Abstract
Cancer stem–like cells (CSLCs) contribute to the initiation and recurrence of tumors and to their resistance to conventional therapies. In this study, small interfering RNA (siRNA)-based screening of ∼4800 druggable genes in 3-dimensional CSLC cultures in comparison to 2-dimensional bulk cultures of U87 glioma cells revealed 3 groups of genes essential for the following: survival of the CSLC population only, bulk-cultured population only, or both populations. While diverse biologic processes were associated with siRNAs reducing the bulk-cultured population, CSLC-eliminating siRNAs were enriched in a few functional categories, such as lipid metabolism, protein metabolism, and gene expression. Interestingly, siRNAs that selectively reduced CSLC only were found to target genes for cholesterol and unsaturated fatty acid synthesis. The lipidomic profile of CSLCs revealed increased levels of monounsaturated lipids. Pharmacologic blockage of these target pathways reduced CSLCs, and this effect was eliminated by addition of downstream metabolite products. The present CSLC-sensitive target categories provide a useful resource that can be exploited for the selective elimination of CSLCs.—Song, M., Lee, H., Nam, M.-H., Jeong, E., Kim, S., Hong, Y., Kim, N., Yim, H. Y., Yoo, Y.-J., Kim, J. S., Kim, J.-S., Cho, Y.-Y., Mills, G. B., Kim, W.-Y., Yoon, S. Loss-of-function screens of druggable targetome against cancer stem–like cells.
Keywords: CSLC sphere culture, lipid profile, network analysis, siRNA screening
A small subpopulation of cells in the tumor mass, cancer stem–like cells (CSLCs), has recently received wide interest because of their suggested roles in tumor initiation, resistance to conventional therapies, and recurrence (1–4). To date, the therapeutic strategies targeting CSLCs that have been evaluated clinically have largely focused on signaling networks important for tissue stem cell biology (5); fewer studies have focused on differential molecular characteristics between CSLCs and bulk cancer cells (6, 7). Regardless, there has been little success regarding the development of new therapeutic strategies selectively targeting CSLCs. Studies suggest that CSLCs should possess unique biologic pathways to mediate their stemlike roles in tumors, and such pathways might have different roles in other cancer cells within the same tumor (6). Accordingly, we aimed to identify a unique biologic process essential for CSLC enrichment in a tumor mass that could be directly used to selectively target CSLCs.
Phenotypic loss-of-function screens using RNAi libraries are a powerful tool for identifying causal genes and their biologic processes in diverse cell-based assays (8). For example, shRNA library screening provided a new clue for improving the outcome of BRAF inhibition in epidermal growth factor (EGF) receptor–positive melanoma cancer cells (9). Although CSLC populations under typical monolayer culture conditions are relatively small among the bulk cells of a given cancer cell line, they can be significantly enriched in 3-dimensional (3-D) sphere culture using well-defined media (10). Thus, in the present study, we attempted to compare the knockdown efficacy of small interfering RNAs (siRNAs) targeting ∼4800 druggable genes on cancer cell growth between 2-dimensional (2-D) monolayer and 3-D sphere culture conditions. On the basis of primary dual screens and secondary validations, knockdown hits with significant inhibitory effects on 2-D- or 3-D–cultured cells were classified into 3 groups of genes essential for the following: survival of the CSLC population only, bulk-cultured population only, or both populations. In this report, we further characterize CSLC selective inhibitory genes and their functional roles, particularly in lipid biosynthesis pathways.
Recent findings show that the cancer cells may reactivate their own lipid synthesis, cholesterols, and fatty acids (11, 12). Although the altered lipid metabolism in cancer cell is now widely accepted (11), the role of self-synthesized lipids in cancer initiation and metastasis remains largely unknown. Moreover, only a few studies have focused on lipid metabolism in CSLCs compared to bulk cells in tumors. In the present study, we demonstrated, using large-scale siRNA library screening and lipid metabolomics approaches, that CSLCs have a lipid metabolomic profile that is distinguishable from that of bulk-cultured cells and that the metabolic pathways responsible for this lipid profile can serve as selective targets for CSLC therapy.
MATERIALS AND METHODS
Cell and sphere culture
Bulk cultures of human cell line U87 (American Type Culture Collection, Manassas, VA, USA), GBM cell line U251 [National Institutes of Health, National Cancer Institute (NCI), Frederick, MD, USA], non-small-cell lung cancer line NCI-H460 (NCI), colon cancer cell line HT-29 (ATCC), and breast cancer cell line MDA-MB-231 (NCI) were grown in RPMI 1640 medium (HyClone Laboratories, Logan, UT, USA) containing 10% fetal bovine serum (HyClone Laboratories), 100 U/ml penicillin, and 100 μg/ml streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). CSLC spheres were cultured in serum-free conditioned medium containing 20 ng/ml EGF, 20 ng/ml basic fibroblast growth factor, and B27 supplemented in DMEM/F-12 (Thermo Fisher Scientific). The cells were maintained in a humidified atmosphere of 5% CO2 and 95% air at 37°C. Culture medium was refreshed every 2 to 3 d. The culture plates for CSLCs were coated with poly-2-hydroxyethyl methacrylate (Sigma-Aldrich, St. Louis, MO, USA) by adding a 5 mg/ml solution in 95% ethanol.
High-throughput siRNA screening
The siRNA screen was performed using 4 pooled siRNAs to target each of the 4786 genes in the human drug target library (On-Target Plus SmartPool; GE Dharmacon, Lafayette, CO, USA). The procedure is shown in Supplemental Fig. S1A. Screening was performed in triplicate for 3 or 5 d in a 384-well plate format (1000 cells per well). Each plate was supplemented with a negative control siRNA, the On-Target Plus NontargetingPool siRNA (siNC; GE Dharmacon), and positive control siRNAs (PLK1; GE Dharmacon). Reverse transfections were performed using a MultiFlo microplate dispenser (BioTek Instruments, Winooski, VT, USA) with siRNAs (final concentration 10 nM) and Lipofectamine RNAiMax (0.05 μl per well; Thermo Fisher Scientific) diluted in Opti-MEM I (Thermo Fisher Scientific) in black 384-well plates (Corning, Corning, NY, USA). After 3 or 5 d, the cells were stained with Hoechst 33342 (Sigma-Aldrich). The plates were imaged using a Cytation 3 (BioTek Instruments) in 2 × 2 montage mode with a ×4 objective to capture the entire well. The number of CSLC spheres with a diameter over 100 μm was counted using Gen5 data analysis software (BioTek Instruments). The control experiment using siRNA against a gene essential for the cell cycle (PLK1) (13) showed that the assay is appropriate for identifying essential genes for bulk culture cells and CSLCs (Supplemental Fig. S1B). Plates that failed quality control (z′ factor < 0.3) were removed (14). Each plate was normalized to the average of 16 siNC wells per plate (Supplemental Fig. S1C).
