Significance
Cobalamin (vitamin B12)-dependent organisms span all domains of life, making procurement of the vitamin from the few prokaryotic producers an essential function in organismal interactions. Yet not all key producers of cobalamin have been identified in the ocean. We show that in the marine environment, select heterotrophic bacteria and Thaumarchaeota produce cobalamin, while Cyanobacteria, the most abundant phytoplankton on earth, supply and use pseudocobalamin. These chemically distinct cofactors support different members of the microbial community because they are not interchangeable as cofactors in enzymes. Our findings identify key organisms supporting cobalamin-based interdependencies that underpin primary production and microbial interactions in the ocean.
Keywords: B12, cobalamin, Thaumarchaeota, pseudocobalamin, Cyanobacteria
Abstract
Organisms within all domains of life require the cofactor cobalamin (vitamin B12), which is produced only by a subset of bacteria and archaea. On the basis of genomic analyses, cobalamin biosynthesis in marine systems has been inferred in three main groups: select heterotrophic Proteobacteria, chemoautotrophic Thaumarchaeota, and photoautotrophic Cyanobacteria. Culture work demonstrates that many Cyanobacteria do not synthesize cobalamin but rather produce pseudocobalamin, challenging the connection between the occurrence of cobalamin biosynthesis genes and production of the compound in marine ecosystems. Here we show that cobalamin and pseudocobalamin coexist in the surface ocean, have distinct microbial sources, and support different enzymatic demands. Even in the presence of cobalamin, Cyanobacteria synthesize pseudocobalamin—likely reflecting their retention of an oxygen-independent pathway to produce pseudocobalamin, which is used as a cofactor in their specialized methionine synthase (MetH). This contrasts a model diatom, Thalassiosira pseudonana, which transported pseudocobalamin into the cell but was unable to use pseudocobalamin in its homolog of MetH. Our genomic and culture analyses showed that marine Thaumarchaeota and select heterotrophic bacteria produce cobalamin. This indicates that cobalamin in the surface ocean is a result of de novo synthesis by heterotrophic bacteria or via modification of closely related compounds like cyanobacterially produced pseudocobalamin. Deeper in the water column, our study implicates Thaumarchaeota as major producers of cobalamin based on genomic potential, cobalamin cell quotas, and abundance. Together, these findings establish the distinctive roles played by abundant prokaryotes in cobalamin-based microbial interdependencies that sustain community structure and function in the ocean.
Cobalamin (vitamin B12) is synthesized by a select subset of bacteria and archaea, yet organisms across all domains of life require it (1–3). In the surface ocean, cobalamin auxotrophs (including most eukaryotic algae) (3) obtain the vitamin through direct interactions with cobalamin producers (3) or breakdown of cobalamin-containing cells (4, 5). Interdependencies between marine cobalamin producers and consumers are critical in surface waters where primary productivity can be limited by the availability of cobalamin and the compound is short-lived (1, 6, 7). The exchange of cobalamin in return for organic compounds is hypothesized to underpin mutualistic interactions between heterotrophic bacteria and autotrophic algae (3, 6, 8, 9). The apparent pervasiveness of cobalamin biosynthesis genes in chemoautotrophic Thaumarchaeota and photoautotrophic Cyanobacteria genomes (1, 10, 11) raises the question of whether cobalamin production by these autotrophs may underlie additional, unexplored microbial interactions.
Cobalamin is a complex molecule with a central cobalt-containing corrin ring, an α ligand of 5,6-dimethylbenzimidizole (DMB), and a β ligand of either OH-, CN-, Me-, or Ado- (12) (Fig. 1). Previous studies have shown that instead of producing cobalamin, Cyanobacteria produce pseudocobalamin (11, 13, 14), an analog of cobalamin in which adenine substitutes for DMB as the α ligand (12) (Fig. 1). Production of pseudocobalamin in a natural marine environment has not been shown, nor have reasons for the production of this compound in place of cobalamin been elucidated.
Fig. 1.
General form of cobalamin analogs. Shown is a schematic of the conserved corrin ring with various upper (β) and lower (α) ligands. Structures of cobalamin analogs monitored in this study (eight total) are shown.
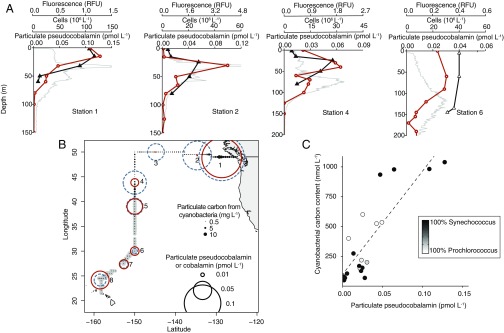
To explore the pervasiveness of cobalamin and pseudocobalamin supply and demand in marine systems, we determined the standing stocks of these compounds in microbial communities from surface waters across the North Pacific Ocean using liquid chromatography mass spectrometry (LC-MS). Our LC-MS method (15) quantifies cobalamin and pseudocobalamin with different β ligands. We found that in the surface ocean, pseudocobalamin and cobalamin concentrations associated with organisms and detritus captured on a 0.2-μm filter (particulate fraction) were often of equal magnitude (Fig. 2B). Pseudocobalamin had peak concentrations within the euphotic zone at each station and was not detected below the euphotic zone. In contrast, cobalamin was measurable throughout the sampled waters and maintained similar or higher concentration from the lower euphotic zone to our deepest samples (Fig. 2A and Fig. S1).
Fig. 2.
Pseudocobalmin and Cyanobacteria cooccurrence. (A) Depth profiles of particulate pseudocobalamin (orange circles), in situ chlorophyll a (gray), and cell abundance of Synechococcus (closed triangles) or Prochlorococcus (open triangles), whichever was the dominant Cyanobacteria at each station. (B) Location of sampling with surface concentration of particulate cobalamin (dashed blue circles) and pseudocobalamin (orange circles). Closed circles are cyanobacterial (Synechococcus, black; Prochlorococcus, gray) carbon content calculated from cell abundance and size estimates (at 5 m via SeaFlow) (16). (C) Correlation between environmental pseudocobalamin and calculated Cyanobacteria carbon content (n = 20, R2 = 0.71, P < 0.001). Cobalamin and pseudocobalamin concentrations are summed values of the detected β ligands for these compounds (Ado-, Me-, and OH- for cobalamin; Me- and OH- for pseudocobalamin); contributions of each beta ligand are provided in Dataset S2. Pseudocobalamin concentrations are presented in cobalamin equivalents (see SI Materials and Methods).
Fig. S1.
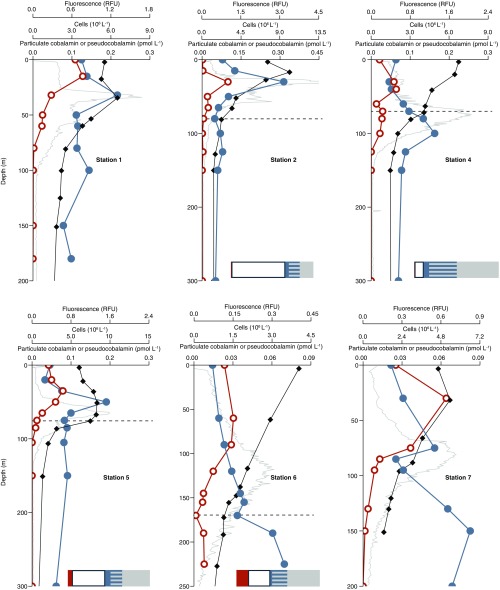
Pseudocobalamin and cobalamin producers at six additional stations. Shown are cobalamin (closed blue circles), pseudocobalamin (orange open circles), prokaryote cell abundance (black diamonds), and in situ fluorescence (gray). Also shown are bar graphs with the percent of cells with the following predicted cobalamin strategies: unlikely producers (gray), bacteria with unknown cobalamin biosynthesis capacity (gray/blue striped), bacteria that likely possess the potential to produce cobalamin (blue), Thaumarchaeota (likely cobalamin producers, white), and Cyanobacteria (likely pseudocobalamin producers, orange) at the corresponding depths. Cobalamin and pseudocobalamin concentrations are summed values of the detected beta ligands as in Fig. 2. Pseudocobalamin concentrations are presented in cobalamin equivalents (see SI Materials and Methods). Station numbers are as shown in Fig. 2; dashed lines indicate the depth at which we analyzed prokaryotic diversity by 16S rRNA genes.
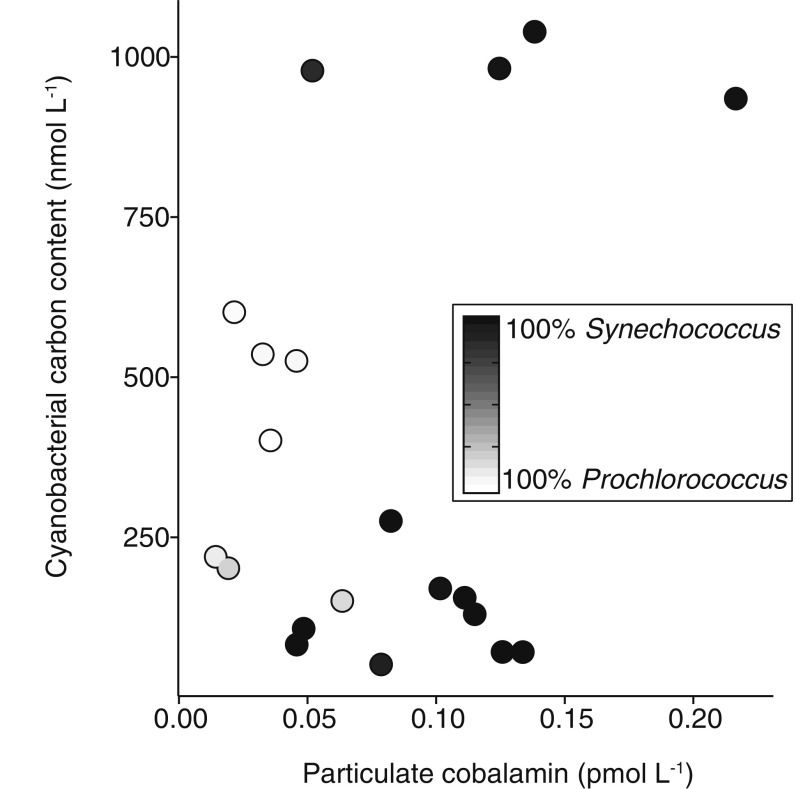
The overlapping spatial distribution of cobalamin and pseudocobalamin suggests that these cofactors are produced in each other’s presence, likely with different sources and sinks. To investigate correlations between Cyanobacteria and pseudocobalamin abundance, we compared observations of Cyanobacteria carbon inferred from flow cytometry with pseudocobalamin measurements taken at the same depth. In cases where we had both continuous measurements (by SeaFlow) (16) and discreet flow cytometry data, we used the discreet measurements, as collection of these samples was closely coupled in time and space to pseudocobalamin sampling (n = 16 for discreet, n = 4 for continuous). Pseudocobalamin concentrations are statistically correlated with carbon from Synechococcus and Prochlorococcus (R2 = 0.71, P < 0.001), both in the surface and into the subsurface ocean (Fig. 2C), suggesting a primarily cyanobacterial source. No significant correlation existed between Cyanobacteria carbon and cobalamin concentrations (Fig. S2).
Fig. S2.
Environmental cobalamin and Cyanobacteria carbon. No significant correlation exists between calculated carbon from Cyanobacteria and cobalamin. Shade of circle is the source of the calculated Cyanobacteria carbon (from 100% Synechococcus to 100% Prochlorococcus). Cobalamin concentrations are summed values of the detected beta ligands for these compounds, as in Fig. 2.
