Disord. 2007;22:2297–305. doi: 10.1002/mds.21644. [DOI] [PubMed] [Google Scholar]
- 72.Minoo P, Hu L, Xing Y, Zhu NL, Chen H, Li M, Borok Z, Li C. Physical and functional interactions between homeodomain NKX2.1 and winged helix/forkhead FOXA1 in lung epithelial cells. Mol Cell Biol. 2007;27:2155–65. doi: 10.1128/MCB.01133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ku MS, Sun HL, Lu KH, Sheu JN, Lee HS, Yang SF, Lue KH. The CC16 A38G polymorphism is associated with the development of asthma in children with allergic rhinitis. Clin Exp Allergy. 2011;41:794–800. doi: 10.1111/j.1365-2222.2010.03679.x. [DOI] [PubMed] [Google Scholar]
- 74.Puthothu B, Krueger M, Heinze J, Forster J, Heinzmann A. Haplotypes of surfactant protein C are associated with common paediatric lung diseases. Pediatr Allergy Immunol. 2006;17:572–7. doi: 10.1111/j.1399-3038.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- 75.Hamvas A, Deterding RR, Wert SE, White FV, Dishop MK, Alfano DN, Halbower AC, Planer B, Stephan MJ, Uchida DA, Williames LD, Rosenfeld JA, Lebel RR, Young LR, Cole FS, Nogee LM. Heterogeneous pulmonary phenotypes associated with mutations in the thyroid transcription factor gene NKX2-1. Chest. 2013;144:794–804. doi: 10.1378/chest.12-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin Z, Gonzales L, Kolla V, Rath N, Zhang Y, Lu MM, Kimura S, Ballard PL, Beers MF, Epstein JA, Morrisey EE. Hop functions downstream of Nkx2.1 and GATA6 to mediate HDAC-dependent negative regulation of pulmonary gene expression. Am J Physiol Lung Cell Mol Physiol. 2006;291:L191–9. doi: 10.1152/ajplung.00385.2005. [DOI] [PubMed] [Google Scholar]
- 77.Barnett CP, Mencel JJ, Gecz J, Waters W, Kirwin SM, Vinette KM, Uppill M, Nicholl J. Choreoathetosis, congenital hypothyroidism and neonatal respiratory distress syndrome with intact NKX2-1. Am J Med Genet A. 2012;158A:3168–73. doi: 10.1002/ajmg.a.35456. [DOI] [PubMed] [Google Scholar]
- 78.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 79.Porcu E, Medici M, Pistis G, Volpato CB, Wilson SG, Cappola AR, Bos SD, Deelen J, den Heijer M, Freathy RM, Lahti J, Liu C, Lopez LM, Nolte IM, O’Connell JR, Tanaka T, Trompet S, Arnold A, Bandinelli S, Beekman M, Bohringer S, Brown SJ, Buckley BM, Camaschella C, de Craen AJ, Davies G, de Visser MC, Ford I, Forsen T, Frayling TM, Fugazzola L, Gogele M, Hattersley AT, Hermus AR, Hofman A, Houwing-Duistermaat JJ, Jensen RA, Kajantie E, Kloppenburg M, Lim EM, Masciullo C, Mariotti S, Minelli C, Mitchell BD, Nagaraja R, Netea-Maier RT, Palotie A, Persani L, Piras MG, Psaty BM, Raikkonen K, Richards JB, Rivadeneira F, Sala C, Sabra MM, Sattar N, Shields BM, Soranzo N, Starr JM, Stott DJ, Sweep FC, Usala G, van der Klauw MM, van Heemst D, van Mullem A, Vermeulen SH, Visser WE, Walsh JP, Westendorp RG, Widen E, Zhai G, Cucca F, Deary IJ, Eriksson JG, Ferrucci L, Fox CS, Jukema JW, Kiemeney LA, Pramstaller PP, Schlessinger D, Shuldiner AR, Slagboom EP, Uitterlinden AG, Vaidya B, Visser TJ, Wolffenbuttel BH, Meulenbelt I, Rotter JI, Spector TD, Hicks AA, Toniolo D, Sanna S, Peeters RP, Naitza S. A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS genetics. 2013;9:e1003266. doi: 10.1371/journal.pgen.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P, Kuhn RM, Zhu J, Smirnov I, Kent WJ, Haussler D, Madden PA, Costello JF, Wang T. The Human Epigenome Browser at Washington University. Nat Methods. 2011;8:989–90. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, Thurman RE, Kaul R, Myers RM, Stamatoyannopoulos JA. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–8. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, Wei CL, Ruan Y, Sung WK. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santagati F, Abe K, Schmidt V, Schmitt-John T, Suzuki M, Yamamura K, Imai K. Identification of Cis-regulatory elements in the mouse Pax9/Nkx2-9 genomic region: implication for evolutionary conserved synteny. Genetics. 2003;165:235–42. doi: 10.1093/genetics/165.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–15. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hamdan H, Liu H, Li C, Jones C, Lee M, deLemos R, Minoo P. Structure of the human Nkx2.1 gene. Biochim Biophys Acta. 1998;1396:336–48. doi: 10.1016/s0167-4781(97)00210-8. [DOI] [PubMed] [Google Scholar]
- 87.Ikeda K, Clark JC, Shaw-White JR, Stahlman MT, Boutell CJ, Whitsett JA. Gene structure and expression of human thyroid transcription factor-1 in respiratory epithelial cells. J Biol Chem. 1995;270:8108–14. doi: 10.1074/jbc.270.14.8108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