Screening data analysis
Raw data were normalized by log2 transformation of percentage of control = xi/average of negative controls per plate, where xi is a measured value of cell or sphere count for each gene (sample) and the negative control is siNC transfection. Statistical significance was calculated by the 2-sample Student’s t test.
Validation screen
A validation screen was performed to identify false-positive findings. siRNAs from each SmartPool were rescreened using the same transfection protocol as the primary screen. siRNAs were classified as hits using the same criteria as in the primary screen (6) (Supplemental Fig. S1D).
Biologic functional analyses and network visualization of gene sets
We collected 1777 gene sets for the biologic processes of gene ontology from the Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/home.jsp). The identified hits from siRNA screening were annotated using GO biologic process terms. The functional enrichment map was graphically organized into a network using Cytoscape 3.2.0 network software (http://www.cytoscape.org/) and the Enrichment Map plugin.
Chemicals
The chemical reagents atorvastatin, lovastatin, fluvastatin, simvastatin, MF-438, diltiazem, carfilzomib, ispinesib, and selinexor were purchased from Selleckchem (Houston, TX, USA). These chemicals were dissolved in DMSO (Calbiochem, San Diego, CA, USA) and diluted to their final concentrations. Mevalonolactone, cholesterol, and oleic acid–albumin were purchased from Sigma-Aldrich; these reagents were dissolved in distilled water.
siRNA transfection
In follow-up siRNA studies, target siRNAs (On-Target Plus SmartPool siRNA; GE Dharmacon) were introduced into cells at a final concentration of 10 nM by reverse transfection using Lipofectamine RNAiMax (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Scanning electron microscopy
Spheres were fixed using 4% formaldehyde for 20 min and washed with PBS. The samples were then immersed in ethanol for dehydration and then hexamethyldisilazane (Sigma-Aldrich) for displacement. The samples were mounted on aluminum stubs with adhesive carbon tapes and sputter coated with gold. The samples were observed by scanning electron microscopy (SEM) (JSM-7600F; Jeol, Akishima, Japan).
Apoptosis analysis
An apoptosis kit (FITC Annexin V Apoptosis Detection Kit I; BD Biosciences, San Jose, CA, USA) was used to detect apoptosis by flow cytometry according to the manuals provided. Briefly, cells and spheres were dissociated, collected, and resuspended in PBS. Cell suspensions were transferred to 0.1 ml of 1× cold binding buffer by pipetting slowly; 5 μl FITC annexin V was added. After incubation for 15 min at room temperature, the cells were resuspended in 1× cold binding buffer, and 5 μl propidium iodide was added. The apoptotic cell percentage was measured using a fluorescence-activated cell sorting (FACS) LSRII flow cytometer (BD Biosciences). FlowJo 10 (Ashland, OR, USA) was used for data analysis.
CD133 expression analysis
To identify the cancer stem cell marker CD133, cells and spheres were dissociated as a single-cell suspension and washed with PBS. The cells were labeled with the CD133-PE antibody (Miltenyi Biotec, San Diego, CA, USA) supplemented with FcR Blocking Reagent (Miltenyi Biotec) and analyzed using a FACS LSRII flow cytometer.
ALDH assay
The Aldefluor kit (Stem Cell Technologies, Durham, NC, USA) was used to detect the cancer stem cell populations with high aldehyde dehydrogenase (ALDH) enzyme activity. Briefly, cells were incubated in ALDH assay buffer containing the ALDH substrate (1 mM) for 30 min at 37°C. As a negative control, cells were stained under identical conditions in the presence of diethylaminobenzaldehyde (15 mM), a specific ALDH inhibitor. A FACS LSRII flow cytometer was used to measure the ALDH-positive cell population.
Limiting dilution assay
Sphere cells were dissociated and placed in 384-well plates at various seeding densities (1–128 cells per well). After a 7-d culture, the percentage of wells not containing spheres for each plating density was calculated and plotted against the number of cells per well.
Rescue assay
CSLCs (2000 cells per well) were seeded in a 96-well black plate (Corning). Cells were reverse transfected with siRNAs against target genes or were treated with the indicated concentration of atorvastatin in the presence or absence of mevalonolactone (1 mM), cholesterol (10 μM), or oleic acid–albumin (200 μM). After 5 d, Hoechst 33342 dye was added to each well at a final concentration of 1 μg/ml, and a Cytation 3 image reader (BioTek Instruments) was used for quantitative sphere counting (≥100 μm).
Tumor xenograft assay
All experiments were preapproved by the Institutional Animal Care and Use Committee of Sookmyung Women’s University. To explore the efficiency of specific siRNAs against target genes on tumor growth, CSLCs were reverse transfected under CSLC sphere culture condition for 3 d as described above. The indicated number of CSLC spheres cells was mixed with 40% Matrigel (Thermo Fisher Scientific) with EGF and basic fibroblast growth factor. The cells were subcutaneously injected into NOD/SCID (Koatech, Pyeongtaek, Korea) or BALB/c Nude (Orient Bio, Seongnam, Korea) mice, and the Matrigel-formed tumor sizes were measured every 2 to 3 d after injection.
To identify the efficiency of atorvastatin treatment, 5 × 105 CSLC sphere cells were mixed with 40% Matrigel (Thermo Fisher Scientific) with EGF, basic fibroblast growth factor, and atorvastatin (10 nM) if needed. Atorvastatin in PBS was administered orally at 20 mg/kg once daily.
