Significance
A great challenge in microbial ecology lies in determining the underlying mechanisms that drive interactions in complex natural communities. In this study we used bacterial isolates from Lake Washington sediment that are important for the utilization of the greenhouse gas methane. We show how simple cocultures can be used to identify mechanisms involved in cross-feeding in microbial communities; these mechanisms cannot be deduced from pure cultures alone. We demonstrate that the presence of one species alters gene expression and metabolism in another species such that the second species excretes a carbon and energy source to sustain the cross-fed species. This mode of cross-feeding based on partner-induced altered gene expression may have important implications for microbial interactions in the environment.
Keywords: synthetic ecology, eco-physiology, flow cytometry, Methylobacter, metabolic transformation
Abstract
The utilization of methane, a potent greenhouse gas, is an important component of local and global carbon cycles that is characterized by tight linkages between methane-utilizing (methanotrophic) and nonmethanotrophic bacteria. It has been suggested that the methanotroph sustains these nonmethanotrophs by cross-feeding, because subsequent products of the methane oxidation pathway, such as methanol, represent alternative carbon sources. We established cocultures in a microcosm model system to determine the mechanism and substrate that underlay the observed cross-feeding in the environment. Lanthanum, a rare earth element, was applied because of its increasing importance in methylotrophy. We used co-occurring strains isolated from Lake Washington sediment that are involved in methane utilization: a methanotroph and two nonmethanotrophic methylotrophs. Gene-expression profiles and mutant analyses suggest that methanol is the dominant carbon and energy source the methanotroph provides to support growth of the nonmethanotrophs. However, in the presence of the nonmethanotroph, gene expression of the dominant methanol dehydrogenase (MDH) shifts from the lanthanide-dependent MDH (XoxF)-type, to the calcium-dependent MDH (MxaF)-type. Correspondingly, methanol is released into the medium only when the methanotroph expresses the MxaF-type MDH. These results suggest a cross-feeding mechanism in which the nonmethanotrophic partner induces a change in expression of methanotroph MDHs, resulting in release of methanol for its growth. This partner-induced change in gene expression that benefits the partner is a paradigm for microbial interactions that cannot be observed in studies of pure cultures, underscoring the importance of synthetic microbial community approaches to understand environmental microbiomes.
Microbial communities and their members are part of every ecosystem and drive important biogeochemical processes on Earth (1). They typically comprise a range of phylogenetically and functionally diverse microbes (2) that are structured by biotic and abiotic factors (3) and are entangled through specific interactions in complex networks (4).
The significance of microbial communities in diverse ecosystems including the human body is now widely accepted in science and has led to a range of initiatives focused on the world’s microbiomes (5, 6). One important goal within these efforts is to understand how microbes interact with each other and the consequences of such interactions at the level of their transcriptomes, proteomes, and metabolomes. For instance, it has been demonstrated that the metabolism of yeast can be transformed by bacteria-induced prions to decrease the release of inhibiting ethanol (7). In another study, an oral biofilm of the genus Streptococcus displayed different transcriptional responses toward the presence of other species in mixed-species cultures (8).
Measuring interactions in complex environmental communities is still a difficult task. Laboratory cocultures represent a simplified approach to assessing cell–cell interactions, allowing controlled manipulation and detailed analysis of the individual strains. Measurement of metabolic interactions in synthetic cocultures addresses a key feature of known interaction patterns (9, 10) that can drive species co-occurrence in microbial communities (11). A model system that is particularly well-suited to such coculture approaches is methane utilization in natural communities.
Microbial methane utilization plays a significant role in global climate and is an important part of the carbon cycle (12, 13). Aerobic and anaerobic methane oxidation are the only two processes known for biotic methane consumption (12, 14). Aerobic methane-oxidizing bacteria (methanotrophs), in particular, play a vital role in the global climate because they can mitigate up to 90% of the biogenically produced methane in soils and sediments (15, 16). To date, aerobic methanotrophs are found in the phyla Verrucomicrobia and Proteobacteria and in the candidate division NC10. Within the proteobacterial aerobic methanotrophs, they belong to the Gammaroteobacteria, which primarily use the ribulose monophosphate cycle for assimilation (type I methanotrophs), and to the Alphaproteobacteria, which use the serine cycle for assimilation (type II methanotrophs) (17).
The application of stable isotope probing (SIP) of 13C-labeled methane to environmental communities has demonstrated that methanotrophs co-occur with other nonmethanotrophic bacteria and function collectively with them as a community to consume methane (18–25). For instance, in the sediment of Lake Washington methane-utilizing communities are not random; they are dominated by methanotrophs within the family Methylococcaceae and nonmethanotrophic methylotrophs within the family Methylophilaceae and also include other specific non–methane-utilizing heterotrophs (18, 19). These studies have shown that the methanotrophs support a community that cannot use methane directly, cross-feeding the nonmethanotrophs with methane-derived carbon. Hence, this group is well-positioned for studying questions of species interactions in microbial communities.
In the aerobic methanotrophs, methane is oxidized to methanol by a monooxygenase, and methanol is further oxidized to formaldehyde by a methanol dehydrogenase (MDH). Traditionally, this reaction was thought to be catalyzed by a calcium-dependent MDH (MxaF). However, recent findings identified an additional, lanthanide-dependent MDH (XoxF), that is widespread in bacterial methylotrophs and dominates under some growth conditions (26–28).
Because methanol is produced in the periplasm and therefore can diffuse out of the cell (29), it is reasonable to suggest that methanol might be cross-fed to nonmethanotrophic methylotrophs involved in community-based methane utilization (18, 30, 31). However, Kalyuzhnaya and colleagues (32) have demonstrated a novel form of fermentation-based methanotrophy that releases compounds such as formate, acetate, or hydrogen as additional possible carbon and energy sources for nonmethanotrophic heterotrophs. Thus, the substrates and mechanisms that drive cross-feeding from methanotrophs and allow nonmethanotrophic heterotrophs to co-occur are still unknown.
In this study we address the question of cross-feeding between methanotrophs and nonmethanotrophic methylotrophs by establishing two-species communities in a microcosm model system using a methanotroph of the genus Methylobacter (Methylococcaceae) and two strains of nonmethanotrophic methylotrophs of the genus Methylotenera (Methylophilaceae). These isolates originated from Lake Washington sediment (33–35), a freshwater ecosystem, which is characterized by a dynamic turnover of methane (16) and strong co-occurrence patterns of methanotrophs and nonmethanotrophic methylotrophs (36, 37). The isolates used are representative of strains that dominate methane-enriched microcosms and 13C SIP studies of sediment (13). We combined a transcriptomics approach, real-time quantitative PCR (qRT-PCR), flow cytometry, and gas chromatography to elucidate the substrates and mechanisms determining the co-occurrence patterns typically observed in natural communities. We demonstrate that in the presence of the co-occurring species the methanotroph metabolism is altered by a change in the predominant MDH expression levels. As a result, methanol is excreted to be used by the co-occurring methylotroph as a carbon and energy source. This study links microbial metabolism and microbial ecology and suggests a mode of metabolic interactions that is likely to be important in environmental microbial communities in lake sediments.
Results
Establishing Cocultures from Isolates as Observed in the Environment.
