Abstract
Overexpression of the multiple myeloma set domain (MMSET) Wolf-Hirschhorn syndrome candidate 1 gene, which contains an orphan box H/ACA class small nucleolar RNA, ACA11, in an intron, is associated with several cancer types, including multiple myeloma (MM). ACA11 and MMSET are overexpressed cotranscriptionally as a result of the t(4;14) chromosomal translocation in a subset of patients with MM. RNA sequencing of CD138+ tumor cells from t(4;14)-positive and -negative MM patient bone marrow samples revealed an enhanced oxidative phosphorylation mRNA signature. Supporting these data, ACA11 overexpression in a t(4;14)-negative MM cell line, MM1.S, demonstrated enhanced reactive oxygen species (ROS) levels. In addition, an enhancement of cell proliferation, increased soft agar colony size, and elevated ERK1/2 phosphorylation were observed. This ACA11-driven hyperproliferative phenotype depended on increased ROS levels as exogenously added antioxidants attenuate the increased proliferation. A major transcriptional regulator of the cellular antioxidant response, nuclear factor (erythroid-derived 2)-like 2 (NRF2), shuttled to the nucleus, as expected, in response to ACA11-driven increases in ROS; however, transcriptional up-regulation of some of NRF2’s antioxidant target genes was abrogated in the presence of ACA11 overexpression. These data show for the first time that ACA11 promotes proliferation through inhibition of NRF2 function resulting in sustained ROS levels driving cancer cell proliferation.—Mahajan, N., Wu, H.-J., Bennett, R. L., Troche, C., Licht, J. D., Weber, J. D., Maggi, L. B., Jr., Tomasson, M. H. Sabotaging of the oxidative stress response by an oncogenic noncoding RNA.
Keywords: ACA11, multiple myeloma, NRF2, ROS, snoRNA
Multiple myeloma (MM) is a malignancy of bone marrow plasma B cells (1), and despite extensive study, its etiology remains unclear. The t(4;14)(p16.3:q32.3) chromosomal translocation is detected in ∼20% of MM cases and is associated with a worse prognosis (1, 2). The t(4;14) translocation transposes the immunoglobulin heavy chain region enhancer on chromosome 4 and the Wolf-Hirschhorn syndrome candidate 1 (WHSC1), multiple myeloma set domain (MMSET) gene on chromosome 14 (2), resulting in the ectopic overexpression of the WHSC1 and FGFR3 genes. Where FGFR3 is deleted in a subset of t(4;14) cases that still have a poor prognosis (3), all cases of t(4;14) myeloma overexpress the MMSET gene, which encodes a H3K36 histone methyltransferase. MMSET knockdown causes cell death in t(4;14)-positive myeloma cell lines (4), but overexpression of MMSET cDNA fails to reliably promote cell growth or transform cells in culture or in transgenic mice (5). These data suggest that there is more to the t(4;14) translocation than simply overexpression of MMSET.
Our group discovered that ACA11 (scaRNA23), an orphan box H/ACA class small nucleolar RNA (snoRNA) encoded within the 18–19 intron of the WHSC1 gene, is cotranscriptionally overexpressed in t(4;14)-positive MM cells and may contribute to cellular transformation (5). snoRNAs are noncoding RNAs that facilitate the pseudouridylation (H/ACA class) or methylation (C/D class) of target RNAs. Additional functions have been described for snoRNAs, including controlling genome organization, gene expression, and epigenetic processes (6–9). Both deletion and up-regulation of snoRNAs have been implicated in cancer (10–12). ACA11 expression promotes cell proliferation and binds a set of proteins distinct from the canonical dyskerin complex (5). However, the mechanism behind ACA11-driven cell proliferation has not been elucidated.
Here we confirmed that ACA11 overexpression drives cellular proliferation in MM cell lines as well as primary mouse splenic B cells. RNA sequencing (RNA-seq) analysis performed with RNA from bone marrow myeloma cells isolated from patients with or without the t(4;14) translocation, combined with pathway analysis of the data, revealed significant dysregulation of genes involved in the cellular oxidative phosphorylation response. Overexpression of ACA11 in a t(4;14)-negative MM cell line, MM1.S, resulted in increased levels of cellular reactive oxygen species (ROS) and correlated with increased ERK1/2 phosphorylation, an indicator of increased oxidative levels in a cell (13). Nuclear factor (erythroid-derived 2)-like 2 (NRF2) or NFE2L2, a master regulator of the ROS stress response (14), translocated to the nucleus, as expected, in response to ACA11-induced increases in ROS. However, ACA11 overexpression in MM1.S cells suppressed the transcriptional activity of NRF2 resulting in a failure of antioxidant target genes to be normally up-regulated. These results support a role for ACA11 as an oncogenic snoRNA activated in t(4;14)-positive myeloma and identify a molecular pathway by which ACA11 contributes to cell proliferation.
MATERIALS AND METHODS
Chemicals, reagents, and antibodies
Chemicals, reagents, and antibodies included the following: anti-NRF2 (1:2000; Abcam, Cambridge, MA, USA); anti-Keap1 (1:2000; Abcam); anti-HDAC2 (1:2000; Cell Signaling Technology, Danvers, MA, USA); anti–heat shock protein (HSP; 1:5000; Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-actin (1:5000; Sigma-Aldrich, St. Louis, MO, USA); anti-phospho-ERK1/2 Kit (Cell Signaling Technology); and antimycin A (Sigma-Aldrich).
RNA-seq analysis and bioinformatics
Patient samples were obtained under an institutional review board–approved protocol. Informed consent from MM patients was obtained in accordance with the Declaration of Helsinki. Total RNA was extracted from CD138+ cells isolated from bone marrow aspirate (tumor) and submitted to the Genome Technology Access Center of the Washington University School of Medicine (St. Louis, MO, USA) for RNA-seq analysis. The raw sequence reads were mapped against the hg19 version of the human reference genome using Tophat2 with default parameters (15). HTSeq was then used to obtain the read counts for each gene or transcript by using the hg19 version of RefSeq annotation (23,836 genes in total) (16). The size factor of each sample was estimated by the DESeq2 software package (17) and was used to generate normalized read counts for each gene or transcript following the DESeq2 pipeline. Differentially expressed genes between t(4;14)-positive and -negative samples were identified by DESeq2 with a false-discovery rate <0.01 and fold change >1.5. Multiple comparisons were corrected by using the method of Benjamini and Hochberg (18). Pathway analysis was performed by MetaCore software (Thomas Reuters, Rochester, NY, USA).