Tumor growth was assessed; the size of the tumor was measured with calipers 3 times per week, and tumor volume was estimated by width × width × length × ½, where width is the short axis and length is long axis. The diets of the mice were as follows: high-fat diet, 60% of calories from fat; low-fat diet, 10% of calories from fat; and high-fat diet + cholesterol (1.25%). All diets were obtained from DooYeol Biotech (Seoul, Korea).
Cholesterol and triglyceride quantification
Cellular lipids were extracted with a mixture of hexane:isopropanol, and the organic layer was dried. The total cholesterol including cholesteryl esters was measured by the Amplex Red Cholesterol Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions, and the signals were detected by fluorometer (Promega, Madison, WI, USA) with excitation at 520 nm and emission at 590 nm. Total cholesterol and triglyceride (TG) from mouse serum were measured by the V-Cholesterol and TG-S assay kits (Asan Pharmaceutical, Seoul, Korea) according to the manufacturer’s instructions.
Neutral lipid droplet analyses
Cells were suspended and then stained for lipid droplets using BODIPY 493/503 (Thermo Fisher Scientific) and Hoechst 33342 (Thermo Fisher Scientific) for 15 min at room temperature and observed with a confocal fluorescence microscope (LSM-700, Carl Zeiss GmbH, Jena, Germany).
Lipid metabolite profiling
Lipid metabolomics was conducted using a quadrupole time-of-flight mass spectrometer (Synapt G2Si; Waters, Milford, MA, USA) connected to an ultraperformance liquid chromatography system (Acquity UPLC; Waters). Chromatographic separation was performed on an CSH C18 column (2.1 × 100 mm, 1.7 μm; Waters) using mobile phase A [10 mM ammonium formate and 0.1% formic acid in acetonitrile/water (60:40)] and mobile phase B [10 mM ammonium formate and 0.1% formic acid in isopropanol/acetonitrile (90:10)]. The flow rate was 0.4 ml/min, and the elution gradients were as follows: 0–2 min, 40–43% B; 2.0–2.1 min, 43–50% B; 2.1–12 min, 50–54% B; 12–12.1 min, 54–70% B; 12.1–18 min, 70–99% B; 18–18.1 min, 99–40% B; and 18.1–20 min, 40% B. The column temperature was 55°C.
Mass acquisition was performed in both positive electrospray ionization (ESI+) and negative ESI (ESI−) modes with the following parameters: capillary voltage of 2.0 kV for positive and 1 kV for negative modes; cone voltage of 30 V; source temperature of 100°C; desolvation temperature of 550°C; and desolvation gas flow of 900 L/h. The mass data were collected in the range of m/z 60 to 1400 with a scan time of 0.25 s. Data were acquired in 2 channels: low-collision energy (6 eV) for molecular ions, and energy ramp (20–60 eV) for product ions. To ensure accuracy of the measured mass, leucine–enkephalin (m/z 556.2771 in ESI+ and m/z 554.2615 in ESI−) was used as a reference lock-mass compound at a concentration of 1 ng/μl and a flow rate of 5 μl/min.
Processing and analysis of mass spectrometry data
Data processing including mass ion alignment, normalization, and peak picking was performed using Progenisis QI software (Waters). The intensities of mass peaks for each sample were normalized according to the total ion intensity and Pareto scaled using SIMCA-P+ 12 software (Umetrics, San Jose, CA, USA).
To discriminate between the intensities of mass peaks of each group of cells, partial least-square discriminant analysis and orthogonal partial least-square discriminant analysis (OPLS-DA) were performed. The reliability correlation [p(corr)] values of all metabolites from the S plot of the OPLS-DA were extracted using the first component. We selected metabolites satisfying the following criteria as potential markers: high confidence (|p(corr)| > 0.6) in discrimination cell groups; mean intensities in a cell group significantly different from that of comparison cell groups (P < 0.05); and fold change of 2 or more between groups. The P value was calculated using the independent 2-sample Student’s t test.
Lipid metabolites were identified by database searches against their accurate masses using databases including Human Metabolomics Database (http://www.hmdb.ca/) (15), MassBank (http://www.massbank.jp/) (16), and Lipid Metabolites and Pathways Strategy (Lipid MAPS; http://www.lipidmaps.org/) (17). Isotope similarity and fragmentation patterns were considered for validation of identified lipids.
Statistical analysis
For all experimental results, the significance of the differences between 2 groups was calculated by Student’s t test or the χ2 test. The data are expressed as the average values and standard error.
RESULTS
Differential efficacy of siRNA treatment on 2-D bulk cell and 3-D CSLC cultures
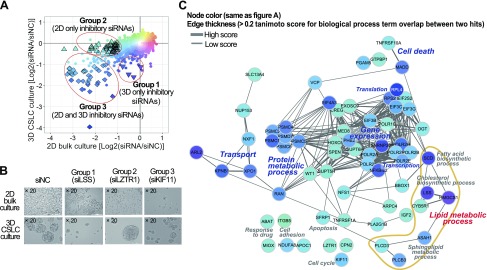
A library of siRNAs targeting ∼4800 druggable genes was screened for effects on the viability of U87, a glioblastoma multiforme (GBM) cell line, CSLC spheres, and bulk cancer cells. (The procedure is shown in Supplemental Fig. S1A.) The inhibitory effect of each siRNA was evaluated by image-based sphere count (>100 µm) and cell count under 3-D CSLC-enriched culture and 2-D bulk culture conditions, respectively (6) (Fig. 1A). A total of 92 siRNA gene probe pools exhibited a significant inhibitory effect (P < 0.01 and >2-fold changes) on the growth of CSLC or bulk-cultured cells (Fig. 1A), with 66 being validated in a secondary assay (Supplemental Fig. S1D and Supplemental Table S1). On the basis of their selective efficacy against CSLC-enriched culture compared to bulk cancer cell culture conditions, we were able to classify the siRNAs into 3 unique groups, as follows: group 1, 5 siRNAs eliminating CSLC spheres only; group 2, 31 siRNAs reducing cell growth of bulk cultures; and group 3, 30 siRNAs required for the survival of both CSLCs and bulk culture cancer cells (Fig. 1A, B).