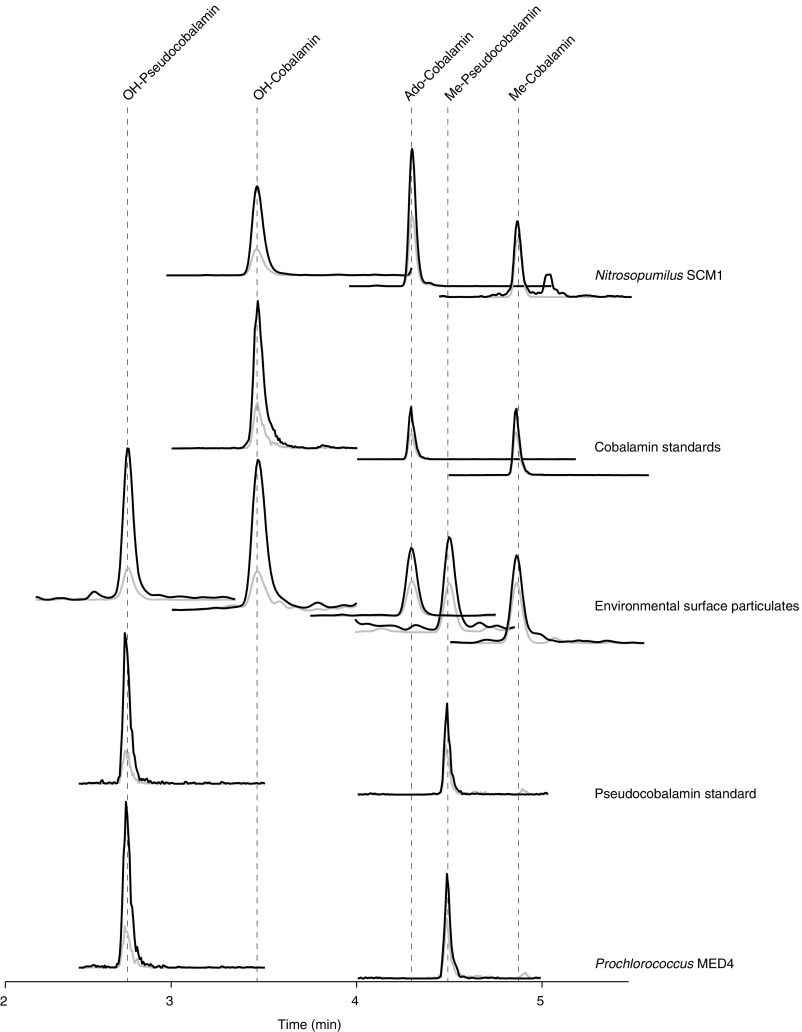
To identify the major producers of cobalamin and pseudocobalamin in the environment, we investigated representative marine isolates and then expanded our search into available genomes that encompass the phylogenetic diversity at our study site. As expected (1, 8), two strains of marine Alphaproteobacteria with cobalamin synthesis genes (Sulfitobacter sp. SA11 and Ruegeria pomeroyi DSS-3) produced cobalamin, whereas the gammaproteobacterium Vibrio fischerii ES114 (which lacks cobalamin biosynthesis genes) did not (Table S1). Four pure strains of marine chemoautotrophic Thaumarchaeota (Nitrosopumilus spp. SCM1, HCE1, HCA1, and PS0) also produced cobalamin (Table S1), confirming earlier suggestions based on the presence of cobalamin biosynthesis genes in Thaumarchaeota genomes (10). Like other Cyanobacteria (11, 13, 14), four axenic strains of marine Cyanobacteria (Prochlorococcus MED4 and MIT9313 and Synechococcus WH8102 and WH7803) produced pseudocobalamin (Table S1). In all of the cobalamin or pseudocobalamin producers, we detected compounds with β ligands Me-, Ado-, and OH- but not CN- (Table S1).
Table S1.
Summary of cultured archaea and bacteria in this study
| Molecules of cobalamin per cell | Molecules of pseudocobalamin per cell | nmole cobalamin analog per mole carbon | |||||||
| Phylum | Strain | n | OH- | Me- | Ado- | OH- | Me- | Ado- | |
| Proteobacteria | V. fischerii | 3 | nd | nd | nd | nd | nd | nd | 0 |
| Sulfitobacter sp. SA11 | 3 | 3 ± 0.1 | 4 ± 0.4 | 7 ± 3 | nd | nd | nd | 1–10 | |
| R. pomeryoi DSS-3 | 3 | 520 ± 290 | 120 ± 100 | 1,200 ± 320 | nd | nd | nd | 240–260 | |
| Thaumarchaeota | Nitrosopumilus sp. HCE1 | 3 | 420 ± 19 | 52 ± 17 | 1,600 ± 140 | nd | nd | nd | 4,200–5,300 |
| Nitrosopumilus sp. HCA1 | 3 | 1,860 ± 14 | 366 ± 55 | 2,252 ± 210 | nd | nd | nd | 9,300–11,600 | |
| Nitrosopumilus sp. PS0 | 3 | 598 ± 65 | 139 ± 20 | 1,548 ± 177 | nd | nd | nd | 4,700–5,900 | |
| N. maritimus SCM1 | 3 | 670 ± 52 | 13 ± 4 | 680 ± 130 | nd | nd | nd | 2,800–3,500 | |
| Cyanobacteria | Prochlorococcus MED4 | 5 | nd | nd | nd | 240 ± 40 | 990 ± 90 | trace | 430–1,190 |
| Prochlorococcus MIT9313 | 5 | nd | nd | nd | 360 ± 57 | 1,600 ± 280 | trace | 130–450 | |
| Synechococcus WH7803 | 3 | nd | nd | nd | 69 ± 13 | 16,600 ± 2,000 | trace | 2,400–3,000 | |
| Synechococcus WH8102 | 3 | nd | nd | nd | 270 ± 43 | 9,170 ± 1,200 | trace | 1,480–1,870 | |
Observed cellular quotas of major forms of cobalamin and pseudocobalamin with SD with n replicates of each strain. nd, not detected; trace, detected a small peak but was unable to quantify (see SI Materials and Methods).
The observed cell quotas of cobalamin or pseudocobalamin per cellular carbon varied greatly among producers (Table S1). Laboratory cultures of Alphaproteobacteria and Procholorococcus strains had lower amounts of cobalamin or pseudocobalamin (less than 1,200 nmol cobalamin analog per mole carbon) than Synechococcus and Thaumarchaeota isolates (1,480–11,600 nmol cobalamin analog per mole carbon). Published values (17) for sea ice bacterial isolates estimated using a bioassay were highly variable (0.6–6,800 nmol cobalamin analog per mole carbon). In our environmental samples, we observed an average stoichiometry of 87 nmol pseudocobalamin per mole cyanobacterial carbon, lower than the cyanobacterial isolates (Fig. 2C). This finding suggests that the cellular stoichiometry of pseudocobalamin is variable and possibly influenced by environmental factors like nutrient availability and growth rate.
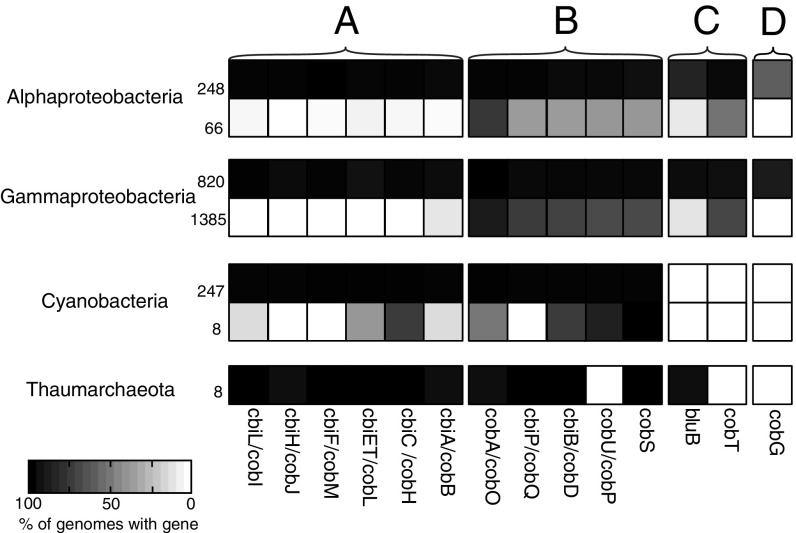
To expand the breadth of our survey beyond laboratory isolates, we inspected publically available whole genome sequences from bacteria and Thaumarchaeota for evidence of cobalamin biosynthesis. This analysis expands on previous work (18) while focusing on the phylogenetic groups present at our study site. We analyzed full genomes from the Integrated Microbial Genomes (IMG) database (https://img.jgi.doe.gov) from phylogenetic groups that encompassed >99.9% of the Bacterial 16S rRNA gene sequences from our environmental samples to develop a systematic inference of cobalamin synthesis capacity (3,410 genomes). Alpha- and Gammaproteobacteria are hypothesized to be major marine cobalamin producers (1, 10), and 94% of the surveyed genomes from these groups that contain genes necessary for corrin biosynthesis (i.e., cbiA/cobB, cbiH/cobJ) (2) also have genes for DMB synthesis and activation (bluB, cobT) (2, 19, 20) (Fig. 3), consistent with the synthesis of cobalamin and the results from our representative cultures. All of the eight available high-quality Thaumarchaeota genomes in the IMG database code for corrin and DMB biosynthesis genes (Fig. 3). Most of the lower quality, incomplete genomes available follow this same pattern (17/19, Dataset S1). No Thaumarchaeota genomes possess the cobT gene and thus must activate DMB through a different pathway. Of the 255 cyanobacterial whole genomes, 247 possessed genes for the synthesis of the corrin ring, but only one genome possessed an annotated bluB or cobT gene (Fig. 3), suggesting the vast majority of Cyanobacteria are unable to produce DMB, in agreement with a recent study that examined a subset of the available Cyanobacteria genomes (11).
Fig. 3.
Presence or absence of annotated cobalamin biosynthesis genes in full-genome representatives. Four major groups of microbes contributing cobalamin biosynthesis genes in marine surface waters (10) are shown; each group is split into potential cobalamin producers (top row) and noncobalamin producers (bottom row), with the number of genomes in each group. All of the high-quality Thaumarchaeota genomes are potential cobalamin producers. The shade of each box indicates the percent of genomes with that gene. Genes are grouped as follows: (A) corrin ring biosynthesis genes that are common to both O2-dependent and -independent pathways in bacteria and archaea, (B) final synthesis and repair genes, (C) genes for DMB synthesis and activation, and (D) O2-dependent cobG in the O2-dependent corrin ring biosynthesis pathway. In Archaea, cobY is used in place of cobU/cobP (10, 55). For the list of genomes in each group, see Dataset S1, summarized in Table S1.
All of the 49 Prochlorococcus genomes have genes for corrin synthesis without genes for DMB synthesis or activation, and our analysis demonstrated that Prochlorococcus MED4, with its highly streamlined genome (21), has maintained these genes to synthesize pseudocobalamin. We propose this originates from an ancient specialization to the production and use of pseudocobalamin in lieu of cobalamin among Cyanobacteria. Biosynthesis of the corrin ring can occur via two separate pathways: an O2-dependent or an O2-independent pathway (2, 18). DMB synthesis can also occur via two separate routes, the O2-dependent BluB (19) or the O2-independent and O2-sensitive Bza pathway (22). Rhodobacters have the O2-dependent corrin ring and DMB pathways (11), whereas Thaumarchaeota likely possess the O2-independent pathway for the corrin ring and the O2-dependent DMB pathway (10). In some bacteria, pseudocobalamin can be produced if DMB synthesis is impaired; this is due to the natural presence of adenine in cells and enzyme substrate promiscuity that allows adenine to substitute for DMB in some organisms’ CobT (22–27). Cyanobacteria use the O2-independent pathway for corrin ring synthesis and neither pathway for DMB synthesis (11, 18) (Fig. 3). The use and production of cobalamin as a cofactor predates oxygenic photosynthesis (28, 29). Possessing an O2-independent mode for producing a cobalamin analog that is not impaired by O2 may have been necessary for the success of oxygenic photosynthetic Cyanobacteria that were largely responsible for the rise of O2 on earth and likely endured variable O2 concentrations over time (30).
