Significance
Influenza A virus (IAV) is a major threat to global public health, and so understanding the biology of IAV is essential to develop antiflu vaccines and therapeutics. Here, we show the links between viral surface glycosylation and IAV function. The glycosylation of HA modulates virus infectivity, and host immune response; the glycosylation of NA affects its structure, activity, specificity, and thermostability to regulate virus release and virulence. In addition, using live attenuated IAV without the stalk and catalytic domains of NA as vaccine can strongly induce IAV-specific CD8+ T-cell responses to various virus strains. Therefore, our findings have clarified the role of glycosylation in IAV and provided a new direction for the development of universal flu vaccines.
Keywords: influenza A virus, glycosylation, vaccine design
Abstract
We have shown that glycosylation of influenza A virus (IAV) hemagglutinin (HA), especially at position N-27, is crucial for HA folding and virus survival. However, it is not known whether the glycosylation of HA and the other two major IAV surface glycoproteins, neuraminidase (NA) and M2 ion channel, is essential for the replication of IAV. Here, we show that glycosylation of HA at N-142 modulates virus infectivity and host immune response. Glycosylation of NA in the stalk region affects its structure, activity, and specificity, thereby modulating virus release and virulence, and glycosylation at the catalytic domain affects its thermostability; however, glycosylation of M2 had no effect on its function. In addition, using IAV without the stalk and catalytic domains of NA as a live attenuated vaccine was shown to confer a strong IAV-specific CD8+ T-cell response and a strong cross-strain as well as cross-subtype protection against various virus strains.
Influenza A viruses (IAV) belong to the Orthomyxoviridae family and can circulate widely and cross interspecies barriers through the highly antigenic drift and shift of the 18 subtypes of HA and 11 subtypes of neuraminidase (NA) (1, 2). In addition, the posttranslational modification of the IAV surface proteins is important to circumvent host defense and support the virus life cycle (3).
HA is a major surface glycoprotein of IAV and is involved in viral infection via binding to sialic acid (SA)-containing glycans on the surface of host cell (3). The other major glycoprotein of IAV, NA, is involved in the cleavage of SA on the host cell receptor to facilitate the release of viral particles to infect other cells (4). M2, the third surface protein of IAV, has ion channel activity to regulate virus penetration and uncoating (1). All surface proteins interact with M1 protein for virus assembly and release (5).
The modification of HA and NA by N-glycosylation is important in the IAV life cycle (1, 6–8). Previously, we have shown that the glycosylation of HA affects its receptor binding, immune response, and structural stability, and glycosite 27 is essential for retaining the structural integrity of HA and its receptor binding (6, 9). In addition, using the monoglycosylated HA as immunogen, it showed a broader protection against various IAV subtypes compared with the fully glycosylated version (10). However, it is not clear whether the other specific glycosites and their glycan structures on HA regulate its functions. With regard to NA, the functional roles of its glycosylation are not well understood, though N-glycosylation was reported to stabilize the protein from protease digestion and may affect the enzyme activity (11, 12). The aim of this study was to understand the functional effects of glycosylation on HA, NA, and M2, and how glycosylation of the surface proteins affects the life cycle of IAV.