Figure 1.
siRNA library screening on 2-D bulk culture and CSLC culture conditions. A) siRNAs against total of 4786 druggable genes were screened in glioma U87 cell lines. Inhibitory effect was compared between 2-D bulk and CSLC cultures by normalized cell count and sphere number, respectively. Open triangles and diamonds indicate 92 inhibitory hits with statistical significance on 2-D bulk and/or CSLC cultured cells. B) Morphologic analysis of 2-D bulk and CSLC cultured cells under light microscopy after siRNA treatment. C) Network-based functional analysis of siRNA hits.
These validated hits in the 3 groups were clustered on the basis of their 3-D vs. 2-D inhibitory effect and their annotated GO (Fig. 1C and Supplemental Fig. S1E). As a result, we observed discriminative functional categories assigned to each groups. Group 2 genes of which knockdown inhibited only bulk-cultured cells, were widely distributed in diverse molecular or biologic events such as cell death, apoptosis, and cell adhesion. In contrast, CSLC-sensitive knockdown hits in groups 1 and 3 were found in limited biologic processes, exhibiting tight clustering patterns in functional categories such as lipid metabolic process, transport, protein metabolism, and gene expression (Fig. 1C and Supplemental Table S1). In particular, group 3 genes of which knockdown inhibited both CSLC and bulk cells were densely distributed among isoforms of proteasome 26S ATPases complex genes (PSMC1, PSMC2, PSMC4, PSMC5, PSMC6, PSMD7), translation initiation factors (EIF3B, EIF3C, EIF3G, EIF4A3), and RNA polymerase II submits (POLR2A, POLR2B, POL2E, POLR2F, POLR2H, POLR2I). Interestingly, the majority of group 1 siRNAs, specific only to CSLC survival, were found to target genes (SCD, LSS, HMGCS1) critical for cholesterol and unsaturated fatty acid synthesis. This biased distribution of group 1 and 3 genes suggests the existence of essential pathways that play a pivotal role in CSLC-specific regulation, while diverse cellular signaling might provide independent mechanisms to achieve inhibitory effects on the growth of bulk cancer cells.
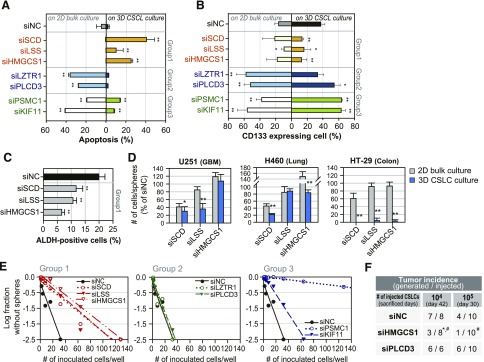
Efficacy of screening hits in maintaining the identity and self-renewal of CSLCs
We further investigated the distinctive effect of the 3 groups of hits on CSLCs and other bulk cancer cells. The 3 siRNAs in group 1, siLSS, siHMGCS1, and siSCD, increased apoptosis only in CSLCs. The siRNAs belonging to group 2, siLZTR1 and siPLCD3, induced apoptotic cells in bulk culture only, whereas those belonging to group 3, siPSMC1 and siKIF11, increased apoptotic cell death in both CSLC and bulk culture (Fig. 2A). Consistently, cells expressing the GBM CSLC marker CD133 were significantly eliminated only by treatment with group 1 siRNAs under CSLC-enriched culture conditions (Fig. 2B and Supplemental Fig. S1F). In contrast, group 2 and 3 siRNAs actually enriched CD133-positive cell populations, regardless of their apoptotic contribution to the total cell mass (Fig. 2B). This observation is consistent with the notion that targeting the growth of the bulk cancer cell population may increase the fraction of CSLCs in surviving populations (18). Cells expressing another CSLC marker, ALDH1, were also decreased by group 1 siRNAs (Fig. 2C). Moreover, the CSLC-selective efficacy of group 1 siRNAs was reproduced in some cell lines from other tissue lineages: CSLC populations of GBM (U251), lung (H460), and colorectal (HT-29) cancer cell lines exhibited increased sensitivity to siHMGCS1, siLSS, and siSCD (Fig. 2D).
Figure 2.
Regulation of self-renewal of CSLC population after diverse gene knockdown. A) Variable apoptotic response between 2-D bulk and 3-D CSLC cultured cells induced by treatment with group 1–3 siRNA hits. Apoptotic cells were detected by FACS using annexin V and propidium iodide, and quantified by annexin V–positive cells. *P < 0.05, **P < 0.01 vs. siNC. B) CSLC population was detected by FACS using CD133 antibodies. Ratio of changes of CD133-positive populations in surviving bulk and CSLC cultured cells after treatment of group 1–3 siRNA hits. *P < 0.05, **P < 0.01 vs. siNC. C) Markers of CSLC and ALDH expressing cells were detected by FACS. Ratio of changes for ALDH-positive cells in CSLC cultures after treatment with group 1 siRNAs. **P < 0.01 vs. siNC. D) Knockdown effect of group 1 genes on 2-D bulk cultured and 3-D CSLC cultured cell lines of diverse cancer lineages. *P < 0.05, **P < 0.01 vs. bulk culture. In vitro (E) and in vivo (F) self-renewal capacity of surviving CSLC cultured cells after treatment of group 1–3 siRNAs from limiting dilution assay. *P < 0.05 vs. siNC, #P < 0.05 vs. siPLCD3.
We also examined the hallmark activities of CSLCs, self-renewal activity, and tumor-forming activity using siRNA-transfected CSLC cells. In vitro limiting dilution assays showed that targeting all of the group 1 lipid biosynthesis genes resulted in the exclusion of self-renewing cells (Fig. 2E). Interestingly, siRNAs against group 3 genes (PSMC1, KIF11) also resulted in serial elimination of self-renewing cells, although they did not decrease the CD133-positive population in 3-D cultures. However, siRNAs against group 2 genes did not alter this activity. Consistently, siRNAs targeting HMGCS1 caused the loss of tumor-forming cells in vivo, though siRNAs targeting a group 2 gene (PLCD3) did not (Fig. 2F).
