Hematopoietic stem cells (HSCs) are responsible for maintaining a sufficient pool of self-renewing cells that can continuously differentiate into lineage-specific hematopoietic cells throughout the lifetime of an individual (1). The choice to self-renew or differentiate is most likely achieved through coordination of extrinsic and intrinsic cell fate signals that drive symmetrical or asymmetrical cell divisions. Several examples of extrinsic signals have been described that regulate HSC self-renewal or differentiation, including Notch signaling in the bone marrow niche (2). Intrinsic mechanisms that regulate these alternative modes of HSC division are still relatively unknown; however, alterations in epigenetic inheritance have been hypothesized as a driving factor. DNA methylation is a heritable epigenetic mark that is highly symmetrical, where 99% of methylated CpG dinucleotides are found to be methylated on both DNA strands (3). DNA methylation patterns are reestablished upon cell division during S phase by the action of DNA methyltransferase 1 (DNMT1). DNMT1 recognizes hemimethylated DNA and remethylates the newly synthesized daughter strand, facilitating epigenetic inheritance in the genome during cell division. In PNAS, a study by Zhao et al. (4) describes how disruption in DNA methylation maintenance has the potential to alter HSC division modes and deregulate lineage-specific gene expression spontaneously, ultimately blocking the self-renewal capacity of HSCs, which leads to their rapid depletion.
The molecular mechanism by which DNA methylation regulates transcription and lineage specification is known to be dependent on the complement of factors that can recognize, and are recruited at, sites of differential methylation. Gene promoters often harbor regions of high CpG density known as CpG islands (CGIs) that are typically hypomethylated. Hypomethylation in promoter CGIs may serve as a recruitment signal for transcriptional activators or CXXC-domain–containing proteins that establish a transcriptionally active chromatin (5). Alternatively, methylated CpGs could prevent binding of transcriptional regulators or attract methyl-CpG–binding proteins that specifically recognize methylation marks and recruit histone-modifying enzymes and chromatin remodeling factors to silence chromatin (6).
Whether changes in DNA methylation patterns are the cause or consequence of self-renewal or differentiation cues in hematopoietic cells has been explored using genetic ablation of regulators of DNA methylation in mice. Deletion or hypomorphic loss of function in Dnmt1 impairs the self-renewal capacity of HSCs, drastically reducing HSC numbers, suggesting that a critical threshold of DNA methylation is required to maintain homeostasis within the HSC pool (7, 8). On the contrary, deficiency in the de novo methyltransferases Dnmt3a and Dnmt3b promotes increased HSC self-renewal, yet blocks differentiation of all mature hematopoietic lineages (9). Similarly, loss of function in members of the Ten-Eleven-Translocation (TET) family of DNA demethylases promotes increased HSC and progenitor cell self-renewal, but also lineage-specific differentiation bias (10).
In the study by Zhao et al. (4), ubiquitin-like with PHD and ring finger domain 1 (UHRF1), a regulator of DNA methylation maintenance, has now been shown to regulate self-renewal and differentiation fate epigenetically in HSCs. Previous studies found that systemic deletion of Uhrf1 is embryonically lethal and Uhrf1-deficient embryonic stem cells exhibit a dramatic loss in DNA methylation (11, 12). UHRF1 (also known as ICBP90 in human and Np95 in mouse) specifically recognizes hemimethylated DNA via a SET and RING-associated (SRA) domain and is an essential cofactor of maintenance methylation by recruiting DNMT1 to replication forks during S phase (11–13). The majority of defects observed in Uhrf1-deficient HSCs in the study by Zhao et al. (4) phenocopy those defects seen with Dnmt1 deficiency. Pan-hematopoietic deletion of Uhrf1 is embryonically lethal due to fetal liver (FL)-HSC depletion, and inducible deletion in adult HSCs also causes a rapid decline in survival and loss of total HSC numbers. In addition, Uhrf1-deficient HSCs lose their capacity for self-renewal in competitive bone marrow transplantation assays.