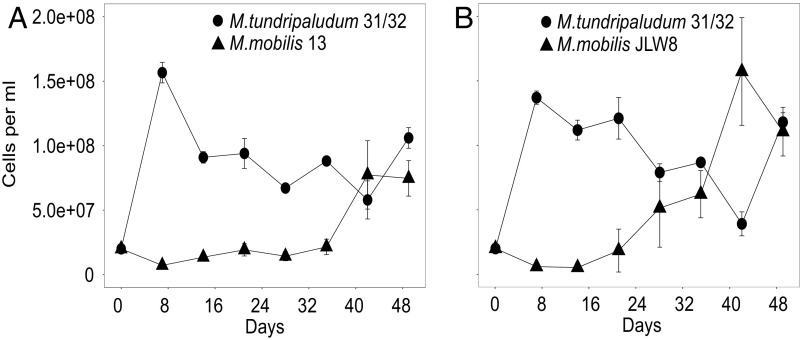
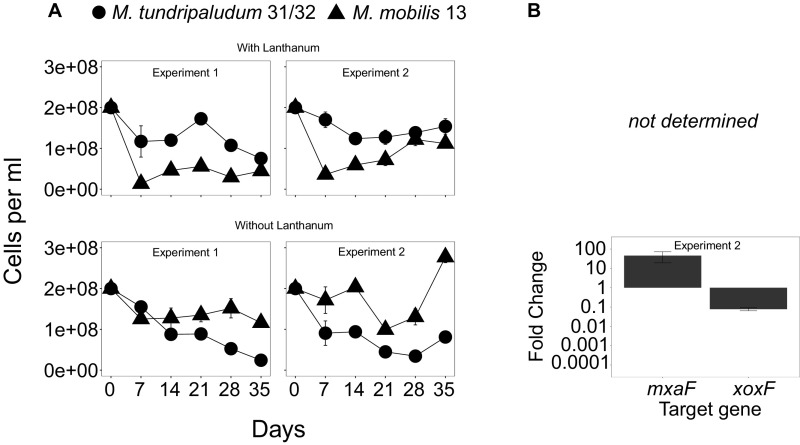
We first verified that it is possible to establish cocultures of a methanotroph from Lake Washington, Methylobacter tundripaludum 31/32 (35), and nonmethanotrophic methylotrophs from Lake Washington, Methylotenera mobilis 13 and JLW8 (33), in the laboratory using a microcosm model system with methane as the sole carbon source (Fig. 1). This microcosm model system is based on that used to enrich methane-consuming communities, which resulted in dominance of M. tundripaludum 31/32 and M. mobilis strains similar to those found in sediment (37). Cells were grown with methane under low initial O2, with daily headspace replacement and weekly culture transfer to new medium (36). In these conditions, the abundances of both populations fluctuated over time but resulted in cocultured populations (Fig. 1 and Fig. S1 A and B) with ratios ranging from 10:90 to 50:50 (M. tundripaludum 31/32:M. mobilis strains) over the course of these experiments, similar to the ratios previously observed in enrichments of natural samples (18, 19, 36). Attempts to coculture the methanotroph with a nonmethanotrophic heterotroph (Escherichia coli) did not result in a stable coculture (Fig. S1C).
Fig. 1.
Cell-abundance dynamics of individual species in model synthetic communities grown on methane as the sole carbon source in 28-mL glass tubes. Different symbols depict individual abundances of M. tundripaludum 31/32 with M. mobilis 13 (A) or M. mobilis JLW8 (B). Individual abundances were determined by flow cytometry. Error bars indicate the SE (n = 3).
Fig. S1.
Cell-abundance dynamics of individual species in model artificial communities grown on methane as the sole carbon source. Individual abundances were determined by flow cytometry. (A and B) Individual abundances of M. tundripaludum 31/32 with M. mobilis 13 (A) or with M. mobilis JLW8 (B) grown in 250-mL glass vials. The gray area depicts the time point that was sampled for RNA extraction and subsequent RNA-seq analysis. Error bars indicate the SE (n = 2). (C) Individual abundances of M. tundripaludum 31/32 with Escherichia coli (strain K-12 MG 1655) grown in 28-mL glass tubes. Error bars indicate the SE (n = 3). (D) Individual abundances of the M. mobilis JLW8 xoxF12 (mmol_1770, mmol_2048) double mutant grown in pure culture (with methylamine) and in the presence of M. tundripaludum 31/31 (with methane). (E) Individual abundances of M. mobilis JLW8 wild type and JLW8 xoxF1 (mmol_1170) and xoxF2 (mmol_2048) single mutants in the presence of M. tundripaludum 31/32 (with methane) in 250-mL glass vials. Error bars indicate the SE (n = 3).
Evidence for Cross-Feeding of Methanol from the Methanotroph to the Nonmethanotroph.
We next analyzed the transcriptome of the methanotroph and the nonmethanotrophic methylotrophs in pure culture and in coculture to identify the key genes expressed exclusively in the presence of the co-occurring species. Samples were taken from cocultures at a ratio of 11:89 (M. mobilis 13 to M. tundripaludum 31/32) and 25:75 (M. mobilis JLW8 to M. tundripaludum 31/32) after 31 d of coculturing (Fig. S1 A and B, gray shaded area). Statistics for the transcriptomics results are shown in Table S1.
Table S1.
Statistics of the transcriptomics run
| Sample ID | Total reads | Reads mapped | Reads mapped to rRNA | Reads mapped to tRNA | Reads mapped to CDS | Reads mapped to hypothetical proteins |
| MB_31_BR1 | 37,578,230 | 0.9649 | 0.0001 | 0.0017 | 0.7805 | 0.0977 |
| MB_31_BR2 | 34,925,111 | 0.9921 | 0 | 0.0014 | 0.8309 | 0.0971 |
| MIX_31_13_BR1 | 35,743,735 | 0.9794 | 0.0001 | 0.0016 | 0.8172 | 0.1016 |
| MIX_31_13_BR2 | 36,988,476 | 0.9747 | 0.0001 | 0.0014 | 0.8224 | 0.095 |
| MIX_31_JLW8_BR1 | 33,031,875 | 0.9915 | 0 | 0.0016 | 0.7756 | 0.0809 |
| MIX_31_JLW8_BR2 | 36,246,187 | 0.9885 | 0 | 0.0018 | 0.8036 | 0.0996 |
| MT_13_BR1 | 37,345,825 | 0.9788 | 0 | 0.0009 | 0.7553 | 0.1212 |
| MT_13_BR2 | 35,935,069 | 0.9672 | 0 | 0.0009 | 0.6927 | 0.1116 |
| MT_JLW8_BR1 | 36,639,981 | 0.9954 | 0 | 0.003 | 0.7802 | 0.0941 |
| MT_JLW8_BR2 | 35,394,607 | 0.957 | 0 | 0.0023 | 0.8018 | 0.095 |
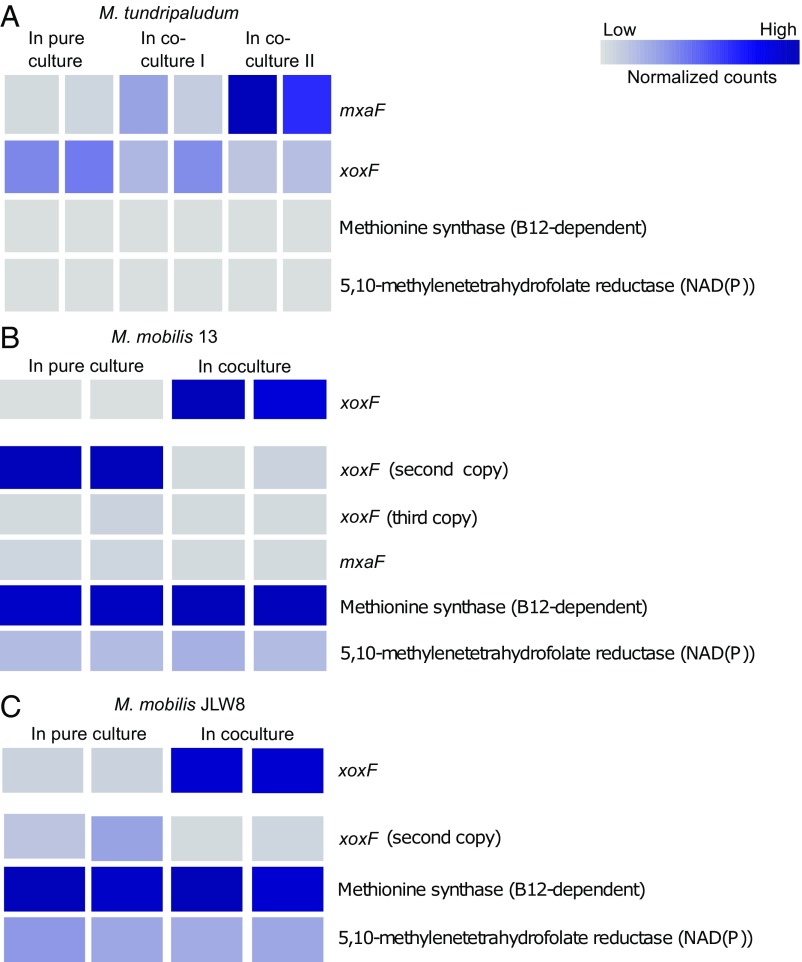
As expected, the key genes involved in methane oxidation were the most highly expressed genes in the methanotroph (Dataset S1). The highly expressed genes in the nonmethanotrophic methylotrophs included the genes for methanol oxidation both in pure cultures grown on methanol as substrates and in cocultures grown with the methanotroph and methane as substrate (Fig. 2 and see Datasets S2 and S3).