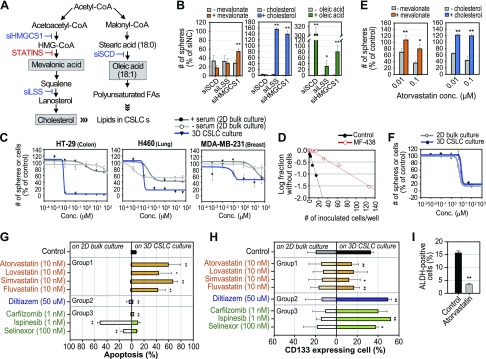
Reversible regulation of CSLCs by targeting lipid biosynthesis
HMGCS1, LSS, and SCD encode key enzymes in the pathways for cholesterol and unsaturated fatty acid biosynthesis (Fig. 3A), and the results presented in Fig. 2 suggest that the reaction products of these pathways might be essential for CSLC enrichment and survival. Indeed, mevalonate, a downstream product of HMGCS1, reversed the inhibitory effect of siHMGCS1 on CSLCs; furthermore, cholesterol rescued CSLCs treated with siHMGCS1 or siLSS (Fig. 3B). Interestingly, although it was most effective against siSCD, oleic acid reversed the effects of all 3 siRNAs (Fig. 3B), suggesting that oleic acid production may be a rate-limiting step for CSLC growth.
Figure 3.
Reversible regulation of CSLC population by inhibitors of lipid biosynthesis pathway. A) Cholesterol biosynthesis (left) and fatty acid biosynthesis (right) pathways. B) Metabolite products reversed effects of siRNAs inhibiting lipid biosynthesis pathways. *P < 0.05, **P < 0.01 vs. without supplement. C) Selective inhibition of CSLC sphere growth by SCD inhibitor MF-438 in diverse cancer lineages. D) In vitro self-renewal capacity of surviving HT-29 cells after treatment of MF-438 from limiting dilution assay. E) Metabolite products reversed inhibitory effect of statin on CSLC growth. *P < 0.05, **P < 0.01 vs. without supplement. F) Comparative efficacy of fatostatin on U87 growth between 2-D bulk cell and 3-D CSLC sphere cultures. G, H) Apoptosis induction (G) and changes in CD133 expressing population (H) by compounds inhibiting group 1–3 genes from FACS analysis. *P < 0.05, **P < 0.01 vs. control. I) Decrease in ALDH-positive cells by atorvastatin in 3-D CSLC culture from FACS analysis.**P < 0.01 vs. control.
Consistently, the SCD inhibitor MF-438 inhibited CSLC sphere formation in diverse cell lines, but its inhibitory effect was insignificant to 2-D bulk-cultured cells with or without serum in the media, indicating that lipids contained in serum were not associated with the resistance of 2-D cultured bulk cells against MF-438 (Fig. 3C). U87 glioma cells did not grow under serum-free 2-D culture condition; they are thus excluded from this validation. In vitro limiting dilution assays showed that MF-438 effectively reduced self-renewing cells from the survived population of HT-29 cells (Fig. 3D). Moreover, the HMG-CoA reductase inhibitor atorvastatin also inhibited CSLC sphere formation; its effect was reversed by the addition of the downstream products mevalonate or cholesterol (Fig. 3E). Consistent with MF-438, statin efficacy was not observed in 2-D cultured cells under serum-free condition, confirming that serum lipids was not the critical factor in the differential efficacy between 2- and 3-D assays (Supplemental Fig. S2A).
We further examined 4 widely prescribed statins (atorvastatin, fluvastatin, lovastatin, and simvastatin) to test their suitability for CSLC-specific targeting. All 4 statins exhibited superior (100 ∼ 1000 times) inhibitory effects on CSLC survival compared to that of cells in bulk culture (Supplemental Fig. S2B). However, pharmacologic intervention of group 2 and 3 gene products with carfilzomib, ispinesib, diltiazem, or selinexor (genes and inhibitors are listed in Supplemental Table S2) had no significant difference on cell growth between the culture conditions (Supplemental Fig. S2B). Furthermore, fatostatin, a sterol regulatory element binding protein inhibitor that also suppresses lipid biosynthesis, did not show differential inhibitory effects between CSLC and bulk culture, suggesting that specific blockade of cholesterol and unsaturated fatty acid synthesis may be important for selective CSLC targeting (Fig. 3F).
The statins tested also increased apoptosis only in CSLCs, whereas the other inhibitors targeting group 2 and 3 gene products decreased or did not change cell death in CSLC culture (Fig. 3G). The statins also selectively decreased the proportion of cells expressing CSLC marker CD133 or ALDH1 (Fig. 3H, I), though the inhibitors of group 2 and 3 gene products increased the percentage of CD133-positive cells (Fig. 3H), which is reminiscent of the knockdown effect of group 2 and 3 genes that we found (Fig. 2B). These data strongly suggest that cholesterol synthesis is an attractive target, and that drugs such as statins that target this metabolic process have a strong potential to be used in therapeutic strategies targeting CSLCs.