Cell culture and plasmid constructs
Myeloma cell line MM1.S [a t(4;14)-negative cell line], H929 [t(4;14)-positive cells], and Arf−/− (alternative reading frame protein of the CDKN2A locus) mouse embryo fibroblasts (MEFs) were cultured as described previously (5). BJ human foreskin fibroblasts were obtained from S. Stewart (Washington University, St. Louis, MO, USA) and were cultured in minimum essential medium with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. All cultures were routinely screened for Mycoplasma and were found to be free of contamination. MM1.S, H929, and BJ human foreskin fibroblast cell lines were originally purchased from American Type Culture Collection (Manassas, VA, USA) and used within 10 to 15 passages of thaw of the original vial. Arf−/− MEFs were used within 10 to 15 passages from isolation. ACA11 was cloned in pLKO.1-puro (Addgene, Cambridge, MA, USA) using AgeI and EcoRI restriction sites. Chemically modified antisense oligonucleotides were used for ACA11 knockdown experiments as previously described (5).
Virus production and infections
For the production of lentivirus, 293T cells were transfected using the calcium phosphate method with pCMV-VSV-G, pCMV-ΔR8.9, and pLKO.1-puro constructs. Viral supernatants were collected 48 h after transfection. Cells were acutely infected with high-titer lentivirus for 12 to 16 h in the presence of 10 μg/ml of polybrene. Puromycin was added to a small aliquot of cells at a concentration of 2 μg/ml; 90% of the cells survived the puromycin selection.
Isolation of mouse splenic B cells
Experiments including animals were performed according to the guidelines established by the Animal Studies Committee at Washington University. Spleen was isolated from mouse; splenocytes were obtained after treating with red blood cell lysing buffer (Sigma-Aldrich), and B cells were isolated by negative selection with CD43 magnetic beads (Mouse B Cell Isolation Kit Miltenyi Biotec, San Diego, CA, USA) with an AutoMacs Pro Separator according to the manufacturer’s instructions. Splenic B cells were cultured as previously described (19) with 10 ng/ml of IL-4 (R&D Systems, Minneapolis, MN, USA) and 10 μg/ml LPS (Sigma-Aldrich) in B cell medium [RPMI 1640 with l-glutamine (Mediatech, Tewksbury, MA, USA), 1% HEPES, 1% penicillin/streptomycin/amphotericin B, and 10% FBS (HyClone Laboratories, Logan, UT, USA)] for the indicated times.
RNA isolation and real-time quantitative PCR
Total RNA was extracted using the RNeasy Plus kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. cDNA was made according to the protocol of the iScript Reverse transcription Supermix for real-time quantitative (qPCR) (Bio-Rad, Hercules, CA, USA), and iQ SYBER Green Supermix (Bio-Rad) was used for amplification. The ACA11 primers have been previously described (5). The primers used were as follows: NRF2 forward: 5′–AGGTTGCCCACATTCCCAAA–3′ and reverse: 5′–AGTGACTGAAACGTAGCCGA-3′; KEAP1 forward: 5′- CGCTCCCCAACCGACAAC–3′ and reverse: 5′–GATAAGCAACACCACCACCTC–3′; HMXO1 forward: 5′–TCCTGGCTCAGACTCAAATG–3′ and reverse: 5′–CACGCATGGCTCAAAAACCA–3′; TXN1 forward: 5′–GTAGATGTGGATGAGTGTCAGGA–3′ and reverse: 5′–ATTCACCCACCTTTTGTCCCT–3′. Fold change was measured using the ΔΔCt method (20).
Quantification of ROS
Fluorogenic substrate carboxy-2′,7′-dicholorodi hydrofluorescein diacetate (carboxy-DCFH-DA; Molecular Probes, Eugene, OR, USA) was used to detect intracellular ROS. Briefly, 2.5 × 104 cells were plated in 96-well plate, 6 wells for each treatment, followed by incubation with 20 μM carboxy-DCFH-DA at 37°C for 30 min in serum-free medium. Cells that were either untreated or treated with different ROS inducers at the indicated final concentrations were incubated at 37°C for the indicated time periods. The dose and time were empirically determined for the various ROS inducers. At the end of the exposure period, cells were washed with PBS (containing 0.5 mM MgCl2 and 0.92 mM CaCl2) twice, and plate was read at 488/530 (emission/excitation) in a plate reader (BioTek Instruments, Winooski, VT, USA). Similarly, MitoSox (Molecular Probes) was used to measure cellular superoxide levels and pates were read at 510/595 (emission/excitation).
Measurement of glutathione/glutathione disulfide ratios
A luminescence-based system was used for detection and quantification of glutathione (GSH)/glutathione disulfide (GSSG) ratios in cultured cells according to manufacturer’s instructions (Promega, Madison, WI, USA).
Western blot analysis
Cell lysates were prepared in radioimmunoprecipitation assay buffer with protease and phosphatase cocktail inhibitor (Thermo Fisher Scientific, Waltham, MA, USA). Cellular debris was cleared from lysates by centrifugation, and the protein concentration was determined by the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL, USA). Samples were separated on precast 4–15% Tris-glycine extended–PAGE (Bio-Rad), transferred to a PVDF membrane (Bio-Rad), blotted with the indicated antibodies, and visualized with enhanced chemiluminescence substrate (Pierce) (21).
Proliferation assay
Cells were seeded in triplicate on 6-well plates the day after the infection. Cell proliferation over time was measured by counting the number of cells by trypan blue exclusion at the indicated time points (22). N-acetyl cysteine (NAC) (Sigma-Aldrich) was freshly added every 48 h to the culture medium at 1 mM final concentration or as indicated.
Bromodeoxyuridine assay
A commercially available kit was used to determine the bromodeoxyuridine (BrdU) incorporation as per the manufacturer’s protocol (Cell Signaling Technology).
Soft agar colony-forming assay
MM.1S cells were infected with control vector or ACA11 virus as described above. Cells (1 × 105 per condition) were plated in triplicate in 60 mm dishes as previously described (22) with the following change: we used high-glucose RPMI 1640 medium (Thermo Fisher Scientific) containing 0.3% Noble agar (Becton Dickinson, San Diego, CA, USA) supplemented with 10% FBS (Thermo Fisher) and 1× penicillin–streptomycin (Thermo Fisher Scientific). Cells were grown at 37°C in 5% CO2 for 21 d and fed with high-glucose RPMI 1640 medium containing 0.3% Noble agar supplemented with 10% FBS and 1× penicillin–streptomycin every 7 d. At the end of 21 d, colonies were counted and representative photographs taken. For counting, the plates were divided into quadrants, and 1 to ×10 visual fields were randomly selected from each quadrant. All colonies through all z planes within the selected ×10 field were counted and averaged for each of 3 plates.