Fig. 2.
Heatmaps depicting selected gene-expression data (normalized counts computed with DESeq2) of the lanthanide-dependent XoxF-type MDH, the calcium-dependent MxaF-type MDH, and two housekeeping genes. Housekeeping genes were selected as predicted to be essential biosynthetic steps not involved in methylotrophy and to represent genes expressed at both low and high levels. (A) M. tundripaludum 31/32 genes grown in the presence or absence of M. mobilis 13 (coculture I) or M. mobilis JLW8 (coculture II). (B) M. mobilis 13 genes grown in the presence or absence of M. tundripaludum 31/32. (C) M. mobilis JLW8 genes grown in the presence or absence of M. tundripaludum 31/32. Expression of the first xoxF gene in B and C had much higher values and is displayed separately. The full gene-expression datasets for all three organisms are in Datasets S1–S3 (n = 2 for each setup).
To test the hypothesis that methanol is a main carbon and energy source cross-fed by the methanotroph, we used mutants of the nonmethanotrophic methylotroph M. mobilis JLW8 (38). This strain can grow on both methylamine and methanol, allowing growth in the absence of the ability to oxidize methanol. M. mobilis JLW8 does not contain the MxaF-type MDH but possesses two gene copies of the XoxF-type MDH, which were deleted by single-knockout (xoxF1, xoxF2) and double-knockout (xoxF12) mutations (38). Although we established cocultures with each single mutant, cocultures were not established with the double mutant, which is unable to grow on methanol (Fig. S1D).
Divergence in Gene Expression of the Two MDHs in the Presence of a Coculture.
The transcriptomics dataset also provided evidence for differential expression in the coculture compared with the single cultures. In the methanotroph, we identified a clear shift in the expression of the two MDH genes in the presence of M. mobilis JLW8 compared with the pure culture (Fig. 2A). Replicates of the methanotroph grown with M. mobilis 13 showed the same shifts but were more moderate (Fig. 2A). Overall, expression of the lanthanide-dependent XoxF-type MDH decreased two- to threefold, whereas that of the calcium-dependent MxaF-type MDH increased five- to 23-fold. Housekeeping genes did not change significantly (Fig. 2A). In addition, one of the XoxF-type MDHs in both nonmethanotrophic methylotrophs was expressed only in the coculture (Fig. 2 B and C).
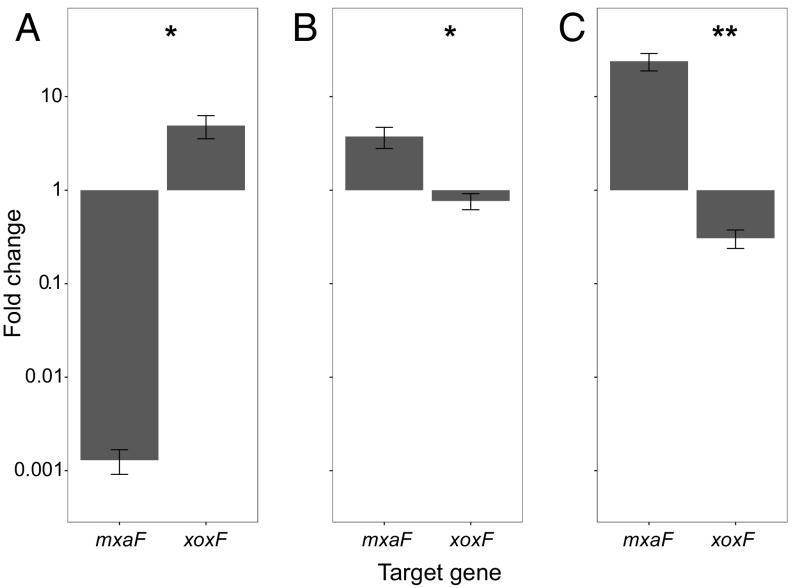
The expression of the methanotroph MxaF-type MDH in the cocultures was unexpected because 30 μM lanthanum was included in all cultures, and recent studies with other methanotrophs have shown that the XoxF-type MDH should be the dominant MDH expressed in the presence of lanthanum (26, 27). To verify these observations, we repeated this experiment with an independent set of cultures and used qRT-PCR to analyze the gene expression of xoxF and mxaF in the methanotroph with and without a co-occurring species (Fig. 3). We first confirmed that in pure cultures of M. tundripaludum 31/32 grown identically to the cocultures, the XoxF-type MDH is the dominant MDH expressed in the presence of lanthanum.
Fig. 3.