Cyanobacteria use pseudocobalamin as a cofactor in two enzymes: methionine synthase (MetH) and class II ribonucleotide reductase (NrdJ). The 3D structure of MetH contains three β pleated sheets and two α helices that form a pocket for the DMB ligand of cobalamin in Escherichia coli (31, 32). Cyanobacterial MetH is predicted to form the same pocket (13). However, conserved amino acids within this pocket in the cyanobacterial MetH differ from sequences of organisms known to use cobalamin (Figs. S3 and S4), suggesting a structure that preferentially binds pseudocobalamin in place of cobalamin as experimentally demonstrated in the Cyanobacteria Arthrospira (13). Many Cyanobacteria also code for O2-independent NrdJ (33), which has limited sequence similarity to noncyanobacterial NrdJ (34, 35). Similar to pseudocobalamin biosynthesis in Cyanobacteria, NrdJ is both O2-independent and O2-insensitive and has been hypothesized as an important bridge between the pre- and postoxygenated oceans (36). The continued maintenance of both the biosynthetic pathway and pseudocobalamin-dependent enzymes throughout the diverse Cyanobacteria phylum (from Arthrospira and Synechocystis to the highly streamlined Prochlorococcus) suggests the production of pseudocobalamin is an ancient relic that persists in the oxic marine environment today.
Fig. S3.
Alignment of cobalamin-binding motif in MetH. Data are from six Cyanobacteria (Synechocystis sp. PCC6803, Prochlorococcus spp. MIT9313 and MED4, Gloeobacter violaceus PCC 7421, Synechococcus WH7803, and Arthrospira platensis NIES-39) and four organisms known to use true cobalamin (H. sapiens, T. pseudonana, Pseudomonas, and E. coli). MetH in A. platensis NIES-39 has been demonstrated to have a higher affinity to pseudocobalamin than cobalamin (13). Black amino acids are part of α-helixes or β-sheets in the pocket housing the base of cobalamin (31); within that pocket, blue highlighted residues are conserved in both Cyanobacteria and the four cobalamin users, and orange residues are conserved in >90% of surveyed Cyanobacteria genomes but are different from the cobalamin-using organisms. These colors are used in Fig. S3 to better visualize the base-housing pocket. *Amino acids that have potential to hydrogen-bond to the pseudocobalamin’s base, adenine.
Fig. S4.
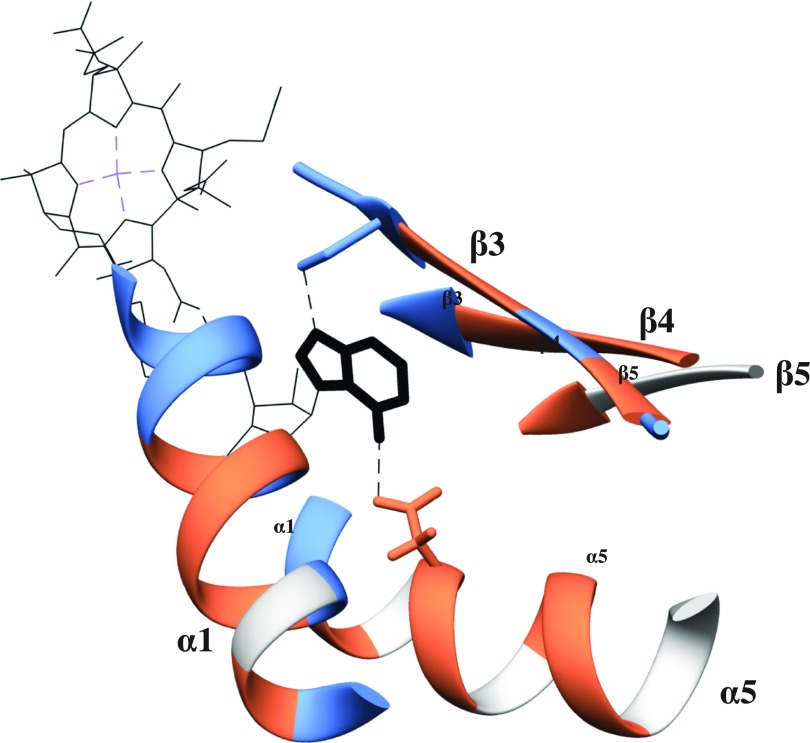
Adenine-housing portion of the cobalamin-binding motif in MetH. The structure shown is a representation of MetH in Prochlorococcus MIT9313 based on the crystal structure of the same enzyme in E. coli (53), with pseudocobalamin (in black, adenine ligand bolded, cobalt in violet) in place of cobalamin. Colors correspond to Fig. S2. Dashed lines are possible hydrogen bonds between the adenine base and the enzyme, a well-conserved serine residue previously suggested to hydrogen bond to DMB (31) (and likely adenine) on β3, and an aspartic acid residue found in all Cyanobacteria that may hydrogen bind to adenine on α5.
For many eukaryotic algae, pseudocobalamin supports lower growth yields than cobalamin (11, 37, 38). We examined the underlying cause of this reduced growth by supplying the model diatom Thalassiosira pseudonana (CCMP 1335) with pseudocobalamin and tracking it into the cells. Like others, we found that growth of T. pseudonana is limited at 1 pM cobalamin (39), and the addition of pseudocobalamin (at 200 pM) is unable to overcome this limitation (11, 38). We observed that T. pseudonana takes up the inactive OH-pseudocobalamin from their growth media and converts it into the cofactor form Ado-pseudocobalamin yet is unable to recover to cobalamin-replete growth rates (Fig. 4, Fig. S5, and Table S2). The role of Ado-cobalamin in diatoms is unclear, although T. pseudonana does code for Ado-cobalamin–dependent methylmalonyl-CoA mutase (MCM) and actively transcribes a putative adenosylcobalamin transferase (which converts OH-cobalamin to Ado-cobalamin) under cobalamin limitation (39). Previous studies suggest that when diatoms are starved for cobalamin, low Me-cobalamin (required for MetH activity) deprives cells of S-adenosylmethionine (SAM) (7, 39). We found that when pseudocobalamin is supplied to cobalamin-limited T. pseudonana, they contain significantly less SAM per cell than cobalamin-replete conditions (Fig. 4, Fig. S5, and Table S2). The depressed levels of SAM and lack of detectable Me-pseudocobalamin within cells suggest that T. pseudonana are unable to efficiently use pseudocobalamin in their MetH. This same phenomenon has been demonstrated physiologically in marine cobalamin-dependent bacteria: Cobalamin or methionine stimulates growth, but pseudocobalamin does not (40). These combined findings demonstrate that marine Cyanobacteria achieve both independence and a competitive advantage by producing and requiring only pseudocobalamin; they meet their own need for their preferred cofactor while also not directly supplying cobalamin to other photoautotrophs.
Fig. 4.
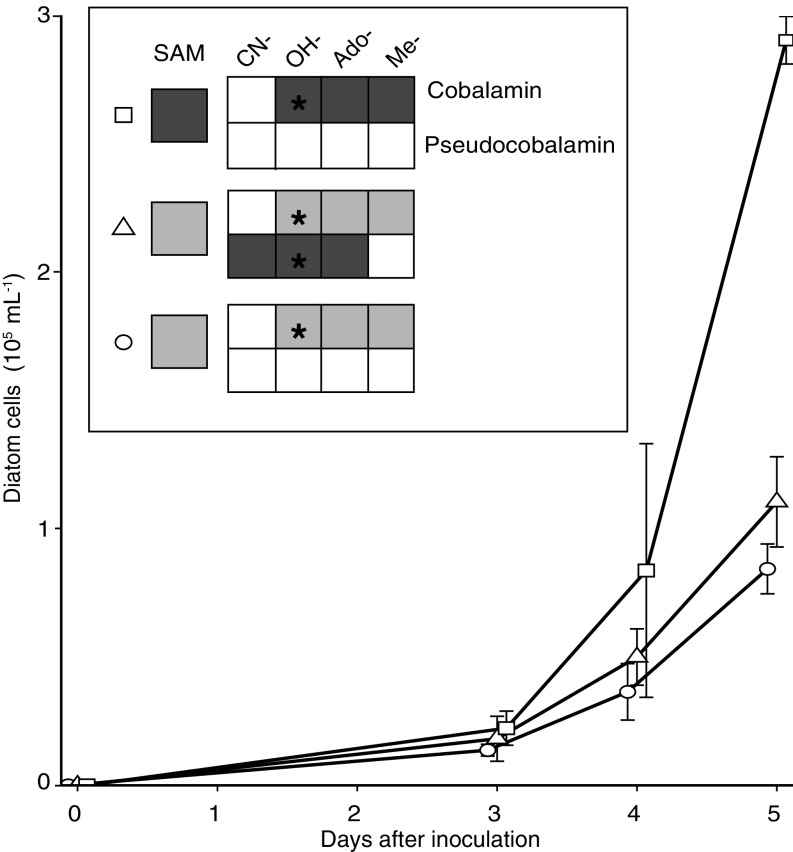
Growth of the diatom T. pseudonana with pseudocobalamin. (A) Growth of diatoms under Control (1 pM cobalamin, circle), +Pseudocobalamin (1 pM cobalamin with 200 pM pseudocobalamin, triangle), and +Cobalamin (200 pM cobalamin, square) treatments over time; error bars, SD from three replicates. Inset shows the final SAM, cobalamin, and pseudocobalamin cellular contents compared with Control treatment on a per cell basis: not detected (white), similar range to control (light gray), or statistically higher than control (Student’s unpaired t test, P < 0.05, dark gray). Black stars indicate what forms of cobalamin or pseudocobalamin were added to each treatment. See Fig. S5 for absolute values of cobalamins and SAM.
Fig. S5.
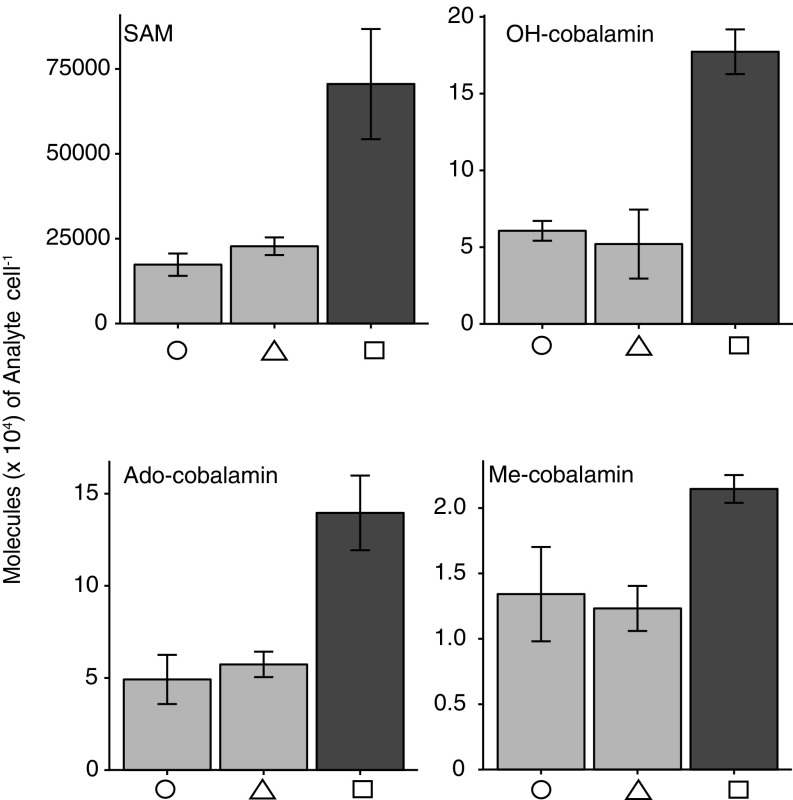
Cellular contents of cobalamin and SAM in the diatom experiment. Shown are molecules per cell of SAM and OH-, Ado-, and Me-cobalamin. Colors and symbols are as in Fig. 4.
Table S2.
Observed intracellular cobalamin and SAM contents of T. pseudonana
| Molecules (×103) of cobalamin per cell | Molecules (×103) of pseudocobalamin per cell | nmole cobalamin per mole carbon | Molecules (×108) SAM per cell | |||||||
| Treatment | OH- | Me- | Ado- | CN- | OH- | Me- | Ado- | CN- | ||
| 200 pM cobalamin | 180 ± 14 | 21 ± 1.1 | 140 ± 20 | nd | nd | nd | nd | nd | 800 ± 85 | 7.1 ± 1.6 |
| 1 pM cobalamin | 61 ± 6.4 | 13 ± 3.6 | 49 ± 13 | nd | nd | nd | nd | nd | 290 ± 74 | 1.7 ± 0.33 |
| 1 pM cobalamin + 200 pM pseudocobalamin | 52 ± 22 | 12 ± 1.7 | 57 ± 6.9 | nd | 990 ± 531 | nd | trace | trace | 290 ± 56 | 2.2 ± 0.26 |
Three different conditions with SD of three replicates of each strain; nd, not detected; trace, detected a peak but was unable to quantify (see SI Materials and Methods).