Cytosolic and nuclear fractionation
The Ne-Per kit (Pierce) was used to isolate the cytosolic and nuclear fractions from cells according to the manufacturer’s protocol. For Western blot analysis, 10 μg of total protein per lane was loaded. HSP90 and HDAC2 were used as loading controls for cytosolic and nuclear fractions, respectively.
Nuclear protein extraction and NRF2 binding activity assay
Nuclei were isolated from the cells using the Nuclear Complex Co-Ip kit (Active Motif, Carlsbad, CA, USA). Protein concentrations in samples were equalized using a BCA calorimetric assay (Bio-Rad). Nuclear protein (10 μg) was used to determine the NRF2 binding activity using the TransAM NRF2 ELISA kit (Active Motif) according to the manufacturer’s guidelines.
Chromatin immunoprecipitation real-time qPCR
Ten million MM1.S cells were cross-linked with 1% formaldehyde at room temperature in 40 ml medium for 10 min. Cross-linking was stopped by the addition of glycine to 120 mM for 5 min. Cells were washed twice with PBS and stored frozen until used for the chromatin immunoprecipitation (ChIP) assay. ChIP was performed using the Chromatin Immunoprecipitation Assay kit from EMD Millipore (Billerica, MA, USA). Briefly, cells were resuspended in SDS lysis buffer and sonicated on ice with 30 cycles of 20 s at 45% power with 1 min between cycles using a Thermo Fisher FB50 sonicator. An Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) was used to evaluate the chromatin shearing efficiency and to confirm that the fragment size was between 200 and 700 bp. Ten percent of each sample was kept as input, while the remainder was diluted 10-fold in ChIP dilution buffer and immunoprecipitated overnight at 4°C using an antibody to NRF2 (Abcam) or rabbit IgG (Santa Cruz Biotechnology). Samples were then washed with 2 washes each of ChIP dilution buffer, low-salt buffer, high-salt buffer, LiCl buffer, and Tris-EDTA buffer. Samples were eluted in freshly prepared 1% SDS, 100 mM NaHCO3 buffer. Cross-links were reversed by a 65°C overnight incubation in the presence of 0.3 M NaCl. DNA was purified by treatment with 1 μg RNase A and 1 μg proteinase K, and the DNA was cleaned up using a Qiagen PCR Purification Kit.
Immunoprecipitated DNAs were amplified by real-time qPCR using the Bio-Rad CFX96 instrument. The reaction mix contained 4 μl DNA, 6 μl SSO Advanced Universal SYBR Green Supermix (Bio-Rad), and 0.6 μl (500 nM final) primers. Primer sequences are as follows: TXN1 forward: 5′–GGAAAAGCTTGAAGCCACCAT–3′, reverse: 5′–GCAGATGGCAACTGGTTATGT–3′; and HMOX1 forward: 5′–CACGGTCCCGAGGTCTATT–3′, reverse: 5′–TAGACCGTGACTCAGCGAAA–3′. Reaction conditions began with a 30 s 95°C hot start, followed by 10 s hybridization and 30 s amplification at 60°C. Samples were analyzed in duplicate, and results from 3 independent experiments are presented as percentage input (percentage input method for ChIP analysis; Thermo Fisher Scientific) and expressed as means ± sem.
Statistical analysis
Quantification of Western blots was performed by ImageJ software (National Institutes of Health, Bethesda, MD, USA). Statistical analysis (2-tailed Student’s t test or ANOVA) was based on a minimum of 3 replicates using Prism 5 for Mac statistical software (GraphPad Software, La Jolla, CA, USA). The differences were considered significant if P < 0.05.
RESULTS
ACA11 overexpression increases cellular proliferation
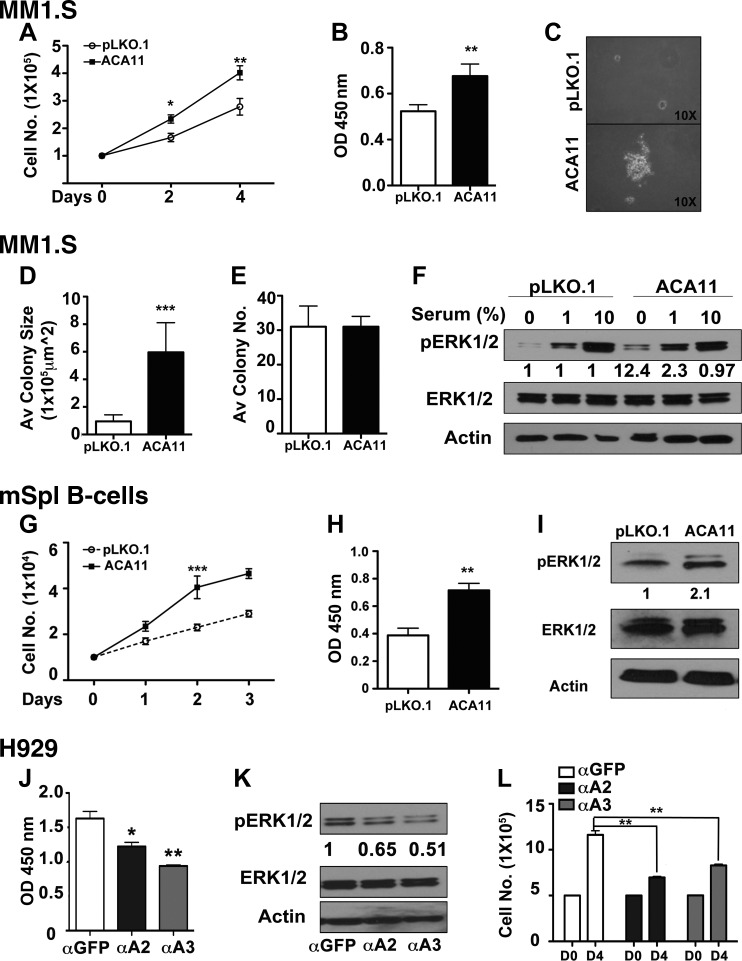
Our previous work had demonstrated that the ACA11 snoRNA is overexpressed in human cancers, including t(4;14)-positive MM, and that it can drive increased proliferation in MM cell lines (5). Given this correlation with increased proliferation, we wanted to understand the potential mechanism of ACA11-driven increases in proliferation. ACA11 increased the growth rate of MM1.S cells as measured by cell-counting assays (Fig. 1A), which do not harbor the t(4;14) translocation and which express relatively low levels of ACA11 (5) (Supplemental Fig. S1A). In addition, ACA11 overexpression drove MM1.S cells into S phase at a faster rate, as demonstrated by incorporation of a 2-h pulse of BrdU into replicating DNA (Fig. 1B). The increase in proliferation coincided with increases in anchorage-independent growth. While the number of colonies grown in soft agar did not change, ACA11 overexpression in MM1.S cells greatly increased the size of the soft agar colonies (Fig. 1C–E). This increase in proliferation was not limited to just MM1.S