Unique lipid metabolomic profile of CSLCs
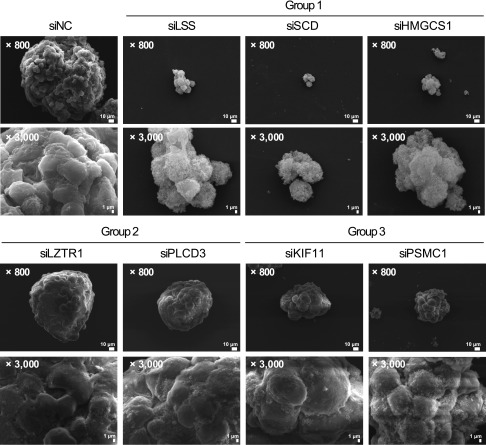
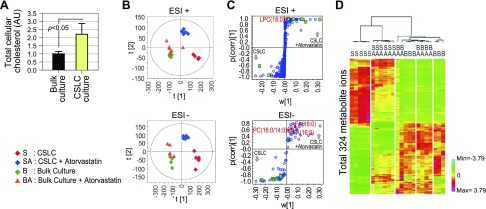
The importance of cholesterol and unsaturated fatty acid synthesis to the CSLC population suggests that the lipid composition of the CSLC plasma membrane may be affected by blocking the lipid synthesis pathway. We examined the surface of CSLC spheres after treatment with group 1–3 siRNAs. Surprisingly, SEM revealed rough irregular structures of the plasma membrane in the CSLC sphere cultures transfected with group 1 siRNAs (targeting HMGCS1, LSS, and SCD; Fig. 4). However, siPSMC1 and siKIF11, which also eliminated the self-renewal activity of CSLCs, did not alter the surface structure of spheres. We hypothesized that these results may be correlated with disrupted membrane compositions due to the blocking lipid anabolism in CSLC spheres. We thus examined the lipid component of CSLCs and bulk-cultured cells in detail. Cholesterol (including its cellular storage form cholesteryl ester) was significantly elevated in CSLCs (Fig. 5A). However, neutral lipid droplets, cellular storage of complex lipids that have been reported to be increased in CSLCs, were not increased in CSLC cells under our conditions and were not significantly altered by statin treatment (19) (Supplemental Fig. S2C). This result suggests that not the total lipid mass but some specific components may contribute to the CSLC sphere integrity. To find out the specific change of lipid components in CSLCs compared to bulk-cultured cells, we analyzed lipidomic profiles in CSLCs and bulk-cultured cells before and after statin treatment using ultraperformance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-qTOF-MS). Unexpectedly, the lipidomic profile was markedly different between CSLCs and bulk cancer cells, and these differences were significantly reduced by statins in CSLCs (Fig. 5B–D). Oleic acid and other unsaturated lipids (mainly monounsaturated phosphatidylcholine and phosphatidylethanolamine) were enriched in CSLCs and were decreased by statins only in these cells (Table 1); in contrast, fully saturated fatty acids were either decreased or did not increase in CSLCs. This result suggests that SCD, which encodes a fatty acid desaturating enzyme that is reported to be increased in stem cells and CSLCs (20), performs an important function in determining the CSLC lipid profile. The levels of increased and decreased lipid components in CSLCs were reversed close to bulk-cultured cells after statin treatment. However, statin treatment did not alter the bulk cell lipidomics, demonstrating that the cholesterol synthetic pathway in bulk cells may not be active. Moreover, statin treatment even decreased the amount of many identified monounsaturated lipids in CSLCs, which is not directly related to the statin mode of action. This may be because statin selectively decreased the cell population that had a CSLC lipidomic profile, and the cells that survived were those having the bulk cultured cell profiles.
Figure 4.
Changes of plasma membrane integrity by inhibition of lipid synthesis in CSLCs. Morphologic analysis of CSLC cultured cells under SEM after siRNA treatment. Disruption of lipid metabolism-related group 1 hits presented modified cell membrane surface and cell–cell interactions.
Figure 5.
Changes of unique lipid profiles by inhibition of lipid synthesis in CSLCs. A) Relative amount of total cholesterol in CSLC and bulk-cultured cells. B) Change of metabolite ion profile (OPLS-DA score plot) in CSLC and bulk-cultured cells by atorvastatin treatment. Multivariate statistical analysis of lipid profiles acquired from UPLC-qTOF-MS data was carried out to define potential markers for each of cell groups. OPLS-DA score plots were derived from metabolite ions acquired from ESI+ and ESI− modes. C) S plot shows covariance w against correlation and p(corr) of variables of discriminating component of OPLS-DA model. Ions of saturated fatty acid (red box) and unsaturated fatty acid (green box) are labeled to be potential markers in S plots. Detailed descriptions are provided in Table 1. D) Heat map for differentially regulated metabolite mass ions between CSLC and bulk-cultured cells by 2-way hierarchical clustering analysis. Each colored cell represents normalized intensity of each mass ion according to color scale.
TABLE 1.
Identified and significantly changed lipid metabolites in CSLC by atorvastatin (10 nM)
| Identified lipid | Fold amount (S/B) | Fold amount (S/SA) |
|---|---|---|
| FA(18:0)c | 0.45 | 0.45 |
| FA(16:0)c | 0.37 | 0.50 |
| PC(16:0/14:0)c | 0.93 | 4.32a,** |
| FA(18:1) (= oleic acid)d | 66.70a,* | 0.43b,* |
| PC(16:0/16:1)d | 3.23a,** | 1.74 |
| PC(18:1/18:1)d | 4.31a,** | 2.11b,** |
| PC(P-18:0/18:1)d | 8.09a,** | 1.55b,* |
| PC(18:1/16:1)d | 17.13a,** | 4.83 b,** |
| LPC(18:0)c | 59.49a,** | 2.80b,** |
| PE[P-18:0/18:1(9Z)] or PE[P-16:0/20:1(11Z)]d | 252.16a,** | 1.72** |
| PE(20:1(11Z)/P-18:0)d | 424.63a,** | 1.79** |
| PE[18:1(9Z)/20:1(11Z)]d | 987.79a,** | 3.02b,* |
| PE[P-16:0/18:1(9Z)]d | 1065.57a,** | 2.38b,** |
| PE(18:1/16:1)d | Undetectable level in Ba,** | 7.06b,** |
| PE(18:1/18:0)d | Undetectable level in Ba,** | 2.18b,* |
| PE(P-16:0/16:1)d | Undetectable level in Ba,** | 4.3 b,** |
| PS(18:1/18:0)d | Undetectable level in Ba,** | 80.24b,** |
S, CSLC spheres; SA, CSLC spheres + atorvastatin (10 nM); B, bulk culture cells; BA, bulk culture cells + atorvastatin (10 nM); FA, fatty acids; LPC, lysophosphatidylcholine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; SM, sphingomyelin. aDetected >2-fold in S than in B. bDecreased >2 fold by statin in S. cSaturated fatty acid. dIncluded unsaturated fatty acid. *P < 0.01, **P < 0.001.