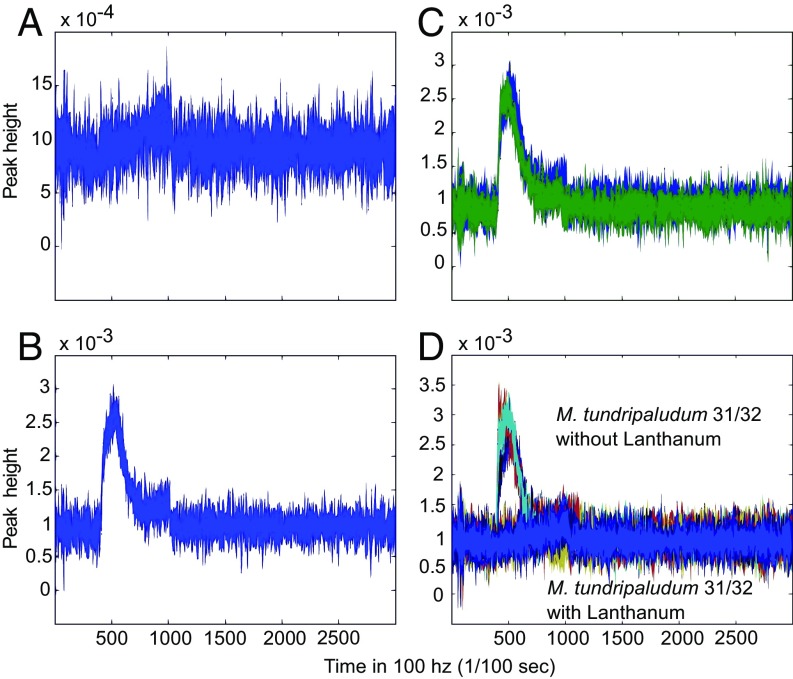
qRT-PCR values depicting the fold change in expression of M. tundripaludum 31/32 mxaF and xoxF genes in pure culture with or without 30 μM lanthanum chloride (A), in the presence or absence of M. mobilis 13 (both with 30 μM lanthanum chloride) (B), or in the presence or absence of M. mobilis JLW8 (both with 30 μM lanthanum chloride) (C). Cell cultures were grown for 3 wk before harvesting as described in Materials and Methods. A t-test was performed to determine the significance of changes in gene-expression levels; **P < 0.01, *P < 0.05. Error bars indicate the SE (n = 4) in A, (n = 8) in B and C.
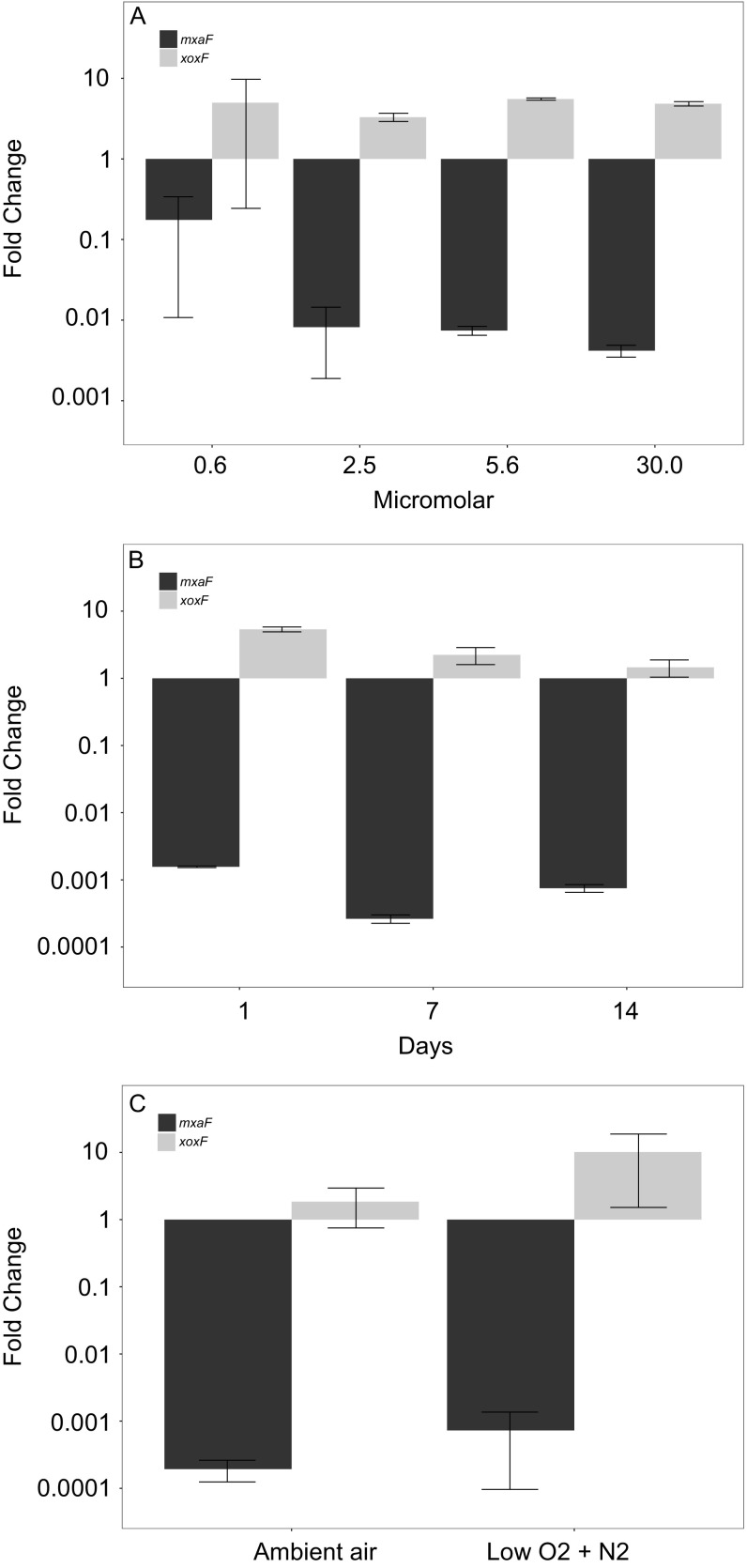
We further verified that in pure cultures of the methanotroph different concentrations of lanthanum in the medium, lanthanum depletion over time, or different O2 levels do not have any notable effect on the gene expression of the dominant MDHs (Fig. S2).
Fig. S2.
qRT-PCR values depicting the fold change in the expression of M. tundripaludum 31/32 mxaF and xoxF genes. (A) In pure culture with different concentrations of lanthanum chloride or without lanthanum (n = 2 for each concentration). (B) In pure culture with or without lanthanum chloride (30 μM) at different time points over 14 d ( n = 2 for each time point). (C) In pure culture with or without lanthanum chloride (30 μM) at 15 μM oxygen tension (low O2 + N2) or 210 μM oxygen tension (ambient air) (n = 2 for each time point). Error bars indicate the SE.
qRT-PCR analysis also was performed on cocultures that were grown for 21 d and with three transfers and dilutions into fresh medium. Cocultures were established with ratios of 30:70 (M. mobilis 13:M. tundripaludum 31/32) or 50:50 (M. mobilis JLW8:M. tundripaludum 31/32). In the presence of a co-occurring nonmethanotrophic methylotroph the expression of the methanotroph XoxF-type MDH decreased and the expression of the methanotroph MxaF-type MDH increased compared with the pure culture grown identically to the cocultures, a finding also seen in the RNA-sequencing (RNA-seq) results (Fig. 3 B and C).
Evidence for MDH-Specific Methanol Release into the Medium by the Methanotroph.
The coculture-dependent change in expression to the MxaF-type MDH suggested that this change might be responsible for methanol cross-feeding. To test this hypothesis, we assessed the supernatant of the pure cultures of the methanotroph used for qRT-PCR (Fig. 3A) for methanol release into the medium (Fig. 4). The results of gas chromatography-flame ionization detector (GC-FID) analysis demonstrated that no methanol could be detected in the presence of lanthanum (Fig. 4A), but in the absence of lanthanum methanol and a shift to mxaF gene expression, methanol was clearly detected with a concentration of 1.24 mM at the end of the incubation (Fig. 4 B and C); this level is sufficient to support growth of the nonmethanotrophic methylotrophs (33). We further show that in cocultures grown without lanthanum, increased expression of the MxaF-type MDH occurs in the methanotroph, and the co-occurring nonmethanotrophic methylotroph increases in abundance (Fig. S3).