cells. Primary splenic B cells also demonstrated an increased growth rate and S-phase entry on ACA11 overexpression (Fig. 1G, H). The increased levels of cellular proliferation were correlated with an increased level of ERK1/2 phosphorylation. In MM1.S cells overexpressing ACA11, the basal levels of ERK1/2 phosphorylation on serum starvation were elevated (Fig. 1F). In addition, as serum was added back to the culture, ACA11 increased ERK1/2 phosphorylation at 1%, but the difference was abrogated at standard 10% serum. Correlating with the MM1.S cells, primary murine splenic B cells overexpressing ACA11 also had increased levels of ERK1/2 phosphorylation (Fig. 1I). While the phosphorylation levels of JNK and p38 MAP kinases were not altered by ACA11 overexpression in any cell types tested (data not shown), inhibition of ERK1/2 phosphorylation with PD98059 (an inhibitor of MEK the upstream kinase that phosphorylated ERK1/2; Supplemental Fig. S1B) blocked ACA11-driven increases in MM1.S cell proliferation (Supplemental Fig. S1C).
Figure 1.
ACA11 overexpression increases cell proliferation and activates ERK1/2. A) ACA11 increases proliferation of MM (MM1S) cells by cell-counting assay. B) Comparison of cellular proliferation pattern as determined by BrdU assay. C) Representative images of soft agar colony-forming assay. D, E) Average (Av) colony size is significantly increased by ACA11 (D), while total colony number (no.) is not changed (E). F) Comparison of pERK1/2 activation in vector (pLKO.1)- and ACA11-overexpressing cells when cultured in presence of reduced serum concentrations. Fold increases are indicated compared to pLKO.1 controls by densitometry of respective bands. Actin is shown as loading control for total ERK levels. G) Cell counts of primary murine B cells overexpressing ACA11 or control retrovirus (pLKO.1). H) Increased cellular proliferation as determined by BrdU incorporation. I) Comparison of pERK1/2 levels in vector (pLKO.1)- and ACA11-overexpressing primary murine B cells. J) Knockdown of ACA11 in H929 MM cells by antisense oligonucleotides (αA2, αA3) reduces BRDU incorporation [α green fluorescent protein (αGFP), antisense oligonucleotide to GFP]. K) Antisense knockdown of ACA11 (αA2, αA3) reduces pERK1/2 levels in H929 cells grown in 1% serum. Fold decreases indicated compared to αGFP control. Actin is shown as loading control for total ERK levels. L) Cell proliferation is also inhibited by ACA11 knockdown in ACA11-overexpressing H929 cells. All data points represent means ± sd (n = 3). Representative figures are shown of experiments completed at least 3 times in triplicate. Asterisks indicate significance according to Student’s t test (2-tailed) or 1-way ANOVA with Bonferroni posttest as required. *P < 0.05, **P < 0.01, ***P < 0.001 compared to respective control cells.
These results were corroborated by complementary knockdown experiments in the t(4;14)-positive cell line, H929, which expresses high levels of ACA11 (5). On knockdown of ACA11 in H929 cells (Supplemental Fig. S1M), BrdU incorporation, phospho-ERK1/2 levels, and growth rate all decreased (Fig. 1J, K, respectively). Taken together, these data demonstrate that ACA11 can drive cellular growth and proliferation, which correlates with increased phosphorylation of the progrowth kinase ERK1/2.
ACA11-driven proliferation is dependent on ROS levels
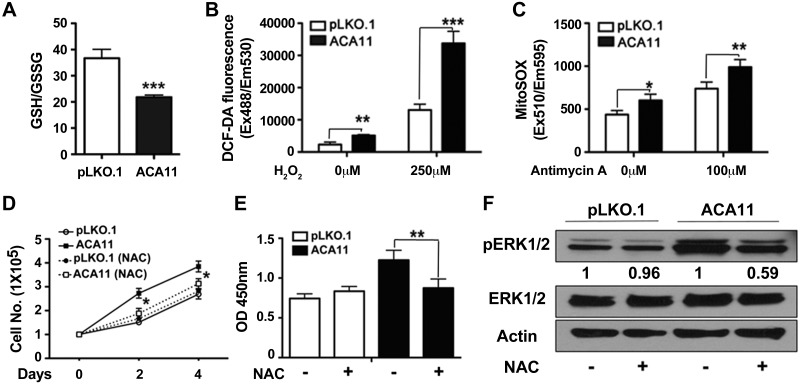
Given the increased proliferation observed on ACA11 overexpression, we next wanted to determine what cellular pathways are altered with ACA11 overexpression. To assay this question in an unbiased fashion, RNA-seq analysis was performed on t(4;14)-positive and t(4;14)-negative MM patient bone marrow samples (Supplemental Fig. S2), which demonstrated an increased oxidative phosphorylation gene signature by MetaCore pathway analysis (Table 1). This correlated with the altered ERK1/2 phosphorylation-driven increase in cellular proliferation observed in our MM cell line data (Fig. 1), as ERK1/2 phosphorylation can be altered by changes in ROS levels in cells (13). One general measure of the oxidative state of a cell is the ratio of reduced (GSH) to oxidized (GSSG) glutathione, with 90% of glutathione existing in the reduced state (GSH) in normal, healthy tissue (23). Cells overexpressing ACA11 had a lower steady-state ratio of GSH/GSSG than vector control cells, indicating an increased oxidative state in cells expressing high ACA11 levels (Fig. 2A).