Change of in vivo growth of CSLCs by modulating lipid metabolism
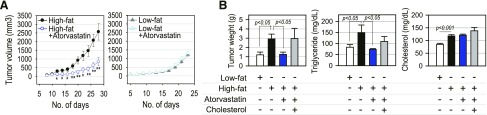
We examined whether lipid metabolism modulation could be the target of CSLC in the effect of statins on CSLCs implanted in immunocompromised mice. In nude mice fed a low-fat diet, daily treatment of atorvastatin did not alter the growth of tumors from the implanted CSLC (Fig. 6A). However, the statin treatment significantly delayed the growth of tumors derived from implanted CSLCs if the mice were fed a high-fat diet (Fig. 6A). These results suggest the statins may better suppress CSLC growth in the enriched lipid availability in circulation. We examined whether the blood lipid concentration and atorvastatin effect on tumor growth were related. The xenograft tumors from CSLC implanted in NOD/SCID mice fed a high-fat (cholesterol-free) diet were larger than those from mice fed a low-fat diet, with significant decreases by atorvastatin (Fig. 6B). However, the inhibitory effect of the statin was not observed when cholesterol was added to the high-fat diet. Circulating TG levels were significantly increased by the high-fat diet and decreased by statin. Although circulating total cholesterol was mildly increased by the high-fat diet, it was not decreased by statin in this study (Fig. 6B), suggesting that the observed in vivo effect of statin might occur through direct effects on cancer cells rather than on a systemic reduction in cholesterol.
Figure 6.
Tumor growth regulation by statins in vivo. A) Effect of atorvastatin on tumor growth of U87 CSLC xenograft in BALB/c Nude mice (high-fat diet, n = 10; high-fat diet and atorvastatin, n = 10; low-fat diet, n = 6; low-fat diet and atorvastatin (n = 6). *P < 0.05, **P < 0.01 vs. high-fat diet. B) In vivo change of tumor growth and circulating blood TG/cholesterol levels in NOD/SCID mice fed by low-fat diet (n = 5), high-fat diet (n = 5), high-fat diet and atorvastatin (n = 5), and high-fat diet plus cholesterol and atorvastatin (n = 4).
DISCUSSION
By performing large-scale loss-of-function screening for druggable genes in CSLC versus bulk cultures of U87 glioma cells, we found that the distribution of CSLC-selective target genes in group 1 and 3 displayed a limited diversity of functional categories. In particular, siRNAs against many isoforms of proteasome 26S ATPase complex genes (PSMC1, PSMC2, PSMC4, PSMC5, PSMC6, PSMD7) were effective on both CSLC and bulk cultured populations. Among validated hits, siPSMC1 showed the best performance in eliminating the self-renewal activity of U87 CSLCs (Fig. 2E), although they did not reduce CD133-expressing populations in the 3-D CSLC culture condition. However, siRNAs targeting the lipid biosynthesis pathway displayed consistent efficacies on reducing self-renewal activity and marker expressing CSLCs. We thus demonstrated that cholesterol and unsaturated fatty acid biosynthesis, which are elevated in CSLCs, were required for CSLC maintenance across multiple cancer lineages. In addition, CSLCs possessed unique lipidomic profiles in comparison to bulk-cultured cells.
The reprogramming of metabolic pathways in cells is now firmly established as a hallmark of cancer (21), and lipid synthesis in individual cancer cells has been shown to be largely modulated (11, 12). Targeting the lipid and cholesterol dependence of cancer cells was recently suggested (11). The higher requirements of cancer cells for fatty acids and cholesterol for proliferation may be fulfilled by reactivating de novo lipid and cholesterol synthesis (11). Interestingly, our results demonstrate that statins do not change the lipid profile in bulk cancer cell, suggesting a specific role of lipid neosynthesis in CSLCs. The lipid rafts that are essential for Notch signaling may be destroyed with a shortage of cholesterol (22). In addition, marked increases in monounsaturated lipid chains may also contribute to the flexibility of the CSLC plasma membrane. The unsaturated fatty acid may enhance the proliferation of embryonic stem cells and myoblasts, suggesting a common metabolism between these stem cells and CSLCs (23). Our SEM observations are interesting because the structural problem in the plasma membrane was generated by blocking the synthesis of cholesterol and many unsaturated lipids. This suggests a correlation of the structural abnormality to the change of lipid composition. The in vivo growth of tumors derived from CSLC was enhanced by a high-fat diet with elevated TG, but statin treatment reversed the growth. It seems that the level of TG in circulation is associated with the in vivo tumor growth.
Our large-scale screening revealed that it was not a specific desaturating enzyme but rather the great change in differences in the lipidome that existed between CSLC and bulk culture cells that was essential for CSLC self-renewal. In addition to genes in lipid biosynthesis, ARL2 and NFKBIL2, associated with another biologic processes, were also found to be CSLC-specific targets in group 1 (Fig. 1C). ASAH1 and PLCB3 in another lipid metabolism pathway were found to be significant targets for both the CSLC and bulk-cultured populations (Supplemental Table S1). Collectively, a total of 35 validated hits from several functional categories provide a useful resource for further studying the molecular mechanism underling CSLC maintenance as well as developing novel approaches targeting CSLCs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation (NRF) of Korea, including the Bio and Medical Technology Development Program (NRF-2012M3A9B6055398 and NRF-2012M3A9B6055466), the Science Research Center Program (NRF-2016R1A5A1011974), and Basic Science Research Program (NRF-2013R1A1A2059115 and NRF-2015R1D1A1A01056594), funded by the Korean government (MEST).
Glossary
- 2-D
2-dimensional
- 3-D
3-dimensional
- ALDH
aldehyde dehydrogenase
- CSLC
cancer stem–like cell
- EGF
epidermal growth factor
- ESI
electrospray ionization
- FACS
fluorescence-activated cell sorting
- GBM
glioblastoma multiforme
- GO
gene ontology
- OPLS-DA
orthogonal partial least-square discriminant analysis
- SEM
scanning electron microscopy
- siRNA
small interfering RNA
- TG
triglyceride
- UPLC-qTOF-MS
ultraperformance liquid chromatography quadrupole time-of-flight mass spectrometry
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
M. Song designed the experimental approach, performed the experimental work, interpreted data, and prepared the article; H. Lee performed in vivo experiments and lipid composition assays, and interpreted associated experiment results; M.-H. Nam performed UPLC-qTOF-MS assay and analyzed the results; M. Song, S. Kim, and H. Y. Yim performed siRNA screening experiments; E. Jeong, Y. Hong, and N. Kim conducted bioinformatics analyses; J. S. Kim and J.-S. Kim performed in vivo experiments; S. Yoon, W.-Y. Kim, Y.-Y. Cho, and G. B. Mills provided scientific direction; M. Song, S. Yoon, and W.-Y. Kim wrote the article; S. Yoon and W.-Y. Kim designed and coordinated the overall research and experiments; and all authors discussed the results and commented on the article.