Corrin synthesis genes in oligotrophic surface water metagenomes are dominated by pseudocobalamin-producing Cyanobacteria (10), suggesting that de novo production of true cobalamin in these regions is limited to a small subset of heterotrophic bacteria, including clades like Rhodobacterales that are commonly associated with eukaryotic algae (9). Cobalamin auxotrophs may foster close physical or chemical relationships with these cobalamin producers, offsetting the expense of cobalamin biosynthesis to secure a consistent cobalamin source. Alternatively, organisms may employ techniques to use closely related compounds like pseudocobalamin. Many organisms without the biosynthetic capacity for de novo production of cobalamin have salvage or remodeling strategies for reactivating cobalamin analogs (11, 20, 41). A recent study has shown that some eukaryotic algae have the genetic capacity to make cobalamin when supplied with pseudocobalamin and DMB, although this phenomenon may be limited by low DMB concentrations in natural seawater (11), and DMB biosynthesis seems to be limited to bacteria and archaea. Of the 3,408 phylogenetically representative prokaryote genomes we surveyed, we found 73 genomes that did not have the biosynthetic pathway for the corrin moiety of cobalamin but did have genetic capacity for cobalamin repair as well as synthesis and activation of DMB, which has no known role in cells beyond α ligand of cobalamin. These organisms include several heterotrophic bacteria known to occur in marine environments such as Methylophaga and Marinobacter (Dataset S1). Previous work has suggested that ligand-bound cobalt in low latitudes may be cobalamin degradation or precursor compounds, which are present at concentrations much higher than has been observed for dissolved cobalamin (1, 15, 42). In these environments, organisms capable of salvaging and repairing cobalamin degradation products or precursors could be major suppliers of cobalamin to the environment and de novo biosynthesis may take a minor role. The prevalence of cobalamin salvage pathway transcripts in marine systems (6) suggests that synthesizing DMB or remodeling cobalamin is a common strategy that plays an important role connecting the production of pseudocobalamin to the growth of cobalamin auxotrophs in the low latitude oceans where Cyanobacteria are abundant.
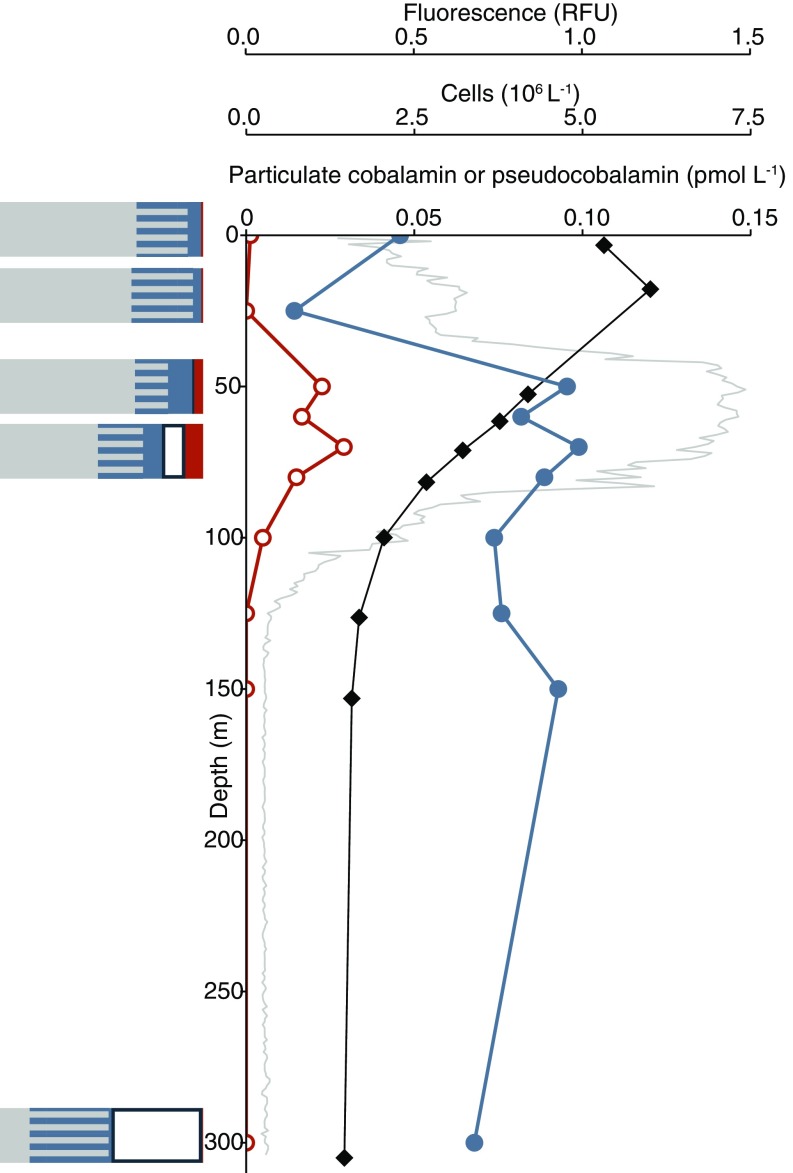
We inspected genomes of marine prokaryotes to identity likely cobalamin producers in our study site. We combined bacterial 16S rRNA gene sequence-derived phylogenetic data with current knowledge of corrin biosynthesis genes (including our analysis) across different phylogenetic groups to infer the cobalamin biosynthetic capacity of organisms in the microbial communities at our study site as likely, unknown, or unlikely (Table S3). We quantified Thaumarchaeota by quantitative PCR (qPCR) (43, 44) and calculated their contribution to the microbial population by comparing this value to direct counts of prokaryotic cells determined for each sample. Our analysis at five targeted locations in the North Pacific suggested that Thaumarchaeota represent 30–80% of prokaryotes having likely or unknown genetic capacity to synthesize cobalamin in the lower euphotic zone and deeper (Fig. 5 and Fig. S1). High cobalamin contents on a per carbon basis in cultured Thaumarchaeota implicate them as a major source of cobalamin in deeper waters (Table S1).
Table S3.
Cobalamin biosynthetic capacity deduced from taxonomy
| Genomes with corrin biosynthetic capacity | ||||||
| Phylum | Class | Order | Clade | This study | 2014 analysis* | Applied category |
| Actinobacter | 1/3 | Unknown | ||||
| Bacteroidetes | Flavobacteriia | 6/342 | 0/25 | Unlikely | ||
| Chlorobi | 5/12 | Unknown | ||||
| Chloroflexi | 9/32 | Unknown | ||||
| Cyanobacteria | Prochlorales | 50/52 | 18/18 | Pseudocobalamin | ||
| Cyanobacteria | Chroococcales | 100/104 | 20/21 | Pseudocobalamin | ||
| Cyanobacteria | All others | 97/99 | Pseudocobalamin | |||
| Deferribacteres | 4/6 | Unknown | ||||
| Planctomycetes | 5/28 | Unknown | ||||
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | 162/200 | 43/50 | Likely | |
| Proteobacteria | Alphaproteobacteria | Rhodospirillales | 83/102 | Likely | ||
| Proteobacteria | Alphaproteobacteria | SAR11 Clade | 0/12 | 0/11 | Unlikely | |
| Proteobacteria | Betaproteobacteria | 0/25 | Unknown | |||
| Proteobacteria | Deltaproteobacteria | 106/159 | Unknown | |||
| Proteobacteria | Gammaproteobacteria | Altermonadales | 12/226 | 3/49 | Unlikely | |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | SAR86 Clade | Unlikely† | ||
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | All others | 36/82 | 6/13 | Unknown |
| Proteobacteria | Gammaproteobacteria | Pseudomondales | 749/790 | 0/6 | Unknown | |
| Proteobacteria | Gammaproteobacteria | Salinisphaerales | 1/2 | Unknown | ||
| Proteobacteria | Gammaproteobacteria | Thiotrichales | 22/117 | 1/9 | Unknown | |
| Proteobacteria | Gammaproteobacteria | Vibrionales | 2/569 | 1/33 | Unlikely | |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | 0/419 | 0/1 | Unlikely | |
| All others | Unknown | |||||
Results are summarized from our full genome analysis (this study) and a previous study (2014 analysis). Applied category is the potential for cobalamin biosynthesis we used for each taxonomic group during our estimation of Thaumarchaeota contribution to biosynthesis of cobalamin.
Sañudo-Wilhelmy et al. (2014) (1).
Dupont et al. (2012) (87).
Fig. 5.
Pseudocobalamin and cobalamin producers at station 3. Shown are particulate cobalamin (closed blue circles), pseudocobalamin (orange open circles), prokaryote cell abundance (black diamonds), and in situ fluorescence (gray). Bar graphs show prokaryote population with the following predicted cobalamin strategies: unlikely producers (gray), bacteria with unknown cobalamin biosynthesis capacity (gray/blue striped), bacteria that are likely to produce cobalamin (blue), Thaumarchaeota (likely to produce cobalamin, white), and Cyanobacteria (likely to produce pseudocobalamin, orange) at the corresponding depths. Cobalamin and pseudocobalamin concentrations are as in Fig. 2.
Like Prochlorococcus, marine Thaumarchaeota have maintained several genes committed to the biosynthesis of cobalamin in their small genomes. Detection of MCM, NrdJ, BluB, and cobalamin biosynthesis proteins in the proteome of an oceanic thaumarchaeote with a highly streamlined genome, “Candidatus Nitrosopelagicus brevis,” implies that cobalamin is actively produced and used in these microbes (45). In Thaumarchaeota, the cobalamin-dependent MCM catalyzes a key step in their exceptionally energy-efficient pathway for carbon fixation (46–48). The scarcity of dissolved cobalamin in the water column (often <1 pM as assessed by bioassay) (1) and enzymatic demands like MCM may have necessitated that Thaumarchaeota retain the ability to synthesize cobalamin. Thaumarchaeota likely play a critical role in the microbial loop in the lower euphotic zone and deeper by providing an essential nutrient to cobalamin auxotrophs. In turn, the auxotrophs provide substrates that promote Thaumarchaeota growth (49, 50)—most critically the ammonia required by the ammonia-oxidizing Thaumarchaeota (46).
Although Thaumarchaeota and select heterotrophic bacteria synthesize cobalamin, undoubtedly benefiting from being their own source of their preferred cofactor, the production of dissimilar cobalamin analogs by marine Cyanobacteria is likely a result of their distinct ecological niches, enzymatic demands, and interactions with other cobalamin-dependent organisms. Producers of cobalamin and related compounds thus play distinct roles in cobalamin-based microbial interdependencies that sustain primary productivity and shape marine community structure.
Materials and Methods
Environmental samples for cobalamin and pseudocobalamin, phytoplankton abundance, archaeal gene quantification, prokaryotic cell abundance, and DNA for 16S rRNA sequencing were collected in August 2013 along the historical transect Line P to ocean station papa (our station 3), then following 150 W to the south into the North Pacific subtropical gyre sampling from the surface down to a maximum depth of 300 m, as shown in Fig. 2. Culture and environmental samples were analyzed for cobalamins, pseudocobalamins, and SAM using an organic solvent extraction (51) paired with LC-MS (15), both modified as described in SI Materials and Methods (Figs. S6 and S7 and Table S4). Phytoplankton abundance was monitored continuously using SeaFlow (16), in addition to discreet samples taken at depth and analyzed by flow cytometry.
Fig. S6.
Example chromatograms. Shown are two transitions from the five most common analytes (OH- and Me-pseudocobalamin and OH-, Me-, and Ado-cobalamin) in standards, cultures (Nitrosopumilus SCM1 and Prochlorococcus MED4), and the environment (surface sample from Station 1). Both the primary (used for quantification, black) and secondary (used for identity confirmation, gray) fragment ions are shown with an arbitrary y axis (ion intensity).