TABLE 1.
Gene expression signatures defined by RNA-seq analysis in t(4;14) MM samples
| Gene ontology process | Gene | P |
|---|---|---|
| Oxidative phosphorylation | NDUFA8, ATP5D, NDUFS7, NDUFA11, COX6C, ATP5J, DAP13 | 2.768E-08 |
| Development—MAG-dependent inhibition of neurite outgrowth | MYH15, MYL6, BDNF | 6.532E-07 |
| Immune response | CD59, CLU, C3 | 1.005E-06 |
| Cytoskeleton remodeling | MYH15, MYL6, RHOB | 5.358E-06 |
| Airway smooth muscle contraction in asthma | MYH15, MYL6, GNAS | 1.402E-04 |
| Regulation of CFTR activity (normal and CF) | GNAS, S100A10, PDE4D, PPP2R5E | 2.678E-04 |
| Transport—role of AVP in regulation of aquaporin 2 and renal water resorption | MYH15, MYL6, GNAS, RYR1 | 5.312E-04 |
MetaCore pathway analysis of significantly altered genes in the data set obtained after RNA-seq analysis of t(4:14) negative (n = 6) and positive (n = 7) MM patient samples. Underlined genes are down-regulated in RNA-seq; genes not underlined are up-regulated. MAG, myelin-associated glycoprotein; CFTR, cystic fibrosis transmembrane conductance regulator; CF, cystic fibrosis.
Figure 2.
Proliferation induced by ACA11 is mediated by increased levels of ROS. A) Ratio of reduced GSH to oxidized GSSG. ACA11 overexpression reduces GSH/GSSG ratio, indicating increased oxidative stress. B) Total intracellular level of ROS at basal state and in response to H2O2 (250 μM) for 1 h were measured by using carboxy-DCFH-DA as fluorogenic substrate in MM1.S cells. C) Levels of mitochondrial superoxide at basal state and in response to antimycin (100 μM) for 1 h were measured by using MitoSOX fluorogenic substrate in MM1.S cells. D, E) Treatment with antioxidant NAC significantly decreased proliferation of ACA11 cells, as determined by cell-counting assay (D) BrdU incorporation (E). F) Comparison of pERK1/2 levels in vector (pLKO.1)- and ACA11-overexpressing MM1.S cells cultured in 1% serum in absence and presence of NAC (1 mM) for 24 h. Actin is shown as loading control for total ERK levels. Representative Western blots are shown. All data points represent means ± sd (n = 3). Representative figures are shown of experiments completed at least 3 times in triplicate. Asterisks indicate significance according to Student’s t test (2-tailed) or 1-way ANOVA with Bonferroni posttest as required. *P < 0.05, **P < 0.01, ***P < 0.001 compared to respective control cells.
We next examined the levels of ROS in MM1.S cells with and without ACA11 overexpression. Measuring ROS levels with a DCF-DA (2′,7′-dichlorofluorescin diacetate) fluorescence–based assay, MM1.S cells overexpressing ACA11 had higher basal levels of cellular ROS (Fig. 2B). In addition, the basal mitochondrial ROS levels were increased on ACA11 overexpression in MM1.S cells, as measured by a MitoSOX fluorescence–based assay (Fig. 2C). Both basal cellular (Fig. 2B) and mitochondrial (Fig. 2C) ROS levels were further enhanced on ROS challenge with H2O2 with ACA11 overexpression. Baseline and challenged cellular and mitochondrial ROS levels were increased in MEFs, human BJ human foreskin fibroblasts, and primary mouse B cells overexpressing ACA11 (Supplemental Fig. S1D–L). In addition, knockdown of ACA11 with antisense oligonucleotides in H929 cells, which have high levels of ACA11, decreased basal and challenged cellular and mitochondrial ROS levels (Supplemental Fig. S1M–O).
To test whether the increased cellular proliferation observed with ACA11 overexpression required increased ROS, MM1.S cells were treated with the antioxidant NAC. As expected, ACA11 overexpression increased cellular proliferation rates (Fig. 2D). However, treatment of MM1.S cells overexpressing ACA11 with NAC returned proliferation rates to vector (pLKO.1) levels (Fig. 2D). In addition, NAC treatment of MM1.S cells attenuated ACA11-driven increases in BrdU incorporation (Fig. 2E) and phopho-ERK1/2 levels (Fig. 2F). Taken together, these data demonstrate that the increase in cell proliferation caused by ACA11 overexpression is mediated by increased ROS levels.
ACA11-driven increases in ROS are driven by an inhibition of NRF2 activity
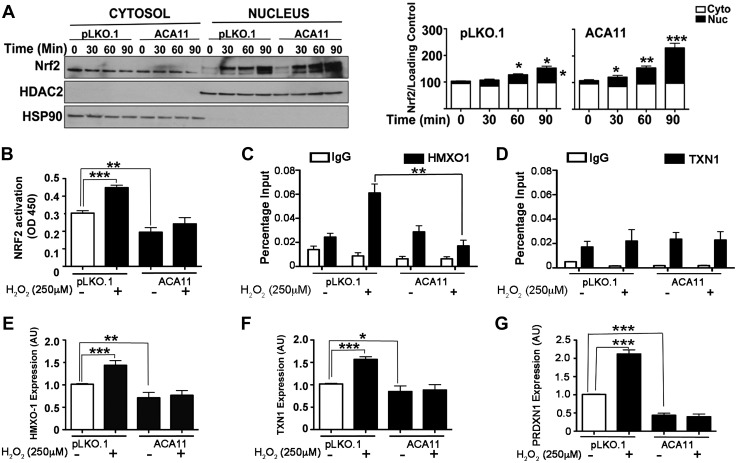
Given that ACA11 is associated with increased ROS production to explore potential mechanisms, we next sought to characterize the cellular antioxidant response in the context of ACA11 expression. NRF2 is a master regulator of the cellular response to increased ROS levels. Normally NRF2 is maintained in the cytoplasm in complex with KEAP1, and when ROS levels increase, NRF2 is released from KEAP1 and translocates to the nucleus to drive transcription of antioxidant genes (14, 24). First we examined the overall levels of NRF2 and KEAP1 in ACA11-overexpressing cells and did not find any changes at the mRNA or protein levels, as measured by qPCR and Western blot analyses, respectively (Supplemental Fig. S3A, B). We next examined the rate at which NRF2 translocates to the nucleus. In unstimulated cells overexpressing ACA11, there was a modest increase in NRF2 protein levels in the nucleus compared to vector control cells (Fig. 3A; cf. pKLO.1 vs. ACA11, nucleus t = 0 min). On stimulation with H2O2, a significant increase was noted in the rate and amount of NRF2 translocated to the nucleus of MM1.S cells overexpressing ACA11 (Fig. 3A). However, this increase in the translocation of NRF2 did not correlate with an increase in transcriptional activity. Using an NRF2 transcriptional reporter assay, ACA11 attenuated NRF2 transcriptional activity at both basal and H2O2-stimulated ROS levels (Fig. 3B). In addition, the NRF2 binding to endogenous promoters of known target genes HMXO1 and TXN1 under basal and H2O2-stimulated states were assessed by ChIP combined with qPCR. H2O2 stimulation induced NRF2 binding to the endogenous HMXO1 promoter (Fig. 3C). ACA11 overexpression leads to a significant attenuation of NRF2 binding to the HMXO1 promoter. We also assessed NRF2 binding to the TXN1 promoter, but interestingly, we observed that NRF2 binding to the TXN1 promoter did not change with H2O2 challenge; nor did ACA11 alter the basal or stimulated binding (Fig. 3D).