Fig. 4.
Chromatograms from GC-FID analysis of M. tundripaludum 31/32 supernatant. (A) Pure culture of M. tundripaludum 31/32 cells grown with 30 μM lanthanum chloride. (B) Pure culture of M. tundripaludum 31/32 cells grown without 30 μM lanthanum chloride for 4 wk. (C) Pure culture of M. tundripaludum 31/32 cells grown without 30 μM lanthanum chloride (blue) compared to a 0.005% methanol standard (green). (D) Overlay of M. tundripaludum 31/32 pure culture samples grown with and without 30 μM lanthanum chloride. Chromatograms shown are representative of duplicate cultures and duplicate technical replicates.
Fig. S3.
(A) Cell-abundance dynamics of individual species in model artificial communities. (B) qRT-PCR values depicting the fold change in mxaF and xoxF gene expression from M. tundripaludum 31/32 with these cocultures. Cocultures were grown on methane as the sole carbon source with or without lanthanum in 250-mL glass vials. Two independent experiments were performed. Different symbols depict individual abundances of M. tundripaludum 31/32 with M. mobilis 13. Individual abundances were determined by flow cytometry. Error bars indicate the SE (n = 3). Please note that we restricted the qRT-PCR analysis to the treatments without lanthanum because the other treatment had already been evaluated (Fig. 3B).
Evidence for a Soluble Factor in the Cocultures That Affects MDH Gene Expression.
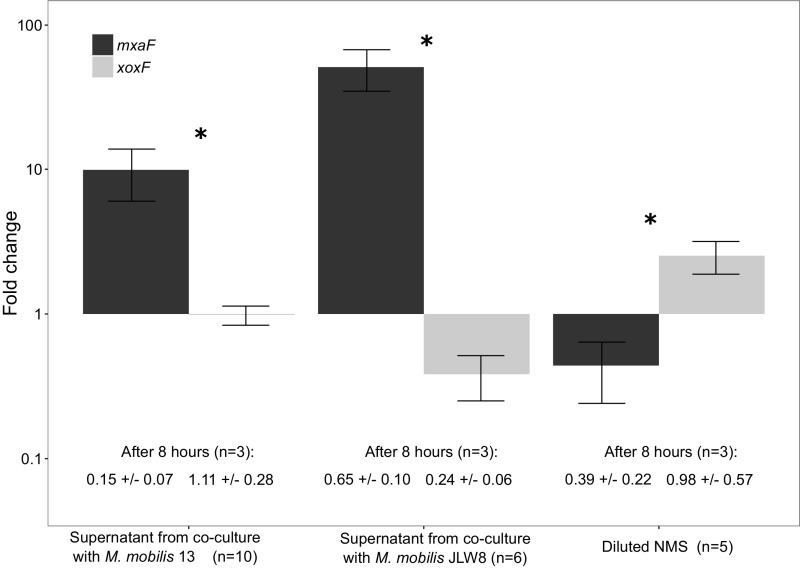
To determine whether coculture-dependent lanthanum depletion might be responsible for the induced shift of the dominant MDH and subsequent methanol release, we measured the lanthanum concentration in the supernatant from pure cultures and cocultures. All supernatants showed large decreases, with a stronger correlation between lanthanum decrease in the supernatant and MxaF-type MDH expression of M. tundripaludum 31/32 in coculture with M. mobilis JLW8 than in coculture with M. mobilis 13 (Figs. 2 and 3 and Table S2). In a second experiment to assess the presence of a soluble factor involved in the expression change, pure cultures of M. tundripaludum 31/32 were incubated with and without supernatant of cocultures. The expression of the MxaF-type MDH in the methanotroph changed only in the presence of the coculture supernatant (Fig. S4).
Table S2.
Lanthanum concentrations in the supernatant of pure cultures and mixed cultures (n = 3) and two sediment samples (n = 2)
| Strains | Setup | Lanthanum, μM |
| M. tundripaludum 31/32 | Pure culture | 0.434 ± 0.29 |
| M. mobilis 13 | Pure culture | 0.183 ± 0.04 |
| M. mobilis JLW8 | Pure culture | 0.06 ± 0.02 |
| M. tundripaludum 31/32 | Coculture | 0.359 ± 0.26 |
| M. mobilis 13 | ||
| M. tundripaludum 31/32 | Coculture | 0.05 ± 0.03 |
| M. mobilis JLW8 |
All cultures were grown for 6 wk as previously described (see details in Materials and Methods).
Fig. S4.
qRT-PCR values depicting the fold change in the expression of M. tundripaludum 31/32 mxaF (black bars) and xoxF (gray bars) genes in pure culture with lanthanum chloride (30 μM) and 10 mL supernatant from cocultures of M. tundripaludum 31/32 and M. mobilis 13 (Left) or M. tundripaludum 31/32, and M. mobilis JLW8 (Center) or 10 mL diluted NMS (Right). A t-test was performed to determine the significance of changes in gene-expression levels (*P < 0.05). Error bars indicate the SE.
Discussion
In this study we used bacterial isolates from Lake Washington sediment that are known to be important in one-carbon utilization in this ecosystem to identify the cross-feeding mechanisms that allow nonmethanotrophic methylotrophs to grow on methane-derived carbon. We established simple synthetic model communities and focused on cross-feeding as a specific mechanism of community interactions that is difficult to determine in complex natural communities. Although synthetic communities do not necessarily reflect the community structure and abundance of natural communities (39), the basic metabolic interactions are hypothesized to be the same in the two systems (10). In the example used here, the species used are representatives of those known to dominate SIP studies and enriched microcosms in Lake Washington sediment (13), suggesting that these metabolic interactions also may occur within the natural community.
Our results add insights into recent discoveries of the effects of rare earth metals on the expression and activities of MDHs. Previous work showed that the lanthanide-dependent XoxF-type MDH enzymes are more widespread than the classical MxaF-type MDH enzymes in known methylotrophs and other organisms (40–42). In line with previous findings with other methanotrophs (26, 27), we provide evidence that in pure culture in the presence of lanthanum the XoxF-type (Fig. 3A) is the predominant MDH in M. tundripaludum 31/32.
However, we discovered that the presence of the nonmethanotrophic methylotroph in the co–culture causes a change in relative expression of the two methanotroph MDHs in the presence of lanthanum and that this change results in the release of methanol into the medium by the methanotroph.
In the methanotroph Methylacidiphilum fumariolicum SolV, the affinity for methanol of XoxF-type MDH was much higher than reported for other MDHs, with a Km of 0.8 μM (40). If the same is true in M. tundripaludum 31/32, the lower affinity of the MxaF-type MDH could result in methanol excretion. Our results suggest that the partner-induced change in gene expression induces the methanotroph to perform a less efficient, leaky function with the result of sustaining the nonmethanotrophic methylotrophic population in the coculture. This methanotroph-based cross-feeding mechanism supports previous results showing that methanotrophs support nonmethanotrophs in natural communities by providing methane-derived carbon (43–45). Hence, this lanthanum-dependent cross-feeding has the potential to be an important mechanism in the environment.