Fig. S7.
Chromatograms and standard curves from pseudocobalamin stock preparation and quantification. Shown are chromatograms of 0.50 µM cobalamin standards (in gray) and pseudocobalamin stocks at (A) 362 nm or (C) 259 nm and standard curves of OH-cobalamin (B, at 362 nm) and Me-cobalamin (D, at 259 nm) 0, 0.2, 0.5, 1, and 2.5 µM.
Table S4.
MS conditions used to monitor for pseudocobalamin and SAM
| Compound | Transitions monitored | CE, V | CV, V |
| OH-pseudocobalamin | 659.8 → 136.1, 348.1 | 30 | 45 |
| CN-pseudocobalamin | 673.3 → 136.1, 348.1, 660.3 | 30 | 45 |
| Me-pseudocobalamin | 668.3 → 136.1, 348.1, 660.3 | 30 | 30 |
| Ado-pseudocobalamin | 785.3 → 136.1, 348.1, 660.3 | 30 | 50 |
| S-adenosyl methionine | 399.258 → 136.1, 250.2 | 26 | 2 |
CE, collision energy in volts (V); CV, cone voltage in volts (V). For the pseudocobalamins, parent masses are all of the [M+2H]2+ except OH-pseudocobalamin, which loses its OH- during ionization as shown previously (15). 136 m/z, α ligand (adenine); 348 m/z, α ligand with sugar and phosphate groups; 660 m/z, doubly charged loss of the β ligand. Bolded transitions were used for quantification.
We investigated 11 axenic strains of marine prokaryotes for demonstrable evidence of cobalamin or pseudocobalamin production: four strains of Nitrosopumilus spp. (SCM1, HCA1, HCE1, and PS0), two strains of Prochlorococcus (MED4 and MIT9313), two strains of Synechococcus (WH7803 and WH8102), Sulfitobacter sp. SA11 (52), R. pomeroyi DSS-3, and V. fischerii ES114. We used the model diatom T. pseudonana to investigate the fate of pseudocobalamin in eukaryotic algae by culturing it under three conditions: cobalamin limited, cobalamin replete, and cobalamin limited with pseudocobalamin. To compare cobalamin- and pseudocobalamin-binding sites in MetH, we gathered MetH amino acid sequences from organisms known to use true cobalamin or pseudocobalamin as their cofactor. We then aligned the sequences and used a known crystal structure (32, 53) to visualize the binding pocket.
We used publically available full genomes from the IMG database that phylogenetically represent organisms at our study site and searched for cobalamin biosynthesis genes in these genomes (genomes and functions we used are listed in Dataset S1). We quantified Thaumarchaeota via qPCR and performed direct cell counts to quantify total prokaryotes as previously described (43, 44, 54). DNA for 16S rRNA sequencing was extracted, amplified, and sequenced as described in SI Materials and Methods. Operational taxonomic units (OTUs) were called using a 97% nucleotide identify threshold, and taxonomic inference was based on the SILVA rRNA gene database (https://www.arb-silva.de). This yielded phylogenetic information we combined with the current knowledge of cobalamin-biosynthesis capacity (from the literature and our whole genome analysis) to estimate the contribution of Thaumarchaeota to the prokaryotic community with the potential for cobalamin biosynthesis capacity at our sampling sites. Further details on all aspects of the methods are given in SI Materials and Methods. Environmental data are supplied in Dataset S2.
SI Materials and Methods
Cultures of Representative Cobalamin Producers.
We analyzed four axenic marine Thaumarchaeota strains in this study: Nitrosopumilus spp. SCM1, HCA1, HCE1, and PS0. All four strains were cultured in artificial medium as previously described (56, 57). Triplicate growth experiments with these strains were carried out in dark without shaking at 25 °C, except SCM1, which was grown at 30 °C. Growth was monitored by NO2− production and microscopic cell counts as previously described (49). Cultures of SCM1 (100 mL) were harvested at five different time points on 0.2 µm Nylon membrane filters (Millipore Co.) to obtain samples representing actively growing cultures at different growth phases. Late exponential phase cells of HCE1, HCA1, and PS0 (100 mL) were harvested on 0.2 µm Durapore membrane filters (Millipore Co.) with a vacuum filter system. To minimize photodegradation, cell harvesting was performed in the dark. All samples were stored at –80 °C until further analysis. For cell quotas reported, we used an estimate of dry weight at 16–20 fg per cell (46) and an estimate of 48% carbon to dry weight to get cobalamin per unit carbon. For reported SCM1 quotas in Table S1, we used the exponentially growing cells; quotas at other growth points are given in Dataset S2.
We analyzed four representative axenic marine Cyanobacteria: Prochlorococcus MED4 and MIT9313 and Synechococcus WH7803 and WH8102. The Prochlorococcus strains were both cultured in natural seawater-based Pro99 medium (58) prepared with 0.2 µm filtered, autoclaved seawater collected from Vineyard Sound, Massachusetts and supplemented with 10 mM sterile sodium bicarbonate. Prochlorococcus were grown in acid-washed polycarbonate carboys under constant light conditions (40 µmol·Q·m−2·s−1 for MED4, 15 µmol·Q·m−2·s−1 for MIT9313) at 24 °C in the Chisholm laboratory (Massachusetts Institute of Technology). Prochlorococcus axenicity was verified by testing for contaminant growth in three purity broths (ProAC, ProMM, and MPTB) (59–61). Synechococcus (WH7803 and WH8102) were cultured in artificial seawater-based Pro99 medium (58) prepared with Turks Island Salt Solution and supplemented with 6 mM sterile sodium bicarbonate and 1 mM N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (62). These strains were grown in combusted borosilicate glass tubes under 16:8 light:dark conditions (20 µmol·Q·m−2·s−1) at 20 °C. We verified that Synechococcus were axenic by testing for contaminant growth in liquid MPTB and on 1/2YTSS plates as well as by SYBR Green staining and observation on Influx high-speed cell sorter flow cytometer (BD Biosciences). All Cyanobacteria cells were collected in the mid- to late exponential growth phase. For each culture, replicate cell biomass filters (five for each Prochlorococcus strain and three for each Synechococcus strain) were collected by gently filtering 10 mL (for Prochlorococcus) or 24 mL (for Synechococcus) of culture onto a 0.2-µm nylon filter and freezing at –80 °C. Cell counts were performed by flow cytometry (for Prochlorococcus, GUAVA 8HT). For per carbon quotas for Cyanobacteria, we used a diameter range of 0.8–1.2 μm for Prochlorococcus MIT9313, 0.5–0.7 μm for MED4 (63), and diameters measured by flow cytometry for the Synechococcus with a conservative estimate of 325 fg·C·μm−3 (64).
Heterotrophic bacterial strains used in this study were Sulfitobacter strain SA11 (52), R. pomeroyi DSS-3, and V. fischerii ES114. All strains were cultured in an artificial seawater–tryptone media (30 g Instant Ocean salts, 5 g tryptone in 1 L MQ water). For cobalamin and pseudocobalamin analysis, cultures were grown overnight at room temperature, with shaking at 250 rpm on a Thermo Scientific Compact Digital Mini Rotator shaker table, using standard aseptic techniques. Overnight cultures were harvested by centrifugation (30 min at 4,100 rcf, 4 °C). Cell density was determined by enumerating plated culture forming units (cfus). R. pomeroyi and Sulfitobacter SA11 were harvested in mid-to late exponential growth phase; V. fischerii were harvested during the stationary phase. For cell quotas, we used a previously measured 142 fg C per cell for R. pomeroyi (65). We do not yet have a cellular carbon estimate for Sulfitobacter SA11, so we used a wide estimate of 32–292 fg C per cell previously measured from 13 species of marine Proteobacteria (65).
Diatom Growth Experiment.
Cultures of T. pseudonana CCMP 1335 were maintained on previously described defined media (66) with f/2 concentrations of nutrients, with the exception of cobalamin, which was added to a final concentration of 1 pM OH-cobalamin (a concentration previously shown to limit the growth rate of T. pseudonana) (39). Although our diatom cultures are not axenic, T. pseudonana could not be maintained on cobalamin-free media (with or without pseudocobalamin), so any bacterial production of or conversion to cobalamin was not sufficient to disrupt our experiment. After observing the high Ado-cobalamin content of our experimental cells, we compared them to an axenic population (axenicity monitored by marine purity test broth) (61) and observed the same pattern in which Ado-cobalamin ∼ OH-cobalamin > Me-cobalamin.
To assess the bioavailability of pseudocobalamin, we used a single culture to inoculate a set of cultures with three different treatments: Control (1 pM OH-cobalamin, same as maintained conditions), +Cobalamin (200 pM OH-cobalamin), and +Pseudocobalamin (1 pM OH-cobalamin with 200 pM OH-pseudocobalamin). Triplicates of each treatment were grown in 50 mL borosilicate culture tubes with 35 mL media at 20 °C with a 12-h light–dark cycle. Cultures were monitored by fluorescence and harvested by gentle vacuum filtration onto a 0.2-µm nylon filter at midexponential phase. We took 1-mL samples (fixed with 1% formaldehyde) at four time points throughout the growth curve for cell counts and used a Beckman Coulter Z2 Particle Count and Size Analyzer (Beckman Coulter) to measure diatom densities and diameters. To estimate carbon content of T. pseudonana in our experiment, we used the average observed diameter (5.36 µm) to calculate carbon using pg C = 0.109V0.991 (17, 67).
Genomic and Phylogenetic Analysis of Cobalamin Biosynthetic Capacity.
We analyzed full genomes from the IMG database (https://img.jgi.doe.gov) from phylogenetic groups that encompassed >99.9% of the Bacterial 16S reads from our environmental samples to develop a systematic inference of cobalamin synthesis capacity. We only included bacterial genomes of high quality (denoted finished or published) and removed all organisms noted as engineered. We included finished or published Thaumarchaeota genomes (including lower quality sequences), excluding Aigarchaeota Caldiarchaeum spp., whose phylogenetic placement is still unknown but likely outside of Thaumarchaeota (68). This resulted in 3,410 full genomes, which we split into six phylogenetic groups: Flavobacteriia, Alphaproteobacteria, Gammaproteobacteria, Cyanobacteria, Thaumarchaeota, and a final group made up of 265 genomes from mixed Bacteria groups with poor phylogenetic coverage in the database such as Chlorobi and Deltaproteobacteria (see Dataset S1 for phylogenetic groups used, functional enzymes searched, and lists of surveyed genomes in each subgroup). Next, we searched these genomes for annotations of functional enzymes related to cobalamin synthesis and clustered them according to the annotated presence or absence of the genes of interest. Each phylogenetic group clustered easily into two groups: genomes with many genes related to the central corrin ring synthesis, and genomes with few genes related to the central corrin ring synthesis.
Beyond relying on annotations, we also searched for potential homologs of genes related to DMB synthesis and activation in Thaumarchaeota and Cyanobacteria genomes. No BLASTp (69, 70) hits below an e-value threshold of 10−5 with bit score greater than 50 could be identified as cobT in the 27 surveyed Thaumarchaeota genomes or 255 Cyanobacterial genomes. Using the same thresholds, we were unable to identify any bluB homologs in any Prochlorococcus or Synechococcus genomes. The newly described anaerobic pathway for synthesizing DMB is not yet annotated, so we searched Prochlorococcus and Synechococcus genomes for evidence of sequence similarity to bzaC, bzaD, and bzaE (22). We were unable to find any BLASTp hits with an e-value threshold of 10−5 with bit score greater than 50 for bzaC. Although there were several BLASTp hits with a lower e-value for bzaD and bzaE, the hits never yielded an alignment that covered greater than 50% coverage of either protein with greater than 30% identical sequences. Given the low sequence similarity and the observation in the obligate anaerobe Eubacterium limosum bzaD and bzaE depend on bzaC to synthesize DMB (22), we concluded that this was not persuasive evidence for DMB synthesis capacity in Prochlorococcus or Synechococcus.
MetH Alignment and Comparison.