Figure 3.
NRF2 response to ACA11 generated ROS and levels of antioxidants. A) Western blot of nuclear and cytosolic fractions showing earlier translocation of NRF2 to nucleus in ACA11 cells in response to H2O2 (left). Bar graphs (right) show densitometry of bands from left panel. B) Activation of NRF2 is abrogated in presence of ACA11. Results of NRF2 reporter assay at baseline and in presence of H2O2 stimulus. C, D) ChIP analysis of 2 NRF2 target genes, HMXO-1 (C) and TXN-1 (D), in MM1.S cells with vector (pLKO.1) and ACA11 overexpression. E–G) qPCR analysis of specific NRF2 target genes HMXO-1 (E), TXN-1 (F), and PRDXN1 (G) in presence or absence of hydrogen peroxide in MM1.S cells, respectively. Representative figures are shown of experiments completed at least 3 times in triplicate. All data points represent means ± sd (n = 3). Asterisks indicate significance according to 1-way ANOVA with Bonferroni posttest. *P < 0.05, **P < 0.01, ***P < 0.001 compared to pLKO.1 in MM1.S.
These promoter binding data correlate with a decreases in the mRNA levels of 3 NRF2 antioxidant target genes, HMXO-1, TNX1, and PRDXN1, at both basal and H2O2-stimulated ROS levels as measured by qPCR (Fig. 3E–G). These data correlated with ACA11 knockdown in the t(4;14)-positive H929 cell line, leading to increases in NRF2-driven gene expression. Knockdown of ACA11 did not alter steady-state levels of NRF2 and KEAP1 mRNA (Supplemental Fig. S3C, D), as overexpression of ACA11 did not alter NRF2 and KEAP1 gene expression (Supplemental Fig. S3A, B). While knockdown of ACA11 in H929 cells does not lead to increased NRF2 reporter gene activity at basal levels, increases in NRF2 transcriptional activity were observed when cells were stimulated with H2O2 (Supplemental Fig. S3E). Conversely, the NRF2 target genes HMXO-1, TXN1, and GCLC at both basal and H2O2-stimulated ROS levels were de-repressed with ACA11 knockdown (Supplemental Fig. S3F–H). In both cell lines, reported NRF2 target genes were not altered by ACA11 expression levels or by H2O2 stimulation (data not shown). Taken together, these data indicated that ACA11 drives increased ROS levels by blocking a set of NRF2-driven antioxidant genes through an inhibition of NRF2 transcriptional activity.
DISCUSSION
The discovery that ACA11 is consistently overexpressed with MMSET in t(4;14) MM offers an explanation for why expression of MMSET alone is unable to drive cellular transformation (5). To obtain further insight into the biology of t(4;14)-positive MM, RNA-seq was performed on RNA isolated from CD138-enriched bone marrow mononuclear cells from t(4;14)-positive and -negative MM patients. Pathway analysis of the RNA-seq data identified biologic processes significantly associated with the t(4;14) translocation in patient samples that included oxidative phosphorylation, immune response, and cytoskeleton remodeling pathways (Table 1). Oxidative phosphorylation was the most significantly altered cellular process in t(4;14)-positive patient samples, suggesting a role for the mitochondria in this subtype of MM. MM is associated with reduced serum antioxidant levels and signs of increased oxidative stress (25), but the mechanism of these changes has not been determined. Besides being a vital part of metabolism, oxidative phosphorylation in the mitochondria produces ROS such as superoxide and hydrogen peroxide. Our results are consistent with our previously published finding that ACA11 affects ROS levels (5).
The increased proliferation of ACA11-expressing cells correlated with increased ROS levels after ACA11 overexpression (Figs. 2 and 3, and Supplemental Fig. S1). Previously we reported that ACA11 increased proliferation while reducing ROS levels in stably transfected cell lines (5), so considerable effort was spent carefully reviewing and verifying the results presented here. ACA11 was overexpressed in multiple cell types including MEFs, BJ human foreskin fibroblasts, human myeloma (MM1.S), and primary mouse splenic B cells, and in all cases, ACA11 overexpression caused a significant increase in ROS (Fig. 2 and Supplemental Fig. S1). Additionally, knocking down ACA11 in the H929 cell line, which naturally overexpresses ACA11, led to a decrease in cell proliferation (Fig. 1J, K) and a concomitant decrease in ROS levels (Supplemental Fig. S1). The biologic readout of ACA11 overexpression resulting in increased proliferation, as observed in t(4;14)-positive MM, is consistent.
The change in the direction of ROS levels is different in this study because of an error in the assay used in the previous study. The previous results were due to ACA11 being cloned into a YFP-expressing vector in the previous experiments. The emission spectra of YFP overlaps with the probe used to measure the ROS (DCFDA), effectively making the reported ROS levels impossible to accurately determine. For the current studies, a redesigned ACA11 expression construct was created using a vector backbone without a fluorescent marker (pLKO.1). In addition, the current study used multiple measures of ROS levels (DCFDA, MitoSOX, and GSH/GSSG levels; Fig. 2 and Supplemental Fig. S1) in our cell lines to ensure that the assays accurately measured ROS levels in the cells. The previous study also used cell lines stably transfected with ACA11 that were selected. The extended passaging of the cells allowed the cells to adapt to ACA11 expression in ways that confounded the previously reported ROS results. A similar discrepancy between stably transfected cell lines and the effects of acute overexpression also occurs with Bcl2, which blocks apoptosis effectively on stable expression but poorly after microinjection (26). Ultimately, the only difference between the current and previous results is the direction of the change in the ROS levels. The biology of increased proliferation due to ACA11 overexpression is not different.