Metabolic interactions and cross-feeding are widespread in microbial communities (9, 11, 46). Those interactions allow microbes to compete for nutrients by scavenging limited nutrients, to cooperate with another species to metabolize substrates they could not metabolize alone, or to cross-feed and support other members in a microbial community (9, 46). In some cases, such as obligate syntrophism, it has been shown that different syntrophic partners result in changes in both growth parameters and gene expression (47). Our study presents a specific mode of carbon and energy source cross-feeding interaction, in which the presence of a co-occurring species alters the metabolism of another species, resulting in the release of a carbon and energy source for the co-occurring species. In addition, we show that the mxaF gene expression in M. tundripaludum 31/32 in coculture is correlated to the level of lanthanum depletion, although it is not expressed in pure culture of M. tundripaludum 31/32 even when lanthanum is depleted to similar levels as in the cocultures. This finding suggests that lanthanum depletion may play a role but is not the sole mechanism whereby the gene expression changes. We found that the supernatant from a coculture causes the gene-expression change, suggesting the presence of an excreted soluble compound involved in altering the methanotroph metabolism. This compound might be a lanthanum sequestration agent, a signal, or a compound that results indirectly in expression change. Future studies are needed to address these factors to determine in more detail the molecular basis of the regulatory mechanism by which the metabolism of the methanotroph is changed by the presence of the nonmethanotroph.
Although two independent methods showed the metabolic alteration, not all cocultures between the methanotroph and the nonmethanotrophic heterotrophs displayed the change in gene expression (Fig. 2A). This variability suggests that additional factors determine when the change in the MDH expression of the methanotroph is activated. In a type II methanotroph, Methylosinus trichosporium OB3b, it has been shown that the presence of copper overrides the lanthanide control (27). In the two type I strains tested so far (ref. 26 and this study), in pure cultures, the amount of lanthanum added was sufficient to repress expression of the genes for the MxaF-type MDH throughout the course of the experiment, even in the presence of copper.
In both M. mobilis JLW8 and M. mobilis 13, the dominant xoxF gene transcripts also change expression in the coculture as compared with the pure cultures. The physiological results of this change are not known, because little is known about the different XoxF isozymes and their biochemical properties. It is possible that this change in gene expression is advantageous in allowing these strains to grow at the low methanol concentrations provided by the methanotroph in coculture, and/or it may reflect the same regulatory mechanism that causes a change in the expression of MDHs in the methanotrophs. However, mutation of the up-regulated xoxF gene did not interfere with coculture formation (Fig. S1E), so this shift in gene expression is not essential for the interaction. This synthetic two-culture system clearly demonstrates an interaction that cannot be deduced from studies in pure cultures and shows the power of such simple systems using strains isolated from a natural microbial community to deduce interaction mechanisms.
Materials and Methods
Strains and Growth Conditions.
The methanotrophic strain M. tundripaludum 31/32 (35) and the nonmethanotrophic methylotrophic strains M. mobilis 13 (33) and M. mobilis JLW8 (34) were isolated within the last few years from Lake Washington sediment. In addition, we used the MDH mutants xoxF1 (mmol_1170), xoxF2 (mmol_2048), and xoxF12 (mmol_1770, mmol_2048) of M. mobilis JLW8, which were created in an earlier study (38). M. tundripaludum 31/32 was pregrown in nitrate mineral salt (NMS) medium (48) and 30 µM lanthanum (III) chloride hydrate (99.9% trace metals basis; Sigma Aldrich) with 25% (vol/vol) methane, 5% (vol/vol) air, and 70% (vol/vol) nitrogen in the headspace. M. mobilis strains and the mutant strains xoxF1 and xoxF2 were pregrown in the same way but with 0.1% methanol instead. The mutant xoxF12 is unable to grow on methanol and was grown with 0.1% methylamine.
Synthetic Communities.
In the first set of experiments, we established cocultures and controls in duplicate or triplicate in autoclaved glass tubes (28-mL volume) and 6 mL of NMS medium. Standard curves of cell numbers and optical density in the pure culture were established by flow cytometry. Different strains then were mixed together in equal starting densities (107 cells/mL). All vials were sealed with rubber stoppers and capped with crimp seals. The headspace was exchanged daily according to the following scheme: (i) flushing with N2 gas for 30 s (flow rate >1.28 L/min); (ii) equalizing pressure by removing the excess volume of N2 gas with a syringe; (iii) removing 6.6 mL of headspace and adding back 5.5 mL of methane and 1.1 mL of air, corresponding to an initial dissolved O2 concentration of ∼15 µM. All vials were incubated in a shaker (200 rpm) at 18 °C.
As shown previously (36), after gas phase replenishment, O2 utilization occurs until the level drops to undetectable amounts. This protocol results in a daily cycle of excess methane but limiting O2 availability. This regime is a representation of O2 and CH4 conditions within the natural gradient of O2 and CH4 concentrations measured in Lake Washington sediments (49). Each vial was diluted weekly to an OD600 of 0.1. Transfer to a new vial ensured that any carbon from inoculum was diluted out and CH4 became the main remaining carbon source.
In a second set of experiments we established two independent setups of cocultures and controls in duplicate in autoclaved serum vials (250-mL volume) and 30 mL of NMS medium to grow sufficient biomass for RNA extraction and subsequent RNA-seq or qRT-PCR analysis. We followed the procedure described above but adjusted the volumes of air and methane to achieve an initial dissolved O2 concentration of 15 µM.
In a third set of experiments, cocultures and controls with M. mobilis JLW8 mutants were set up in triplicate in autoclaved serum vials (250-mL volume) and 30 mL of NMS medium following the procedure described above. In addition, cultures of M. mobilis JLW8 mutants were washed twice by centrifuging at 4,500 × g for 10 min and were resuspended in fresh NMS medium to remove remaining methylamine or methanol.
Flow Cytometry.
To obtain real-time abundances, cell numbers of individual strains in cocultures and pure cultures were determined by flow cytometry. We used the 15:1 difference in the size of M. tundripaludum 31/32 and M. mobilis 13 or JLW8 and a nucleic acid dye to distinguish different species in a mixed sample. Before dilution, 900-μL samples were removed from the culture, were fixed immediately with 100 μL of a mixture of glutaraldehyde and paraformaldehyde [1.6% (vol/vol) and 0.1% final concentrations, respectively], and were stored at 4 °C. For the analysis 1–10 μL of fixed sample was mixed with 10 μL of SYBR Green dye (1:100 in DMSO) (Thermo Scientific) and 0.22 μm filtered NMS medium to a final volume of 830 μL per sample. These samples were incubated for 30 min in the dark at room temperature. Cells were measured with a CyFlow space flow cytometer (Partec) with the following parameters: triggering on green fluorescence; all measured parameters SSC, FSC, green fluorescence analyzed and displayed in log3 or 4; flow rate between 4 and 6 μL/s; particle analysis rate below 1,000 particles/s.
Methanol Detection in Supernatant of Methylobacter Pure Cultures.
Duplicate 30 mL M. tundripaludum 31/32 cultures were grown as described above with and without lanthanum for 4 d. All culturing glassware for experiments performed without lanthanum was acid-washed overnight in 1 M hydrochloric acid before use to remove trace amounts of lanthanum adhering to the glass. Before the experiment a pure culture of M. tundripaludum 31/32 cells was pregrown without 30 μM lanthanum chloride for 4 wk to remove any traces of lanthanum in the cells. Then 10-mL samples were taken and centrifuged for 10 min at 20,913 × g. Subsequently, 1-mL samples were filtered through a 0.22-μm filter and were stored at 4 °C for further analysis. In addition 30-mL samples were used for subsequent RNA extraction and qRT-PCR.