We gathered MetH amino acid sequences from the Cyanobacteria genomes in our surveyed full genomes. We added in MetH sequences from organisms known to use true cobalamin as their cofactor: E. coli, Pseudomonas, T. pseudonana, and Homo sapiens (12, 37, 71). Sequences were aligned using MAFFT v6.864 (www.genome.jp/tools/mafft/) (72) and trimmed to include only the cobalamin-binding motif (31, 32). A selection of aligned sequences is shown in Fig. S3. Using the cobalamin-binding motif of Prochlorococcus MIT9313, we used SWISS-MODEL (73) to visualize a rough proposed structure of MetH from Prochlorococcus MIT9313 based on the same enzyme in E. coli (53) (37.6% identical in cobalamin binding domain). For protein graphics, we used the University of California, San Francisco (UCSF) Chimera Package (74) (Resource for Biocomputing, Visualization, and Informatics at UCSF, www.cgl.ucsf.edu/chimera) to edit the crystal structure to include pseudocobalamin in place of cobalamin. Using the built-in parameters on the UCSF Chimera Package, we elucidated possible hydrogen bonds between adenine and the protein as shown in Fig. S4.
Field Sampling.
At sea, samples for particulate cobalamin and pseudocobalamin, archaeal gene quantification, prokaryotic cell abundance, and phytoplankton abundance were collected using Niskin bottles mounted onto a rosette system equipped with a conductivity–temperature–depth sensor package (CTD, Seabird SBE). Cobalamin and pseudocobalamin sampling was performed in a single cast at ∼11 AM local time at each location; fluorescence profiles are reported as uncorrected relative fluorescence units (RFUs) for qualitative purposes only. Other reported measurements were taken no more than 7 h earlier. Samples for DNA were collected using in situ McLane large volume pumps collecting particles from the 0.2–1.6-µm size range by using a GF/A prefilter (Whatman) and a 0.2 µm Supor (Pall) filter.
Cobalamin, Pseudocobalamin, and SAM Extraction and Quantification.
At sea, 2 L samples for particulate analysis of cobalamins and SAM were filtered onto 0.2 µm Nylon membrane filters and frozen in combusted Al foil at –20 °C (preliminary tests showed no difference between samples frozen at –20 °C or –80 °C). Culture samples were processed in the same way as environmental samples but with smaller volumes as noted. For the archaea and bacteria cultures, we processed and analyzed blank media from all cultures according to previously described methods (15) and verified that there was no detectable cobalamin or pseudocobalamin in the starting media.
To extract metabolites from cellular matter in culture and environmental samples, we used an organic solvent extraction that has previously been used for marine environmental and culture samples (51, 75) paired with physical bead beating. We placed filters into bead beating tubes containing 100 and 400 µm beads of equal volume, added 1 mL of cold (–20 °C) acidic acetonitrile:methanol:water mixture (40:40:20 with 0.1% formic acid) (51). Over the course of 20 min, we bead beat the samples for 40 s three times and kept the samples in a –20 °C freezer when possible. After centrifugation, the supernatant was removed and the filter was rinsed once with the 40:40:20 solvent and twice with methanol, centrifuging and combining the supernatants after each step. The supernatant and rinses were dried down under clean N2 or under vacuum with minimal heat (less than 40 °C). Due to the light-sensitive nature of these compounds, extractions were performed under dimmed lights, and samples were protected from light exposure whenever possible. Without a consensus standard, it is difficult to ascertain if this is the most effective extraction, but our reproducibility suggests it is a consistent technique that yields comparable data between samples.
Samples were reconstituted with an internal standard mixture and quantified by standard addition curves using ultra-high pressure LC coupled to a triple quadrupole mass spectrometer (Waters I-Class Aquity UPLC coupled to a Waters Xevo TQS) as previously described (15) but altered to include four additional transitions corresponding to the CN-, OH-, Me-, and Ado-pseudocobalamin as well as SAM (Table S4). Internal standard concentrations were identical to previous work on the same LC-MS system (15). Injection volumes were 40 µL for all environmental samples. Total cobalamin or pseudocobalmin values presented in all figures are the summed total of all detected β ligands for the corresponding cobalamin analog. Transitions, collision energies and cone voltages for the pseudocobalamins were optimized using a quantified pseudocobalamin stock from a recognized pseudocobalamin source, obtained and quantified as described in the following section. These quantified pseudocobalamin stocks were also used for the standard addition curves for quantification. We used a standard of SAM to optimize transitions, collision energies, and cone voltages for this compound. See Fig. S6 for example chromatograms.
Limits of detection in LC-MS analyses can vary among sample matrices (affecting background or analyte ion suppression), injection volume, chromatography quality, and instrument performance. Instead of using a preset concentration as our limit of detection, we assessed data quality on a batch-by-batch basis (i.e., environmental samples and culture samples were run at separate times and therefore treated as separate batches). Each peak is subject to the following criteria during our quality control: Peaks must be at the same retention time (±0.2 min) as a standard, at least two daughters must be present, signal-to-noise ratio greater than 5, and the integrated peak area at least five times greater than any peak found in the blank in the appropriate retention time window.
Preparation of Pseudocobalamin Stocks.
Pseudocobalamin is not available for purchase in its most ecologically relevant forms (with β ligands of OH-, Me-, or Ado-). To quantify the compound, we concentrated and quantified pseudocobalamins from a known pseudocobalamin source: Spirulina powder (GNC SuperFoods Spirulina) (76). First we extracted pseudocobalamin from ∼6 g of Spirulina powder using the extraction method described in this publication. The extract was dried and reconstituted in 20 mL Milli-Q water (>18.2 MΩ·cm, MQ). A clean-up step aimed at obtaining a pseudocobalamin-enriched fraction of the extract was performed by loading the aqueous mixture onto a conditioned C18 solid-phase extraction column (Waters, 35 mL capacity, 10 g resin). The column was rinsed with 40 mL MQ water, 20 mL 5% (vol/vol) MeOH in MQ, and 20 mL 30% (vol/vol) MeOH in MQ, before eluting the pseudocobalamin-containing fraction with 60 mL 50% (vol/vol) MeOH in MQ.
Further purification of the pseudocobalamin-containing fraction was achieved by a two-step HPLC method with fraction collection (HPLC, Agilent 1100). For purification, we carried out 50 µL injections onto a C18 column (4.6 × 150 mm Agilent Eclipse). Initial conditions were 92% (vol/vol) solvent A (water with 20 mM ammonium formate and 0.1% formic acid) and 8% (vol/vol) solvent B (acetonitrile). The gradient ramped from 8% to 25% (vol/vol) solvent B over 25 min with a flow rate of 2 mL·min−1. Fractions were collected in 5-mL aliquots, and each aliquot was analyzed for Me- and Ado-pseudocobalamin by ultra-performance liquid chromotography (UPLC) MS/MS as described for sample quantification in this text (Waters Aquity UPLC coupled to Xevo TQS mass spectrometer). Pseudocobalamin-containing fractions were dried down and subjected to a second round of HPLC purification consisting of an isocratic HPLC run of 8% (vol/vol) B and 13% (vol/vol) B to isolate Ado- and Me-pseudocobalamin, respectively (modeled after previous purifications) (77). Fractions containing each analyte were collected and pooled. At this point, our fractions are not pure (bulk UV-VIS does not show characteristic corrin ring peaks), but UPLC MS/MS analysis (the most sensitive analysis available to us) confirmed that fractions contained only the pseudocobalamin of interest and no detectable cobalamin forms, imperative to our diatom experiment. To make OH-pseudocobalamin, we exposed glass vials containing an aqueous solution of the Me-pseudocobalamin containing fraction to full spectrum light for 10 min (78). Throughout the preparation of pseudocobalamin stocks, we took great care to protect the quantified stocks from light to avoid degradation. To make CN-pseudocobalamin, we exposed this fraction to 50:50 acetonitrile:water in full spectrum light for 10 min. This produced a mixture of both CN-pseudocobalamin and OH-pseudocobalamin.
Quantification of Pseudocobalamin Stocks.
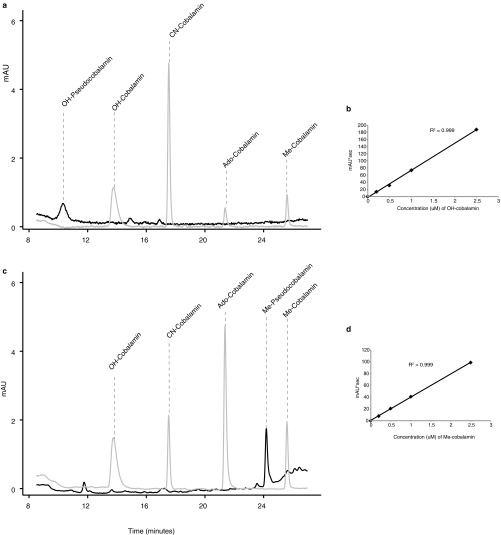
OH- and Me-pseudocobalamin were analyzed spectrophotometrically by HPLC–diode array detection (DAD) using 362 nm for OH- and 259 nm for Me-pseudocobalamin detection. To our knowledge, no UV-VIS data exist for Me- or OH-pseudocobalamin, so we used the observed extinction coefficients of Me- and OH-cobalamins (39,700 and 74,600 cm−1·mol−1, respectively) to quantify the corresponding pseudocobalamins as previously done for cobalamin analogs with CN β ligands (77), with chromatograms shown in Fig. S7. All data are therefore presented as cobalamin equivalents as noted in the figure legends. Although cobalamin and pseudocobalamin likely have different extinction coefficients, this approach provides a first attempt to compare concentrations of these analogs. Furthermore, if UV-VIS–based extinction coefficients become available in the future for Me- or OH-pseudocobalamin, a simple conversion at the wavelengths measured will provide true pseudocobalamin concentrations. Although we were able to confirm the structure and continue to monitor for Ado- and CN-pseudocobalamin by UPLC MS/MS, we were unable to purify enough Ado- or CN-pseudocobalamin to quantify by DAD due to the much lower concentration in Spirulina; we were only able to detect these forms in the Prochlorococcus and Synechococcus cultures (where the peaks were 50–100 times smaller than the OH- or Me-pseudocobalamin peaks) and in the diatom experiment. The HPLC–DAD mobile phases and column used for quantification of pseudocobalamin standards mobile phases were the same as for purification but with a flow rate of 0.3 mL·min−1 and a gradient from 9% to 32% (vol/vol) B over 30 min.
Confirmation of Pseudocobalamin Structure.
To confirm the presence of OH-, Me-, CN-, and Ado-pseudocobalamin, we obtained accurate mass data for our analytes using an LC-MS system consisting of a Waters Acquity UPLC coupled to a quadrupole time-of-flight mass spectrometer (Waters Xevo G2-S QTOF) in positive ion mode. We used observed accurate masses, fragmentation patterns, and retention times to confirm compound identities. Electrospray ionization source conditions for the QTOF method were the same as those used during sample quantification, as previously described (15). We used MS/MS conditions and extracted the predicted accurate mass from associated spectra of each parent without collision energy (doubly charged; OH-, 659.7750; Me-, 667.7907; Ado-, 784.8220; CN-, 673.2805) and corresponding daughters (described in Table S4: adenine, 136.0623; adenine with sugar and phosphate, 348.0709; loss of β ligand, 660.2789 m/z). For each compound, we detected the doubly charged parent and each of its daughters at the same retention time with a 0.01 Dalton mass tolerance. Retention times matched those shown in Fig. S6. As expected from previous work with CN-pseudocobalamin (22, 77), each of the pseudocobalamins eluted earlier than its corresponding cobalamin (same β ligand) in both the HPLC (purification and stock quantification) and UPLC (for stock confirmation and sample quantification, Figs. S6 and S7) analyses.
Phytoplankton Abundance and Composition.
While at sea, continuous measurements of picophytoplankton abundance and cell size were made using SeaFlow (16). The instrument was equipped with a 457-nm, 300-mW laser (Melles Griot). Forward light scatter (a proxy for cell size) and orange and red fluorescence were collected using a 457–50 bandpass filter, 572–27 bandpass filter, and 692–40 bandpass filter, respectively. Seawater was prefiltered through a 100-µm stainless steel mesh (to eliminate large particles) before analysis. The flow rate of the water stream was set at 15 mL·min−1 through a 200-µm nozzle for field and laboratory experiments; this corresponded to an analysis rate of 15 µL·min−1 by the instrument (16). A programmable syringe pump (Cavro XP3000, Hamilton Company) continuously injected fluorescent microspheres (1 µm, Polysciences) into the water stream as an internal standard. Data files were created every 3 min. Data were analyzed using the R package Popcycle version 0.2, which uses a SQLite relational database management system to retrieve flow cytometry data (https://github.com/uwescience/popcycle).