Increased ROS levels driven by ACA11 overexpression also correlate well with the clinical outcome of MM patients. t(4;14)-positive MM patients have a better response to bortezomib (27). It has also been shown that there is a direct correlation between increased levels of ROS in MM cell lines and an increased sensitivity to bortezomib treatment (28). Taken together with the data presented here, a model of ACA11-driven increases in ROS due to the t(4;14) translocation in a subset of MM patients would explain their response to bortezomib treatment.
The current results verify the previous finding that ACA11 promotes growth; they also extend our understanding of this proproliferative function. ACA11-overexpressing cells consistently proliferate at an accelerated rate (Fig. 1) and form larger colonies in soft agar (Fig. 1). Furthermore, NAC treatment abrogated proliferation in ACA11-overexpressing cells, indicating that proliferation is a result of increased ROS (Fig. 2). We observed higher levels of pERK1/2 in ACA11-overexpressing cells compared to controls (Fig. 1). Antioxidant pretreatment led to a decrease in pERK1/2 expression in these cells (Fig. 2). Finally, inhibition of ERK1/2 phosphorylation with a MEK inhibitor, PD98059, leads to an attenuation of ACA11-driven increases in ERK1/2 phosphorylation and cell proliferation (Supplemental Fig. S1B, C). These results collectively substantiate the fact that ACA11-overexpressing cells have higher ROS levels, with increased phosphorylated pERK1/2 levels driving increased proliferation rates compared to controls. B cells lacking antioxidant enzymes have increased both ROS levels and proliferation rates (29). Our data agree with this in that ACA11-overexpressing cells have a lower GSH/GSSG ratio (Fig. 2A) as well as lower levels of antioxidant enzymes (Fig. 3 and Supplemental Fig. S3) resulting in increased ROS levels. Therefore, these data suggest that overexpression of ACA11 by the t(4;14) translocation inactivates antioxidant enzymes, lowers the GSH/GSSG ratio, and stimulates proliferation. Together, our data demonstrate that ACA11-induced ROS are necessary to promote cell proliferation.
A tight regulation of the intracellular redox status is critical for cellular homeostasis, and several enzymatic (e.g., catalase, peroxidases, superoxide dismutase) and nonenzymatic protective mechanisms (e.g., glutathione, ascorbic acid) have evolved to carefully modulate cellular ROS levels (14, 24, 30). The coordinated action of various cellular antioxidants in cells is critical for maintaining a steady redox state. The transcription factor NRF2 is a master regulator of the antioxidant stress response. In response to increased cellular redox, the cytoplasmic KEAP1-NRF2 complex is disrupted; NRF2 is stabilized and translocates to the nucleus, where it normally up-regulates numerous antioxidant genes. In response to ACA11-driven increases in ROS levels, NRF2 efficiently translocates to the nucleus (Fig. 3A). Interestingly, increased nuclear translocation of NRF2 failed to lead to increased NRF2 transcriptional activity by reporter gene assay or by ChIP-qPCR analysis of 2 endogenous NRF2-driven promoters (Fig. 3C, D). Measurement of expression of NRF2 target genes in ACA11-overexpressing cells (Fig. 3E–G and data not shown) demonstrated that only a subset of NRF2-regulated genes responds to H2O2 stimulation in MM1.S cells, but those that do are blocked by ACA11 expression (Fig. 3E–G). Correlating with these findings, there are a subset of NRF2 target genes in H929 ACA11-overexpressing cells that are sensitive to H2O2 stimulation, and again, ACA11 knockdown in these cells increases their expression (Supplemental Fig. S3E–H and data not shown). These results indicate that the NRF2-driven antioxidant pathway in response to increased ROS levels is compromised in ACA11-overexpressing cells.
ACA11 alone does not explain the complex biology of the t(4;14) translocation. Cells with t(4;14) consistently overexpress both MMSET and ACA11, and knockdown of either MMSET or ACA11 inhibits the growth of t(4;14)-positive MM cell lines (Fig. 1) (5, 31). Overexpression of MMSET alone fails to promote cell growth in nontransformed cells (5), which led to our discovery of a role for ACA11, a snoRNA cotranscriptionally expressed with MMSET, in promoting transformation. The present study confirms that ACA11 is an oncogenic snoRNA and improves our understanding of how this small noncoding RNA contributes to a progrowth phenotype. Further, our results demonstrate that ACA11 compromised the ability of NRF2 to induce target antioxidant genes, which we hypothesize is the cause of increased oxidative stress and cell proliferation.
In summary, we demonstrate here for the first time that ACA11 overexpression increases proliferation of primary B cells and results in dramatically increased colony size in soft agar assays. In multiple cell types including primary B cells, ACA11-driven hyperproliferation is associated with and dependent on increased cellular ROS. Additional studies are required to identify the protein components of the ACA11 snRNPs in vivo and to determine the mechanism by which ACA11 inhibits NRF2 transcriptional activity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported, in part, by the U.S. National Institutes of Health, National Cancer Institute (Grants R01-CA174743 to M.H.T. and L.B.M., and R01-CA-175349 to M.H.T.). The authors thank M. Bates (University of Iowa, Iowa City, IA, USA) for her critical review. The authors also thank all the members of the M.H.T. laboratory for discussion and suggestions, and the Genome Technology Access Center (Department of Genetics, Washington University School of Medicine) for help with genomic analysis.