GC-FID was used for methanol detection with a 6890 Gas Chromatograph equipped with a flame ionization detector (Agilent). Data were collected and converted into Matlab input files with LabVIEW 2010. Data analysis was done later in Matlab. One microliter of supernatant was injected (split with a ratio of 1:20) and separated with a SLB IL-15 column (Supelco) with a 5 m × 0.1 mm i.d. and a thickness of 0.8 μm. The oven temperature was set at 120 °C and was held for 0.5 min to evaporate the sample. The FID was operated at 220 °C. Methanol standards were diluted in water from 99.9% (vol/vol) methanol.
RNA Extraction and qRT-PCR.
RNA was extracted from 30-mL cultures grown with and without lanthanum. Separate experiments were carried out for transcriptomics and for qRT-PCR. Cells were grown to an OD600 of ∼0.5 for extracting RNA. Before RNA was extracted, samples were flushed and incubated as described above for 2 h to ensure that samples were obtained at the onset of the micro-oxic phase of the experiment. The procedure was performed as described (26) except that FastPrep Lysing Matrix E tubes (MP Biomedicals) were used. The purified RNA was tested for DNA contamination using 16S PCR. Afterwards, samples were stored at −80 °C for subsequent analyses.
cDNA was generated from ∼500 ng of isolated RNA following the procedure described earlier (26). PCR reactions for qRT-PCR were performed as described in ref. 26 using a LightCycler 2.0 (Roche Diagnostics) and LightCycler Software Version 3.5 (Roche). We used the 16S gene to normalize expression values. Primers are described in Table S3.
Table S3.
Primers used for qRT-PCR
| Name | Sequence | Target gene |
| xoxF_fw | AATTCACGCCAACACAAGATGCTG | Lanthanide-dependent methanol dehydrogenase |
| xoxF_rev | CCAAATTCACCACCGGAAATACCAG | |
| mxaF_fw | ATGCCGACACAGGCTAACAAAG | Calcium-dependent methanol dehydrogenase |
| mxaF_rev | GTTATTAGGGAAGGGCGTATGC | |
| 16S_mbac31_fw | AATTACTGGGCGTAAAGCGTGC | 16S rRNA gene |
| 16S_mbac31_rev | CTTTCGTACCTCAGCGTCAGTTCT |
Transcriptomics and Data Analysis.
Fifty base pairs were sequenced in single-read configuration in Rapid Run mode on a HiSeq 2500 System by GENEWIZ. The raw reads were aligned to the combined isolate genomes, as appropriate for each sample, using Burrows–Wheeler Aligner version 0.7.12-r1044 with default parameters (50). The alignments were postprocessed into sorted BAM files with SAMtools version 1.2-232-g87cdc4a (51). Reads were attributed to ORFs using the htseq-count tool from the HTseq framework version 0.5.4p5 in the intersection-nonempty mode (52). Differential abundance analysis was performed with DESeq2 1.2.10 (53, 54). Sequences have been archived with the National Center for Biotechnology Information in the Gene Expression Omnibus database (accession no. GSE85736).
Significant differences between samples were tested by using Student’s t-test. Unless otherwise specified, all data analysis and graphics were performed using R 3.2.4 (55).
SI Materials and Methods
Lanthanum Measurement in Supernatant.
To determine the lanthanum concentration in the supernatant of pure cultures and cocultures, we set up a new experiment in which cultures were grown for 4 wk as previously described. All culturing glassware was acid-washed overnight in 1 M hydrochloric acid before use to remove trace amounts of lanthanum adhering to the glass. At the end of the final incubation cycle (i.e., 7 d), 10 mL of supernatant was collected for lanthanum analysis. For this purpose, cell cultures were centrifuged at 4,500 × g for 10-min at 4 °C. The supernatant was transferred into sterile 15-mL tubes and stored at −80 °C for further analysis. In addition, two samples from frozen (−80 °C) sediment were analyzed. These samples were thawed on ice, centrifuged at 4,500 × g for 10 min, and then the upper phase was transferred to clean tubes. To remove any residuals from the sediment, the supernatant was centrifuged again for 10 min and then transferred to clean tubes. Samples were sent for analysis to the inductively coupled plasma mass spectrometry laboratory at Michigan State University. Before analysis samples were diluted 100 times and acidified in 2% (vol/vol) HNO3.
Growth of M. tundripaludum with Supernatant from Cocultures.
In a new experiment, pure cultures and cocultures were grown as previously described for 6 wk. Afterward pure cultures of M. tundripaludum 31/32 were set up in autoclaved serum vials (250-mL volume) with 30 mL NMS medium at a final optical density of 0.1 following the procedure described above. Four treatments were established: (i) 30 mL NMS; (ii) 20 mL NMS and 10 mL supernatant from coculture with M. mobilis 13; (iii) 20 mL NMS and 10 mL supernatant from coculture with M. mobilis JLW8; and (iv) 20 mL NMS and 10 mL 5× diluted NMS. Supernatant was centrifuged for 10 min and filtered through a 0.22-μm nylon filter unit (Nalgene filtration unit; Thermo Scientific) before use. Different treatments were incubated as described above for 8 h and were sampled after 4 and 8 h followed by RNA extraction and qRT-PCR as described above.
Supplementary Material
Acknowledgments
This study was supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research under Award DE-SC-0010556.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE85736).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619871114/-/DCSupplemental.