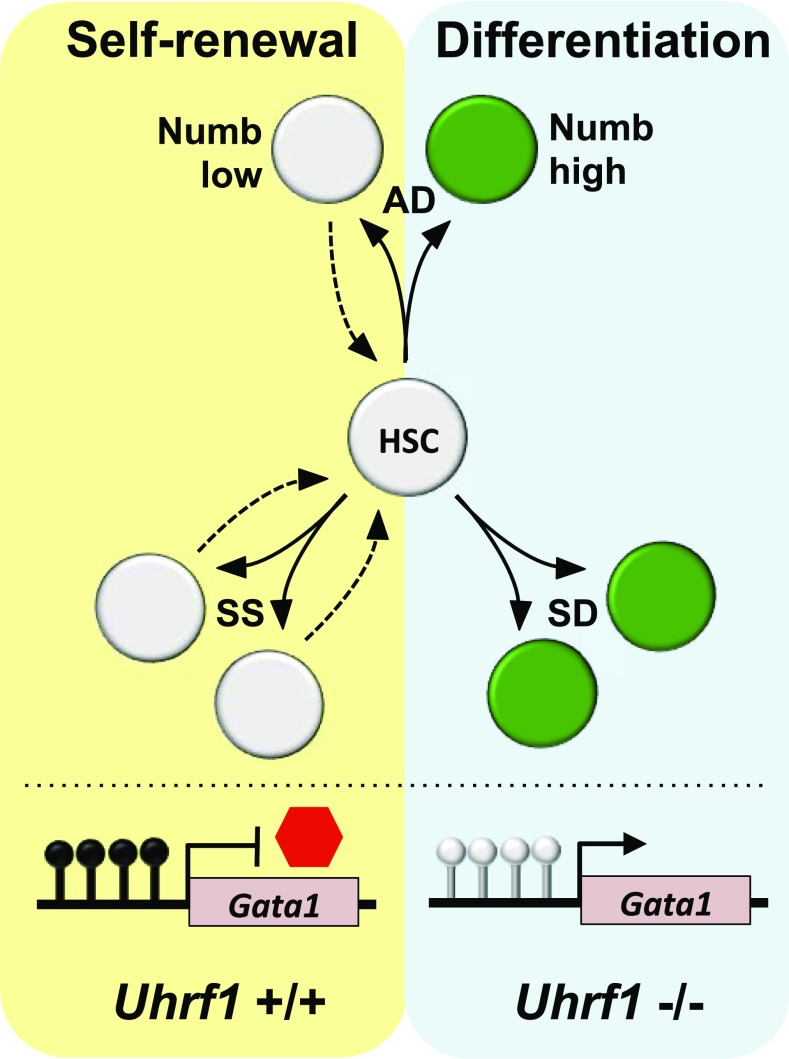
The mechanism behind the altered HSC homeostasis upon Uhrf1 deletion was found to be independent of changes in cell cycle and apoptosis, which is consistent with previous reports for Dnmt1-deficient HSCs (7, 8). Both FL and adult hematopoietic stem and progenitor cells (HSPCs) produce fewer total colony-forming units in methylcellulose assays upon conditional loss of Uhrf1 and are almost exclusively erythroid lineage-restricted colonies. To understand the decreased output and biased differentiation of Uhrf1−/− HSCs, the expression of the endogenous Notch inhibitor Numb was used to trace the cell fate of Uhrf1-deficient HSPCs in single-cell division studies with regard to Notch signaling potential. Asymmetrical segregation of Numb protein expression in mother/daughter cell doublets provides a readout for alterations in notch signaling potential, suggestive of asymmetrical distribution of differentiation potential in HSPCs (14). Zhao et al. (4) monitored the three distinct outcomes of Numb distribution in both fetal and adult HSCs to assess symmetrical self-renewal (SS; Numb low), symmetrical differentiation (SD; Numb high), or asymmetrical division (AD; Numb low and high) among cell progeny (Fig. 1). Strikingly, it was found that SD cell division events increased significantly in Uhrf1-deficient HSCs, whereas SS divisions decreased. Using a division-sensitive dye to follow proliferation and maintenance of HSCs after Uhrf1 deletion, it was confirmed that Uhrf1 loss promotes a nonrenewable cell division bias that leads to depletion of the HSC pool but an increase in Kit+ cells. These Kit+ cells, however, were found to be unable to differentiate into common myeloid progenitor or granulocyte and myeloid-restricted progenitor cells and, instead, formed exclusively myeloerythroid progenitor (MEP) cells in vitro in keeping with the substantial retention in MEP numbers observed in Uhrf1−/− mice in vivo despite a differentiation block in all other progenitors and mature lineages.
Fig. 1.
Uhrf1 regulates the modes of HSC division, leading to self-renewal or differentiation. Expression of the endogenous Notch inhibitor Numb was used to trace the cell fate of Uhrf1+/+ or Uhrf1−/− HSPCs in single-cell division studies. Low Numb expression is associated with maintenance of self-renewal, whereas high Numb expression suggests an acquisition in differentiation potential. SD cell division events, which display higher Numb expression in mother/daughter cell pairs, increases significantly in dividing Uhrf1−/− HSCs, whereas SS divisions with low Numb expression in mother/daughter cell pairs decreases. The frequency of AD in HSCs does not significantly change in the absence of Uhrf1. The promoter of Gata1, a master regulator of erythroid lineage commitment, is methylated in Uhrf1+/+ fetal and adult HSCs (black lollipops); however, these modifications are lost in Uhrf1−/− HSCs (gray lollipops), causing derepression of Gata1 expression and the activation of an erythroid-specific gene expression program. Hypomethylation of the Gata1 promoter was also shown to be enriched in daughter cells displaying high Numb expression, a hallmark of HSCs undergoing differentiation. Loss of DNA methylation maintenance in Uhrf1−/− HSCs upon methylation causes a rapid depletion of the HSC pool, with a bias toward SD into erythroid lineage cells.