Glossary
- Arf
alternative reading frame protein of the CDKN2A locus
- BCA
bicinchoninic acid
- BrdU
bromodeoxyuridine
- carboxy-DCFH-DA
carboxy-2′,7′-dicholorodi hydrofluorescein diacetate
- ChIP
chromatin immunoprecipitation
- FBS
fetal bovine serum
- GSH
glutathione
- GSSG
glutathione disulfide
- HSP
heat shock protein
- MEF
mouse embryo fibroblast
- MM
multiple myeloma
- MMSET
multiple myeloma set domain
- NAC
N-acetyl cysteine
- NRF2
nuclear factor (erythroid-derived 2)-like 2
- qPCR
quantitative PCR
- RNA-seq
RNA sequencing
- ROS
reactive oxygen species
- snoRNA
small nucleolar RNA
- WHSC1
Wolf-Hirschhorn syndrome candidate 1
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
N. Mahajan, L. B. Maggi, Jr., and M. H. Tomasson conceived the study, designed and interpreted the experiments, and wrote the article; N. Mahajan, L. B. Maggi Jr., R. L. Bennett, and C. Troche performed the experiments; H.-J. Wu helped analyze the RNA-seq data and edit the article; and J. D. Weber and J. D. Licht suggested and helped design experiments and edit sections of the article.
REFERENCES
- 1.Kuehl W. M., Bergsagel P. L. (2002) Multiple myeloma: evolving genetic events and host interactions. Nat. Rev. Cancer 2, 175–187 [DOI] [PubMed] [Google Scholar]
- 2.Chesi M., Nardini E., Lim R. S., Smith K. D., Kuehl W. M., Bergsagel P. L. (1998) The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92, 3025–3034 [PubMed] [Google Scholar]
- 3.Santra M., Zhan F., Tian E., Barlogie B., Shaughnessy J. Jr (2003) A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood 101, 2374–2376 [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Garcia E., Popovic R., Min D.-J., Sweet S. M. M., Thomas P. M., Zamdborg L., Heffner A., Will C., Lamy L., Staudt L. M., Levens D. L., Kelleher N. L., Licht J. D. (2011) The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 117, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu L., Su M. Y., Maggi L. B. Jr., Lu L., Mullins C., Crosby S., Huang G., Chng W. J., Vij R., Tomasson M. H. (2012) Multiple myeloma–associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J. Clin. Invest. 122, 2793–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova J. A., Ivanova S. M., Tonevitsky A. G., Grigoriev A. I. (2013) New functions of small nucleolar RNAs. Biochemistry (Mosc.) 78, 638–650 [DOI] [PubMed] [Google Scholar]
- 7.Bratkovič T., Rogelj B. (2014) The many faces of small nucleolar RNAs. Biochim. Biophys. Acta 1839, 438–443 [DOI] [PubMed] [Google Scholar]
- 8.Mannoor K., Liao J., Jiang F. (2012) Small nucleolar RNAs in cancer. Biochim. Biophys. Acta 1826, 121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cech T. R., Steitz J. A. (2014) The noncoding RNA revolution—trashing old rules to forge new ones. Cell 157, 77–94 [DOI] [PubMed] [Google Scholar]
- 10.Siprashvili Z., Webster D. E., Johnston D., Shenoy R. M., Ungewickell A. J., Bhaduri A., Flockhart R., Zarnegar B. J., Che Y., Meschi F., Puglisi J. D., Khavari P. A. (2016) The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat. Genet. 48, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams G. T., Farzaneh F. (2012) Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 12, 84–88 [DOI] [PubMed] [Google Scholar]
- 12.Su H., Xu T., Ganapathy S., Shadfan M., Long M., Huang T. H.-M., Thompson I., Yuan Z.-M. (2014) Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 33, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 13.Wang X., Martindale J. L., Liu Y., Holbrook N. J. (1998) The cellular response to oxidative stress: influences of mitogen-activated protein kinase signalling pathways on cell survival. Biochem. J. 333, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sporn M. B., Liby K. T. (2012) NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. Cancer 12, 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anders S., Pyl P. T., Huber W. (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love M. I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 19.He Z., O’Neal J., Wilson W. C., Mahajan N., Luo J., Wang Y., Su M. Y., Lu L., Skeath J. B., Bhattacharya D., Tomasson M. H. (2016) Deletion of Rb1 induces both hyperproliferation and cell death in murine germinal center B cells. Exp. Hematol. 44, 161–5.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 21.Mahajan N., Shi H. Y., Lukas T. J., Zhang M. (2013) Tumor-suppressive maspin functions as a reactive oxygen species scavenger: importance of cysteine residues. J. Biol. Chem. 288, 11611–11620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forys J. T., Kuzmicki C. E., Saporita A. J., Winkeler C. L., Maggi L. B. Jr., Weber J. D. (2014) ARF and p53 coordinate tumor suppression of an oncogenic IFN-β-STAT1-ISG15 signaling axis. Cell Reports 7, 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halprin K. M., Ohkawara A. (1967) The measurement of glutathione in human epidermis using glutathione reductase. J. Invest. Dermatol. 48, 149–152 [DOI] [PubMed] [Google Scholar]
- 24.Schieber M., Chandel N. S. (2014) ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma A., Tripathi M., Satyam A., Kumar L. (2009) Study of antioxidant levels in patients with multiple myeloma. Leuk. Lymphoma 50, 809–815 [DOI] [PubMed] [Google Scholar]
- 26.Desbarats L., Schneider A., Müller D., Bürgin A., Eilers M. (1996) Myc: a single gene controls both proliferation and apoptosis in mammalian cells. Experientia 52, 1123–1129 [DOI] [PubMed] [Google Scholar]
- 27.Hervé A. L., Florence M., Philippe M. (2011) Molecular heterogeneity of multiple myeloma: pathogenesis, prognosis, and therapeutic implications. J. Clin. Oncol. 29, 1893–1897 [DOI] [PubMed] [Google Scholar]
- 28.Nerini-Molteni S., Ferrarini M., Cozza S., Caligaris-Cappio F., Sitia R. (2008) Redox homeostasis modulates the sensitivity of myeloma cells to bortezomib. Br. J. Haematol. 141, 494–503 [DOI] [PubMed] [Google Scholar]
- 29.Bertolotti M., Yim S. H., Garcia-Manteiga J. M., Masciarelli S., Kim Y.-J., Kang M.-H., Iuchi Y., Fujii J., Vené R., Rubartelli A., Rhee S. G., Sitia R. (2010) B- to plasma-cell terminal differentiation entails oxidative stress and profound reshaping of the antioxidant responses. Antioxid. Redox Signal. 13, 1133–1144 [DOI] [PubMed] [Google Scholar]
- 30.Sullivan L. B., Chandel N. S. (2014) Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2, 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirabella F., Wu P., Wardell C. P., Kaiser M. F., Walker B. A., Johnson D. C., Morgan G. J. (2013) MMSET is the key molecular target in t(4;14) myeloma. Blood Cancer J. 3, e114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.