Discrete 2-mL samples for cytometry analysis were collected at different depths and fixed to a final concentration of 1% paraformaldehyde and 0.01% glutaraldehyde, frozen in liquid nitrogen, and analyzed onshore by an Influx high-speed cell sorter (BD Biosciences). A sequential bivariate manual gating scheme was used to cluster the different phytoplankton populations in both continuous and discrete samples. Phycoerythrin (PE)-positive cells (Synechococcus) were classified based on orange fluorescence. Light scattering and red fluorescence were used to distinguish Prochlorococcus and picoeukaryotes.
To estimate carbon content of environmental Prochlorococcus and Synechococcus populations, we first estimated Prochlorococcus and Synechococcus cell volumes using an empirical relationship between light scatter measured by the flow cytometer and cell volume measured by a Coulter Counter for different exponentially growing phytoplankton cultures of cell sizes with diameters ranging from 1 to 10 μm (79) and again used a conservative estimate of 325 fg C·μm−3 (64). The carbon per cell was multiplied by cell abundance to obtain the carbon content (μg·L−1) and averaged and binned into 0.5 × 0.5 degree bins.
qPCR Analysis of amoA.
The archaeal ammonia monooxygenase subunit A (amoA) gene abundances were estimated by qPCR analysis. Environmental samples (1 L) for qPCR were filtered on 0.2-µm Sterivex-GP filters (Millipore Co.) with a peristaltic pump, immediately frozen in liquid nitrogen, and stored in a –80 °C freezer until analysis. DNA was extracted in the laboratory using the modified phenol-chloroform protocol and quantified using the ND-1000 spectrophotometer (NanoDrop Technologies) as described previously (43, 44, 80). qPCR targeting archaeal amoA gene copy number was performed using the Roche LightCycler FastStart DNA Master SYBR Green I kit and Roche LightCycler capillary system with the cycling conditions described previously (43, 44). The primer sets used for targeting archaeal amoA genes were CrenAmoAQ-F and CrenAmoAModR (81). The amplification efficiencies for qPCR reactions were in the range of 96.6–97.4%.
Prokaryotic Cell Counts.
Environmental water samples for total prokaryotic cell counts (50 mL) were fixed with 2% (vol/vol) glutaraldehyde and filtered (1 mL) onto a 0.02-µm Anodisc 25 filter (Whatman). Cells were stained with Moviol-SybrGreen mix and counted with an Olympus BHS/BHT system microscope on board (54). At least 20 random fields of view with 10–100 SybrGreen stained cells per field were counted.
Environmental 16S Sequencing and Analysis.
DNA extraction methods were adapted from the spin-column protocol for the Qiagen DNeasy Blood & Tissue Kit. A section from each 0.2-μm Supor filter (collected with large volume in situ McClane pumps) was placed in a falcon tube, submerged in 4 mL sucrose lysis buffer, and flash frozen in liquid nitrogen. After thawing, 200 μL of 1 mg/mL lysozyme was added, and the tube was incubated at 4 °C for 1 h, shaking every 10 min. Next, 930 μL 10% (vol/vol) SDS and 500 μL proteinase K were added, and the tube was placed in a shaking incubator at 55 °C for 2 h. After the incubation, 5 mL AL buffer and 5 mL 100% ethanol were added to the tube, and the solution was centrifuged through three Mini Spin Columns for 1 min at 6,973 × g. Wash buffers AW1 and AW2 were then applied as described in the Qiagen Blood & Tissue Kit Protocol. The DNA was eluted with 30 μL water, which was added to the spin column membrane for 2 min at room temperature and centrifuged 1 min at full speed into microfuge tubes. The elution step was repeated with another 30 μL of water to yield 60 μL of eluent.
Genomic DNA was amplified for 16S rRNA genes with previously described primer sets (82), independently followed by sample-specific barcoding using a microfluidic array (Fluidigm Access Array TM, Fluidigm INC). Barcoded samples were quantified using pico-green, diluted to equal-molar concentrations, pooled, and sequenced in paired-end mode on an Illumina MiSeq. Fastq files were trimmed (200 bp) and quality filtered in QIIME with default setting adjustments according to previous methods (83). Quality sequences were then analyzed using the usearch software as suggested in “UPARSE” (84). Briefly, OTUs were called using a 97% nucleotide identity threshold and dereplicated sequences mapped to OTUs using “usearch_global” with the default settings.
Taxonomic inference was used based on comparison of OTU sequences to the SILVA rRNA gene database (version 119) using nucleotide basic-local alignment searches (i.e., BLASTN and BLAST 2.2.28+) (85). Sample matrices were converted to biom format, and taxonomic metadata were incorporated from SILVA and summarized at varying resolutions using “summarize_taxa.py” in QIIME. Data from this study can be found in the European Nucleotide Archive under accession no. PRJEB10943 (ERP012248) (Project Title “Variable Influence of Light on the Activity of Thaumarchaea”).
Estimation of Thaumarchaeota Contribution to the Microbial Community with Potential for Cobalamin Synthesis.
We summarized the potential for cobalamin synthesis in important marine prokaryotes by using our full genome analysis (grouped taxonomically) and previous literature (1, 8, 10, 86, 87) (results summarized in Table S3). Although inferences from phylogeny are most valuable when done using specific phylogenetic classification, many groups have few high-quality genomes available (i.e., the Actionobacter and Planctomycetes phyla). For groups with few genomes available, we used any genomes available at the wider classifications (Phylum and Class). When possible, we collected genomes at the Order level. Groups were generally considered unlikely, likely, or unknown to have cobalamin biosynthesis capacity if less than 10%, greater than 80%, or between 10% and 80% of the genomes had corrin biosynthetic capacity, respectively. Most of the genomes from the Pseudomonadales order with cobalamin biosynthetic capacity were from one species (Dataset S1), which is not marine. Of the marine genomes available (six genomes), none have the biosynthetic capacity (1), thus we categorized them as unknown rather than likely cobalamin producers. Similarly, no high-quality genomes of the abundant SAR86 Clade are available, but other studies have shown that they are unlikely to have the genetic capacity for cobalamin biosynthesis (87), so we separated these organisms from the remaining Oceanospirillales.
We then applied the categories to our genomic 16S rRNA-derived taxonomy (after filtering out mitochondria and chloroplast reads). This step assumes that the categories of cobalamin biosynthesis capacity derived from high-quality genomes in the IMG database reflect the microbial population present in our study site and yields the percentage of bacteria with the likely, unlikely, and unknown genomic potential for cobalamin biosynthesis. Using 16S rRNA-based abundances imperfectly describes bacterial community composition due to variable copy numbers of 16S among bacteria (88). In the ocean, this can lead to an overestimation of taxa with high 16S rRNA copy numbers like Alteromonadales and a slight underestimation of taxa with low 16S rRNA copy numbers like Rhodobacterales (89).
To investigate the Thaumarchaeal contribution to cobalamin biosynthetic capacity, we first used the qPCR amoA data to calculate an abundance of ammonia-oxidizing archaea in the water column (43, 44), as there is one copy of amoA per cell in all characterized Thaumarchaeota (90). We assumed that the abundance of ammonia-oxidizing archaea was equal to the abundance of archaeal cells capable of cobalamin biosynthesis [marine group II euryarchaea are unlikely to have cobalamin biosynthesis capacity (10, 91) and are a small percentage of total prokaryotes (92)].
We used the difference between the prokaryote cell counts and qPCR amoA gene counts to estimate the bacterial abundance at each location; this may be a slight underestimation of bacterial contribution in the surface ocean as other archaeal groups are likely present. For the cyanobacterial contribution, we multiplied the contribution of 16S reads attributed to Cyanobacteria (but not chloroplasts) by the bacterial cell abundance. In two instances, we had both 16S data and flow cytometry data (St. 4, 70 m; St. 2, 80 m); cyanobacterial contributions to the microbial population were low (0.5–3%) by direct observations and 16S data. For all other bacterial contributions, we took the percentage of bacteria with the different genomic potentials (likely, unlikely, unknown) for cobalamin biosynthesis and multiplied it by our cellular abundance. All data used in these calculations are reported in Dataset S2.
Supplementary Material
Acknowledgments
We thank L. T. Carlson, R. Lionheart, A. Weid, R. Morales, and D. Rico for laboratory assistance; L. T. Carlson, D. French, R. Horak, A. Nelson, S. Amin, H. van Tol, and the captain and crew of the R/V Kilo Moana for sampling support; S. Chisholm, J. Becker, and S. Biller for axenic Prochlorococcus cells; and B. Durham for axenic Synechococcus cells. This work was supported by National Science Foundation (NSF) Awards OCE1228770 (to A.E.I. and D.A.S.) and OCE1046017 (to A.E.I., E.V.A., D.A.S., A.H.D., and J.W.M.). This work was supported by the Simons Foundation SCOPE Award ID 329108 (to A.E.I., E.V.A., and S.C.) and NSF Research Experience for Undergraduates Award OCE-1046017AM004 (to W.C-M.). K.R.H. was partially supported by an NSF Graduate Research Fellowship Program and University of Washington's Program on Climate Change graduate student fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive [accession no. PRJEB10943 (ERP012248) (project title “Variable Influence of light on the activity of Thaumarchaea”)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608462114/-/DCSupplemental.