HSC self-renewal has been shown to require constitutive DNA methylation, whereas erythroid lineage commitment and differentiation are associated with widespread progressive hypomethylation (15, 16). Drastic changes in DNA methylation precede transcriptional alterations, which begin with promoter demethylation of erythroid lineage-specific transcription factors that then reinforce commitment by binding to additionally hypomethylated regions of the erythroid progenitor genome. The study by Zhao et al. (4) provides a direct link between maintenance of DNA methylation and its role in suppressing erythroid differentiation, promoting gene expression programs during cell division to control cell fate decision making. The master regulator of erythroid lineage commitment, Gata1, loses methylation in the proximal promoter in Uhrf1−/− fetal and adult HSCs, which becomes exacerbated after successive cell divisions in vitro. Hypomethylation of the Gata1 promoter was also shown to be enriched in daughter cells displaying high Numb expression, a hallmark of HSCs undergoing differentiation. The increased expression of Numb was not found to be a direct consequence of Uhrf1 loss, but rather a marker of a switch in the proportion of HSC division modes that produce daughter cells through SD. In this way, the stem cell pool rapidly becomes depleted while spontaneously biasing them toward erythroid lineage differentiation.
These studies lead to a greater understanding of how readers of DNA methylation influence cell fate and lineage choices. UHRF1, in addition to binding 5-methylcytosine (5mC), was also the first protein identified as a binder of 5-hydroxymethylcytosine (5hmC) (17, 18). Structural studies have also shown that the SRA domain of UHRF1 can bind both hemimethylated and hemihydroxymethylated CpG sites with the same affinity (17). Hydroxylation of 5mC is regulated by the TET DNA demethylases to generate 5hmC, 5-formylcytosine, and 5-carboxylcytosine (5caC) through iterative oxidation reactions. These oxidized methylcytosines act as unique epigenetic marks in the genome, and their roles in the regulation of transcription and DNA demethylation are still being elucidated (18, 19). Both UHRF1 and DNMT1 can specifically interact with additional oxidized forms of cytosine such as 5caC; however, although UHRF1 binds 5mC and 5hmC with similar high affinity, DNMT1 has a low binding affinity for 5hmC (17, 18). The inability to recognize 5hmC by DNMT1 may by one mechanism by which locus-specific DNA demethylation can be achieved upon DNA replication. UHRF1 is also involved in cross-talk between DNA and histone methylation to direct the maintenance of DNA methylation (20). UHRF1-chromatin–binding profiles have not yet been reported in hematopoietic cells but would be invaluable for future studies that should focus on mapping the global methylation changes in Uhrf1-deficient mother/daughter HSC divisions in the context of 5mC and 5hmC distribution to help understand how this adaptor of DNA methylation maintenance relies on locus-specific cues in the genome to regulate self-renewal or differentiation.
This study highlights how the inheritance of DNA methylation can exert a cell-intrinsic influence over the mode of HSC division. Excessive SS may lead to the inappropriate expansion of stem and progenitor cell compartments, an underlying feature of malignant hematopoiesis. Excessive AD or, in the case of Uhrf1 deficiency, elevated SD rates in daughter cells would rapidly lead to stem cell exhaustion. Promoting SD may therefore be an important tumor-suppressive mechanism and provide a potential therapeutic strategy to restore the imbalance in lineage fate determination and self-renewal associated with leukemic stem cells.
Footnotes
The author declares no conflict of interest.
See companion article on page E142.
References
- 1.Bryder D, Rossi DJ, Weissman IL. Hematopoietic stem cells: The paradigmatic tissue-specific stem cell. Am J Pathol. 2006;169(2):338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132(4):583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao J, et al. Uhrf1 controls the self-renewal versus differentiation of hematopoietic stem cells by epigenetically regulating the cell-division modes. Proc Natl Acad Sci USA. 2017;114:E142–E151. doi: 10.1073/pnas.1612967114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464(7291):1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fournier A, Sasai N, Nakao M, Defossez PA. The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief Funct Genomics. 2012;11(3):251–264. doi: 10.1093/bfgp/elr040. [DOI] [PubMed] [Google Scholar]
- 7.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bröske AM, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 9.Challen GA, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15(3):350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillamot M, Cimmino L, Aifantis I. The impact of DNA methylation in hematopoietic malignancies. Trends Cancer. 2016;2(2):70–83. doi: 10.1016/j.trecan.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 12.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 13.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455(7214):818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 14.Wu M, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1(5):541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y, et al. High resolution methylome analysis reveals widespread functional hypomethylation during adult human erythropoiesis. J Biol Chem. 2013;288(13):8805–8814. doi: 10.1074/jbc.M112.423756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shearstone JR, et al. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334(6057):799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frauer C, et al. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6(6):e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Ko M, et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263(1):6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothbart SB, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19(11):1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]