REFERENCES
- 1.Beck B., Blanpain C. (2013) Unravelling cancer stem cell potential. Nat. Rev. Cancer 13, 727–738 [DOI] [PubMed] [Google Scholar]
- 2.Liu S., Wicha M. S. (2010) Targeting breast cancer stem cells. J. Clin. Oncol. 28, 4006–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matchett K. B., Lappin T. R. (2014) Concise reviews: cancer stem cells: from concept to cure. Stem Cells 32, 2563–2570 [DOI] [PubMed] [Google Scholar]
- 4.Wicha M. S., Liu S., Dontu G. (2006) Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 66, 1883–1890 [DOI] [PubMed] [Google Scholar]
- 5.Jung Y., Kim W. Y. (2015) Cancer stem cell targeting: are we there yet? Arch. Pharm. Res. 38, 414–422 [DOI] [PubMed] [Google Scholar]
- 6.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N. M., Pastorino S., Purow B. W., Christopher N., Zhang W., Park J. K., Fine H. A. (2006) Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 [DOI] [PubMed] [Google Scholar]
- 7.Ginestier C., Monville F., Wicinski J., Cabaud O., Cervera N., Josselin E., Finetti P., Guille A., Larderet G., Viens P., Sebti S., Bertucci F., Birnbaum D., Charafe-Jauffret E. (2012) Mevalonate metabolism regulates basal breast cancer stem cells and is a potential therapeutic target. Stem Cells 30, 1327–1337 [DOI] [PubMed] [Google Scholar]
- 8.Song M., Jeong E., Lee T. K., Tsoy Y., Kwon Y. J., Yoon S. (2015) Analysis of image-based phenotypic parameters for high throughput gene perturbation assays. Comput. Biol. Chem. 58, 192–198 [DOI] [PubMed] [Google Scholar]
- 9.Sun C., Wang L., Huang S., Heynen G. J., Prahallad A., Robert C., Haanen J., Blank C., Wesseling J., Willems S. M., Zecchin D., Hobor S., Bajpe P. K., Lieftink C., Mateus C., Vagner S., Grernrum W., Hofland I., Schlicker A., Wessels L. F., Beijersbergen R. L., Bardelli A., Di Nicolantonio F., Eggermont A. M., Bernards R. (2014) Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508, 118–122 [DOI] [PubMed] [Google Scholar]
- 10.Gilbert C. A., Ross A. H. (2009) Cancer stem cells: cell culture, markers, and targets for new therapies. J. Cell. Biochem. 108, 1031–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. (2016) Lipid metabolic reprogramming in cancer cells. Oncogenesis 5, e189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos C. R., Schulze A. (2012) Lipid metabolism in cancer. FEBS J. 279, 2610–2623 [DOI] [PubMed] [Google Scholar]
- 13.Van Vugt M. A. T. M., Medema R. H. (2005) Getting in and out of mitosis with Polo-like kinase-1. Oncogene 24, 2844–2859 [DOI] [PubMed] [Google Scholar]
- 14.Mazur S., Kozak K. (2012) Z′ factor including siRNA design quality parameter in RNAi screening experiments. RNA Biol. 9, 624–632 [DOI] [PubMed] [Google Scholar]
- 15.Wishart D. S., Jewison T., Guo A. C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E., Bouatra S., Sinelnikov I., Arndt D., Xia J., Liu P., Yallou F., Bjorndahl T., Perez-Pineiro R., Eisner R., Allen F., Neveu V., Greiner R., Scalbert A. (2013) HMDB 3.0—the Human Metabolome Database in 2013. Nucleic Acids Res. 41, D801–D807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., Suwa K., Ojima Y., Tanaka K., Tanaka S., Aoshima K., Oda Y., Kakazu Y., Kusano M., Tohge T., Matsuda F., Sawada Y., Hirai M. Y., Nakanishi H., Ikeda K., Akimoto N., Maoka T., Takahashi H., Ara T., Sakurai N., Suzuki H., Shibata D., Neumann S., Iida T., Tanaka K., Funatsu K., Matsuura F., Soga T., Taguchi R., Saito K., Nishioka T. (2010) MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714 [DOI] [PubMed] [Google Scholar]
- 17.Ivanova P. T., Milne S. B., Myers D. S., Brown H. A. (2009) Lipidomics: a mass spectrometry based systems level analysis of cellular lipids. Curr. Opin. Chem. Biol. 13, 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidal S. J., Rodriguez-Bravo V., Galsky M., Cordon-Cardo C., Domingo-Domenech J. (2014) Targeting cancer stem cells to suppress acquired chemotherapy resistance. Oncogene 33, 4451–4463 [DOI] [PubMed] [Google Scholar]
- 19.Tirinato L., Liberale C., Di Franco S., Candeloro P., Benfante A., La Rocca R., Potze L., Marotta R., Ruffilli R., Rajamanickam V. P., Malerba M., De Angelis F., Falqui A., Carbone E., Todaro M., Medema J. P., Stassi G., Di Fabrizio E. (2015) Lipid droplets: a new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells 33, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-David U., Gan Q. F., Golan-Lev T., Arora P., Yanuka O., Oren Y. S., Leikin-Frenkel A., Graf M., Garippa R., Boehringer M., Gromo G., Benvenisty N. (2013) Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell 12, 167–179 [DOI] [PubMed] [Google Scholar]
- 21.Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 22.Mollinedo F., Gajate C. (2015) Lipid rafts as major platforms for signaling regulation in cancer. Adv. Biol. Regul. 57, 130–146 [DOI] [PubMed] [Google Scholar]
- 23.Das U. N. (2011) Essential fatty acids and their metabolites as modulators of stem cell biology with reference to inflammation, cancer, and metastasis. Cancer Metastasis Rev. 30, 311–324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.