References
- 1.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320(5879):1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 2.Fierer N, Lennon JT. The generation and maintenance of diversity in microbial communities. Am J Bot. 2011;98(3):439–448. doi: 10.3732/ajb.1000498. [DOI] [PubMed] [Google Scholar]
- 3.Torsvik V, Øvreås L, Thingstad TF. Prokaryotic diversity--magnitude, dynamics, and controlling factors. Science. 2002;296(5570):1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 4.Faust K, Raes J. Microbial interactions: From networks to models. Nat Rev Microbiol. 2012;10(8):538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 5.Alivisatos AP, et al. Unified Microbiome Initiative Consortium MICROBIOME. A unified initiative to harness Earth’s microbiomes. Science. 2015;350(6260):507–508. doi: 10.1126/science.aac8480. [DOI] [PubMed] [Google Scholar]
- 6.Dubilier N, McFall-Ngai M, Zhao L. Microbiology: Create a global microbiome effort. Nature. 2015;526(7575):631–634. doi: 10.1038/526631a. [DOI] [PubMed] [Google Scholar]
- 7.Jarosz DF, et al. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell. 2014;158(5):1083–1093. doi: 10.1016/j.cell.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Wu C, Huang IH, Merritt J, Qi F. Differential response of Streptococcus mutans towards friend and foe in mixed-species cultures. Microbiology. 2011;157(Pt 9):2433–2444. doi: 10.1099/mic.0.048314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponomarova O, Patil KR. Metabolic interactions in microbial communities: Untangling the Gordian knot. Curr Opin Microbiol. 2015;27:37–44. doi: 10.1016/j.mib.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Grosskopf T, Soyer OS. Synthetic microbial communities. Curr Opin Microbiol. 2014;18(0):72–77. doi: 10.1016/j.mib.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelezniak A, et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci USA. 2015;112(20):6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knief C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol. 2015;6:1346. doi: 10.3389/fmicb.2015.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chistoserdova L. Methylotrophs in natural habitats: Current insights through metagenomics. Appl Microbiol Biotechnol. 2015;99(14):5763–5779. doi: 10.1007/s00253-015-6713-z. [DOI] [PubMed] [Google Scholar]
- 14.Ruff SE, et al. Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci USA. 2015;112(13):4015–4020. doi: 10.1073/pnas.1421865112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad R, Rothfuss F. Methane oxidation in the soil surface layer of a flooded ride field and the effect of ammonium. Biol Fertil Soils. 1991;12:28–32. [Google Scholar]
- 16.Kuivila KM, Murray JW, Devol AH, Lidstrom ME, Reimers CE. Methane cycling in the sediments of Lake Washington. Limnol Oceanogr. 1988;33(4):571–581. [Google Scholar]
- 17.Trotsenko YA, Murrell JC. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol. 2008;63:183–229. doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- 18.Beck DAC, et al. A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae. PeerJ. 2013;1:e23. doi: 10.7717/peerj.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalyuzhnaya MG, et al. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol. 2008;26(9):1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- 20.Saidi-Mehrabad A, et al. Methanotrophic bacteria in oilsands tailings ponds of northern Alberta. ISME J. 2013;7(5):908–921. doi: 10.1038/ismej.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen S, Neufeld JD, Birkeland NK, Hovland M, Murrell JC. Methane assimilation and trophic interactions with marine Methylomicrobium in deep-water coral reef sediment off the coast of Norway. FEMS Microbiol Ecol. 2008;66(2):320–330. doi: 10.1111/j.1574-6941.2008.00575.x. [DOI] [PubMed] [Google Scholar]
- 22.Cébron A, et al. Nutrient amendments in soil DNA stable isotope probing experiments reduce the observed methanotroph diversity. Appl Environ Microbiol. 2007;73(3):798–807. doi: 10.1128/AEM.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lueders T, Wagner B, Claus P, Friedrich MW. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ Microbiol. 2004;6(1):60–72. doi: 10.1046/j.1462-2920.2003.00535.x. [DOI] [PubMed] [Google Scholar]
- 24.Radajewski S, Ineson P, Parekh NR, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403(6770):646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 25.Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol. 2004;6(2):111–120. doi: 10.1046/j.1462-2920.2003.00543.x. [DOI] [PubMed] [Google Scholar]
- 26.Chu F, Lidstrom ME. XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol. 2016;198(8):1317–1325. doi: 10.1128/JB.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu W, Farhan Ul Haque M, DiSpirito AA, Semrau JD. Uptake and effect of rare earth elements on gene expression in Methylosinus trichosporium OB3b. FEMS Microbiol Lett. 2016;363(13):fnw129. doi: 10.1093/femsle/fnw129. [DOI] [PubMed] [Google Scholar]
- 28.Taubert M, et al. XoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ Microbiol. 2015;17(10):3937–3948. doi: 10.1111/1462-2920.12896. [DOI] [PubMed] [Google Scholar]
- 29.Corder RE, Johnson ER, Vega JL, Clausen EC, Gaddy JL. Biological production of methanol from methane. American Chemical Society Energy and Fuels Division. 1986;33(3):469–478. [Google Scholar]
- 30.Antony CP, et al. Active methylotrophs in the sediments of Lonar Lake, a saline and alkaline ecosystem formed by meteor impact. ISME J. 2010;4(11):1470–1480. doi: 10.1038/ismej.2010.70. [DOI] [PubMed] [Google Scholar]
- 31.He R, et al. Diversity of active aerobic methanotrophs along depth profiles of arctic and subarctic lake water column and sediments. ISME J. 2012;6(10):1937–1948. doi: 10.1038/ismej.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalyuzhnaya MG, et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun. 2013;4:2785. doi: 10.1038/ncomms3785. [DOI] [PubMed] [Google Scholar]
- 33.Beck DAC, et al. The expanded diversity of methylophilaceae from Lake Washington through cultivation and genomic sequencing of novel ecotypes. PLoS One. 2014;9(7):e102458. doi: 10.1371/journal.pone.0102458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalyuzhnaya MG, Bowerman S, Lara JC, Lidstrom ME, Chistoserdova L. Methylotenera mobilis gen. nov., sp. nov., an obligately methylamine-utilizing bacterium within the family Methylophilaceae. Int J Syst Evol Microbiol. 2006;56(Pt 12):2819–2823. doi: 10.1099/ijs.0.64191-0. [DOI] [PubMed] [Google Scholar]
- 35.Kalyuzhnaya MG, et al. Draft genome sequences of gammaproteobacterial methanotrophs isolated from Lake Washington sediment. Genome Announc. 2015;3(2):e00103–15. doi: 10.1128/genomeA.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez ME, Beck DAC, Lidstrom ME, Chistoserdova L. Oxygen availability is a major factor in determining the composition of microbial communities involved in methane oxidation. PeerJ. 2015;3:e801. doi: 10.7717/peerj.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oshkin IY, et al. Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J. 2015;9(5):1119–1129. doi: 10.1038/ismej.2014.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mustakhimov I, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Insights into denitrification in Methylotenera mobilis from denitrification pathway and methanol metabolism mutants. J Bacteriol. 2013;195(10):2207–2211. doi: 10.1128/JB.00069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Z, Krause SMB, Beck DAC, Chistoserdova L. A synthetic ecology perspective: How well does behavior of model organisms in the laboratory predict microbial activities in natural habitats? Front Microbiol. 2016;7:946. doi: 10.3389/fmicb.2016.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keltjens JT, Pol A, Reimann J, Op den Camp HJM. PQQ-dependent methanol dehydrogenases: Rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98(14):6163–6183. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 41.Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13(10):2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 42.Chistoserdova L. Lanthanides: New life metals? World J Microbiol Biotechnol. 2016;32(8):138. doi: 10.1007/s11274-016-2088-2. [DOI] [PubMed] [Google Scholar]
- 43.Manefield M, et al. Insights into the fate of a 13C labelled phenol pulse for stable isotope probing (SIP) experiments. J Microbiol Methods. 2007;69(2):340–344. doi: 10.1016/j.mimet.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Murase J, Frenzel P. A methane-driven microbial food web in a wetland rice soil. Environ Microbiol. 2007;9(12):3025–3034. doi: 10.1111/j.1462-2920.2007.01414.x. [DOI] [PubMed] [Google Scholar]
- 45.Dumont MG, Pommerenke B, Casper P, Conrad R. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ Microbiol. 2011;13(5):1153–1167. doi: 10.1111/j.1462-2920.2010.02415.x. [DOI] [PubMed] [Google Scholar]
- 46.Seth EC, Taga ME. Nutrient cross-feeding in the microbial world. Front Microbiol. 2014;5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer B, Kuehl JV, Deutschbauer AM, Arkin AP, Stahl DA. Flexibility of syntrophic enzyme systems in Desulfovibrio species ensures their adaptation capability to environmental changes. J Bacteriol. 2013;195(21):4900–4914. doi: 10.1128/JB.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dedysh S, Dunfield P. 2014. Cultivation of Methanotrophs. Springer Protocols Handbooks (Humana, Berlin), pp 1–17.
- 49.Auman AJ, Stolyar S, Costello AM, Lidstrom ME. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol. 2000;66(12):5259–5266. doi: 10.1128/aem.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 55.R Core Team. 2016 R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna), Version 3.2.4. Available at https://cran.r-project.org/. Accessed 11 November 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.