References
- 1.Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. The role of B vitamins in marine biogeochemistry. Annu Rev Mar Sci. 2014;6(1):339–367. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 2.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19(4):390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 3.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438(7064):90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 4.Droop MR. Vitamins, phytoplankton and bacteria: Symbiosis or scavenging? Plankton Res. 2007;29(2):107–113. [Google Scholar]
- 5.Azam F. Microbial control of oceanic carbon flux: The plot thickens. Science. 1998;280(5364):694–696. [Google Scholar]
- 6.Bertrand EM, et al. Phytoplankton-bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc Natl Acad Sci USA. 2015;112(32):9938–9943. doi: 10.1073/pnas.1501615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand EM, Allen AE. Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol. 2012;3:375. doi: 10.3389/fmicb.2012.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durham BP, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;112(2):453–457. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper MB, Smith AG. Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr Opin Plant Biol. 2015;26:147–153. doi: 10.1016/j.pbi.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Doxey AC, Kurtz DA, Lynch MDJ, Sauder LA, Neufeld JD. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J. 2015;9(2):461–471. doi: 10.1038/ismej.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helliwell KE, et al. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol. 2016;26(8):999–1008. doi: 10.1016/j.cub.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renz P. Biosynthesis of the 5, 6-dimethylbenzimidazole moiety of cobalamin and of the other bases found in natural corrinoids. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley and Sons; New York: 1999. pp. 557–575. [Google Scholar]
- 13.Tanioka Y, et al. Methyladeninylcobamide functions as the cofactor of methionine synthase in a Cyanobacterium, Spirulina platensis NIES-39. FEBS Lett. 2010;584(14):3223–3226. doi: 10.1016/j.febslet.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Tanioka Y, et al. Occurrence of pseudovitamin B12 and its possible function as the cofactor of cobalamin-dependent methionine synthase in a cyanobacterium Synechocystis sp. PCC6803. J Nutr Sci Vitaminol (Tokyo) 2009;55(6):518–521. doi: 10.3177/jnsv.55.518. [DOI] [PubMed] [Google Scholar]
- 15.Heal KR, et al. Determination of four forms of vitamin B12 and other B vitamins in seawater by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2014;28(22):2398–2404. doi: 10.1002/rcm.7040. [DOI] [PubMed] [Google Scholar]
- 16.Swalwell JE, Ribalet F, Armbrust E. SeaFlow: A novel underway flow‐cytometer for continuous observations of phytoplankton in the ocean. Limnol Oceanogr Methods. 2011;9(10):466–477. [Google Scholar]
- 17.Taylor GT, Sullivan CW. Vitamin B12 and cobalt cycling among diatoms and bacteria in Antarctic sea ice microbial communities. Limnol Oceanogr. 2008;53(5):1862–1877. [Google Scholar]
- 18.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278(42):41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 19.Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature. 2007;446(7134):449–453. doi: 10.1038/nature05611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escalante-Semerena JC. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J Bacteriol. 2007;189(13):4555–4560. doi: 10.1128/JB.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Partensky F, Garczarek L. Prochlorococcus: Advantages and limits of minimalism. Annu Rev Mar Sci. 2010;2:305–331. doi: 10.1146/annurev-marine-120308-081034. [DOI] [PubMed] [Google Scholar]
- 22.Hazra AB, et al. Anaerobic biosynthesis of the lower ligand of vitamin B12. Proc Natl Acad Sci USA. 2015;112(34):10792–10797. doi: 10.1073/pnas.1509132112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crofts TS, Seth EC, Hazra AB, Taga ME. Cobamide structure depends on both lower ligand availability and CobT substrate specificity. Chem Biol. 2013;20(10):1265–1274. doi: 10.1016/j.chembiol.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Cheong C-G, Escalante-Semerena JC, Rayment I. Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica. J Biol Chem. 2001;276(40):37612–37620. doi: 10.1074/jbc.M105390200. [DOI] [PubMed] [Google Scholar]
- 25.Cheong C-G, Escalante-Semerena JC, Rayment I. The three-dimensional structures of nicotinate mononucleotide:5,6- dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella typhimurium complexed with 5,6-dimethybenzimidazole and its reaction products determined to 1.9 A resolution. Biochemistry. 1999;38(49):16125–16135. doi: 10.1021/bi991752c. [DOI] [PubMed] [Google Scholar]
- 26.Anderson PJ, et al. One pathway can incorporate either adenine or dimethylbenzimidazole as an α-axial ligand of B12 cofactors in Salmonella enterica. J Bacteriol. 2008;190(4):1160–1171. doi: 10.1128/JB.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keck B, Renz P. Salmonella typhimurium forms adenylcobamide and 2-methyladenylcobamide, but no detectable cobalamin during strictly anaerobic growth. Arch Microbiol. 2000;173(1):76–77. doi: 10.1007/s002030050011. [DOI] [PubMed] [Google Scholar]
- 28.Benner SA, Ellington AD, Tauer A. Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci USA. 1989;86(18):7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazcano A. Planetary change and biochemical adaptation: Molecular evolution of corrinoid and heme biosyntheses. Hematology. 2012;17(Suppl. 1):s7–s10. doi: 10.1179/102453312X13336169155015. [DOI] [PubMed] [Google Scholar]
- 30.Holland HD. The oxygenation of the atmosphere and oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361(1470):903–915. doi: 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig ML, Evans PR. X-ray crystallography of B12 enzymes: Methylmalonyl-CoA mutase and methionine synthase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley and Sons; New York: 1999. pp. 595–632. [Google Scholar]
- 32.Drennan CL, Matthews RG, Ludwig ML. Cobalamin-dependent methionine synthase: The structure of a methylcobalamin-binding fragment and implications for other B12-dependent enzymes. Curr Opin Struct Biol. 1994;4(6):919–929. doi: 10.1016/0959-440x(94)90275-5. [DOI] [PubMed] [Google Scholar]
- 33.Sintchak MD, Arjara G, Kellogg BA, Stubbe J, Drennan CL. The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer. Nat Struct Biol. 2002;9(4):293–300. doi: 10.1038/nsb774. [DOI] [PubMed] [Google Scholar]
- 34.Lundin D, Gribaldo S, Torrents E, Sjöberg B-M, Poole AM. Ribonucleotide reduction - Horizontal transfer of a required function spans all three domains. BMC Evol Biol. 2010;10(1):383. doi: 10.1186/1471-2148-10-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gleason FK, Olszewski NE. Isolation of the gene for the B12-dependent ribonucleotide reductase from Anabaena sp. strain PCC 7120 and expression in Escherichia coli. J Bacteriol. 2002;184(23):6544–6550. doi: 10.1128/JB.184.23.6544-6550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole AM, Logan DT, Sjöberg B-M. The evolution of the ribonucleotide reductases: Much ado about oxygen. J Mol Evol. 2002;55(2):180–196. doi: 10.1007/s00239-002-2315-3. [DOI] [PubMed] [Google Scholar]
- 37.Guillard RRL. B12 specificity of marine centric diatoms(1, 2) J Phycol. 1968;4(1):59–64. doi: 10.1111/j.1529-8817.1968.tb04677.x. [DOI] [PubMed] [Google Scholar]
- 38.Provasoli L, Carlucci AF. Vitamins and growth regulators. In: Stewart WDP, editor. Algal Physiology and Biochemistry, Botanical Monographs. University of California Press; Berkeley: 1974. pp. 741–787. [Google Scholar]
- 39.Bertrand EM, et al. Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc Natl Acad Sci USA. 2012;109(26):E1762–E1771. doi: 10.1073/pnas.1201731109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayers WA. Specificity of the vitamin B12 requirement in certain marine bacteria. J Bacteriol. 1960;80(6):744–752. doi: 10.1128/jb.80.6.744-752.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seth EC, Taga ME. Nutrient cross-feeding in the microbial world. Front Microbiol. 2014;5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito MA, Rocap G, Moffett JW. Production of cobalt binding ligands in a Synechococcus feature at the Costa Rica upwelling dome. Limnol Oceanogr. 2005;50(1):279–290. [Google Scholar]
- 43.Horak REA, et al. Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J. 2013;7(10):2023–2033. doi: 10.1038/ismej.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urakawa H, Martens-Habbena W, Stahl DA. High abundance of ammonia-oxidizing Archaea in coastal waters, determined using a modified DNA extraction method. Appl Environ Microbiol. 2010;76(7):2129–2135. doi: 10.1128/AEM.02692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santoro AE, et al. Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: An ammonia-oxidizing archaeon from the open ocean. Proc Natl Acad Sci USA. 2015;112(4):1173–1178. doi: 10.1073/pnas.1416223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urakawa H, Martens-Habbena W, Stahl DA. Physiology and genomics of ammonia-oxidizing archaea. In: Ward BB, Klotz MG, Arp DJ, editors. Nitrification. ASM Press; Washington, DC: 2011. pp. 117–156. [Google Scholar]
- 47.Walker CB, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107(19):8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Könneke M, et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA. 2014;111(22):8239–8244. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin W, et al. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA. 2014;111(34):12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J-G, et al. Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc Natl Acad Sci USA. 2016;113(28):7888–7893. doi: 10.1073/pnas.1605501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rabinowitz JD, Kimball E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal Chem. 2007;79(16):6167–6173. doi: 10.1021/ac070470c. [DOI] [PubMed] [Google Scholar]
- 52.Amin SA, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522(7554):98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 53.Datta S, Koutmos M, Pattridge KA, Ludwig ML, Matthews RG. A disulfide-stabilized conformer of methionine synthase reveals an unexpected role for the histidine ligand of the cobalamin cofactor. Proc Natl Acad Sci USA. 2008;105(11):4115–4120. doi: 10.1073/pnas.0800329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lunau M, Lemke A, Walther K, Martens-Habbena W, Simon M. An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ Microbiol. 2005;7(7):961–968. doi: 10.1111/j.1462-2920.2005.00767.x. [DOI] [PubMed] [Google Scholar]
- 55.Woodson JD, Peck RF, Krebs MP, Escalante-Semerena JC. The cobY gene of the archaeon Halobacterium sp. strain NRC-1 is required for de novo cobamide synthesis. J Bacteriol. 2003;185(1):311–316. doi: 10.1128/JB.185.1.311-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 57.Qin W, et al. Confounding effects of oxygen and temperature on the TEX86 signature of marine Thaumarchaeota. Proc Natl Acad Sci USA. 2015;112(35):10979–10984. doi: 10.1073/pnas.1501568112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moore LR, et al. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Methods. 2007;5(10):353–362. [Google Scholar]
- 59.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol. 2008;74(14):4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berube PM, et al. Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J. 2015;9(5):1195–1207. doi: 10.1038/ismej.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saito MA, Moffett JW, Chisholm SW, Waterbury JB. Cobalt limitation and uptake in Prochlorococcus. Limnol Oceanogr. 2002;47(6):1629–1636. [Google Scholar]
- 62.Jeffrey Morris J, Zinser ER. Continuous hydrogen peroxide production by organic buffers in phytoplankton culture media. J Phycol. 2013;49(6):1223–1228. doi: 10.1111/jpy.12123. [DOI] [PubMed] [Google Scholar]
- 63.Thompson AW, Huang K, Saito MA, Chisholm SW. Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J. 2011;5(10):1580–1594. doi: 10.1038/ismej.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DuRand MD, Olson RJ, Chisholm SW. Phytoplankton population dynamics at the Bermuda Atlantic Time-series station in the Sargasso Sea. Deep Sea Res Part II Top Stud Oceanogr. 2001;48(8):1983–2003. [Google Scholar]
- 65.Zimmerman AE, Allison SD, Martiny AC. Phylogenetic constraints on elemental stoichiometry and resource allocation in heterotrophic marine bacteria. Environ Microbiol. 2014;16(5):1398–1410. doi: 10.1111/1462-2920.12329. [DOI] [PubMed] [Google Scholar]
- 66.Sachs JP, Kawka OE. The influence of growth rate on 2H/1H fractionation in continuous cultures of the coccolithophorid Emiliania huxleyi and the diatom Thalassiosira pseudonana. PLoS One. 2015;10(11):e0141643. doi: 10.1371/journal.pone.0141643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montagnes DJS, Berges JA, Harrison PJ, Taylor F. Estimating carbon, nitrogen, protein, and chlorophyll a from volume in marine phytoplankton. Limnol Oceanogr. 1994;39(5):1044–1060. [Google Scholar]
- 68.Beam JP, et al. Ecophysiology of an uncultivated lineage of Aigarchaeota from an oxic, hot spring filamentous ‘streamer’ community. ISME J. 2016;10(1):210–224. doi: 10.1038/ismej.2015.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altschul SF, et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272(20):5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lengyel P, Mazumder R, Ochoa S. Mammalian methylmalonyl isomerase and vitamin B(12) coenzymes. Proc Natl Acad Sci USA. 1960;46(10):1312–1318. doi: 10.1073/pnas.46.10.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 74.Pettersen EF, et al. UCSF Chimera--A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 75.Soule MCK, Longnecker K, Johnson WM, Kujawinski EB. Environmental metabolomics: Analytical strategies. Mar Chem. 2015;177(2):374–387. [Google Scholar]
- 76.Watanabe F, et al. Pseudovitamin B(12) is the predominant cobamide of an algal health food, spirulina tablets. J Agric Food Chem. 1999;47(11):4736–4741. doi: 10.1021/jf990541b. [DOI] [PubMed] [Google Scholar]
- 77.Allen RH, Stabler SP. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr. 2008;87(5):1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Juzeniene A, Nizauskaite Z. Photodegradation of cobalamins in aqueous solutions and in human blood. J Photochem Photobiol B. 2013;122:7–14. doi: 10.1016/j.jphotobiol.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 79.Ribalet F, et al. Light-driven synchrony of Prochlorococcus growth and mortality in the subtropical Pacific gyre. Proc Natl Acad Sci USA. 2015;112(26):8008–8012. doi: 10.1073/pnas.1424279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martens-Habbena W, et al. The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol. 2015;17(7):2261–2274. doi: 10.1111/1462-2920.12677. [DOI] [PubMed] [Google Scholar]
- 81.Mincer TJ, et al. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9(5):1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- 82.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10(1):57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 85.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 86.Tripp HJ, et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452(7188):741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 87.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6(6):1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crosby LD, Criddle CS. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques. 2003;34(4):790–794, 796, 798 passim. doi: 10.2144/03344rr01. [DOI] [PubMed] [Google Scholar]
- 89.Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8(2):e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stieglmeier M, Alves RJE, Schleper C. The phylum Thaumarchaeota. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes. Springer; 2014. pp. 347–362. [Google Scholar]
- 91.Iverson V, et al. Untangling genomes from metagenomes: Revealing an uncultured class of marine Euryarchaeota. Science. 2012;335(6068):587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 92.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409(6819):507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.