Results
Glycosylation of HA at N-142.
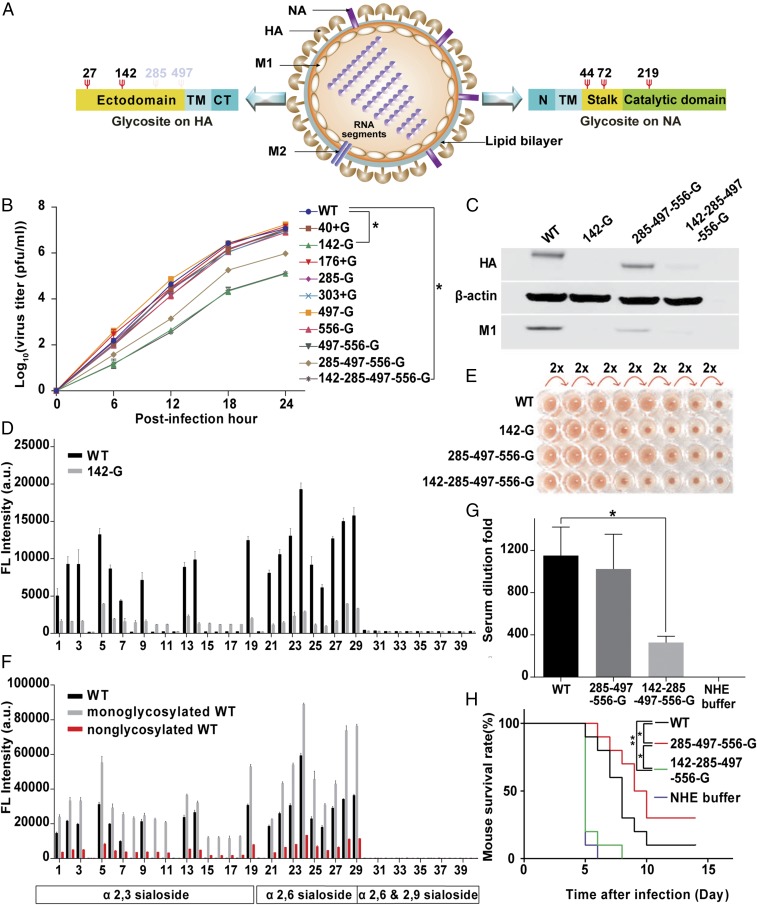
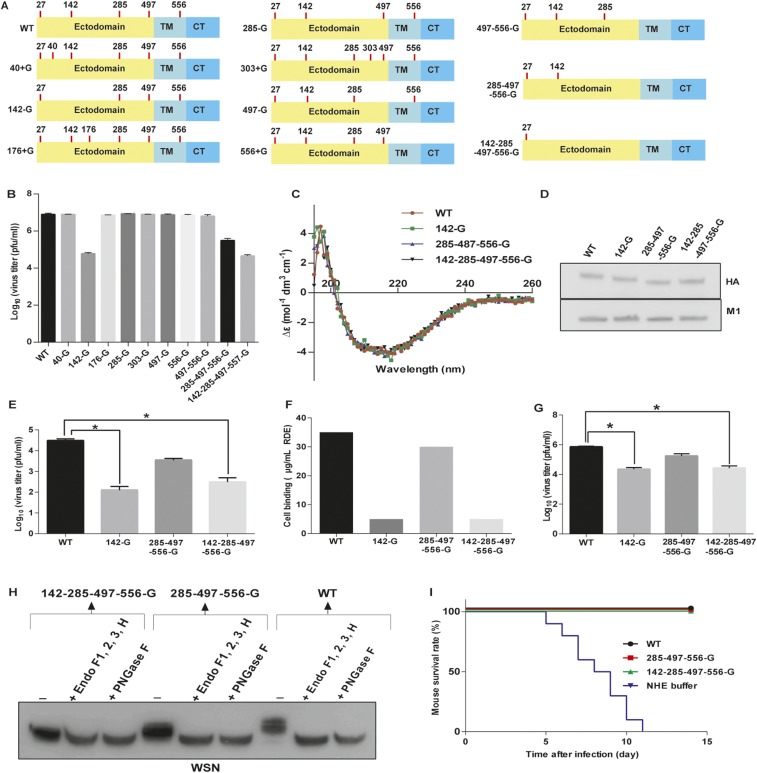
Because several glycosylation sites (glycosites 27, 40, 176, 303, and 497) on HA are highly conserved among the H1, H3, and H5 subtypes (9), we used wild-type H1N1 A/WSN/33 (WSN) as a model to create or delete specific glycosites to address the effect of glycosylation by using reverse genetics (Fig. 1A and Fig. S1A). We found that the virus could not survive with glycosite-27 deletion (i.e., N27A mutation designated as 27-G), consistent with our previous finding that glycosylation at glycosite 27 played a key role in HA folding (6). Similarly, we also mutated the highly conserved and potential glycosite at position 40, 176, or 303 to Asn (designated as 40 + G, 176 + G, and 303 + G) and evaluated the effect on replication. The results showed that the replication rates of variants with mutation at the highly conserved or potential glycosite (40 + G, 176 + G, 303 + G, or 497 − G) and WT were similar, but the replication rate of glycosite 142-deleted virus (142-G and 142-285-497-556-G virus) was two orders of magnitude lower than that of the WT virus in both Madin-Darby canine kidney cells (MDCK) and A549 cells, suggesting that glycosite 142 plays an important role in IAV replication (Fig. 1B and Fig. S1B). In a circular dichroism study, the glycans at glycosite 142 did not affect the secondary structure of HA (Fig. S1C), and the ratios of HA to M1 among the glycosite 142-deleted variants were similar, suggesting that glycosite 142 is not essential for virus assembly and/or maturation (5) (Fig. S1D).
Fig. 1.
HA glycosylation and IAV. (A) Schematic overview of glycosites (red Ψ) on IAV surface proteins. CT, C-terminal cytoplasmic domain; N, N-terminal cytoplasmic domain; TM, transmembrane domain. (B) Comparison of virus replication rates in MDCK cells. (C) Western blot analysis of A549 cells infected with four variants of IAV and the total Iysates were collected to run SDS/PAGE at 10 hpi. The filter was probed with anti-HA, anti-M1, and anti–β-actin antibodies. (D) HA binding patterns in a glycan array from WT and 142-G. (E) Hemagglutination assay of lAV variants. (F) Glycan array analysis of deglycosylated HA WT viruses. (G) Mice were immunized with inactivated viruses as indicated in the figure, and the sera of immunized mice were then analyzed using hemagglutination inhibition assay. (H) The mouse survival rate was measured after challenge with a lethal dose of H5N1 viruses. (B, D, and F) Mean ± SEM for three independent experiments. (G and H) Mean ± SEM for 10 independent experiments. *P < 0.001. **P < 0.05.
Fig. S1.
HA glycosylation and IAV. (A) Schematic overviews of 11 IAVs with different glycosites on HA highlighted with red lines; C, C-terminal cytoplasmic domain; TM, transmembrane domain. All recombinant viruses were confirmed by genome sequencing. (B) Comparison of virus production in A549 cells infected with 11 variants of virus at 48 hpi. (C) Circular dichroism spectra of HA variants as indicated. (D) Western blot analysis of four variants of IAV at the same amount. The filter was probed with anti-HA and anti-M1 monoclonal antibodies. (E) The infectivity of viruses with changes of HA glycosylation. MDCK cells were infected with the same amount of viruses as indicated in the figure and then the virus titer was determined by plaque assay. (F) Cell receptor binding avidities were determined by agglutination of turkey erythrocytes pretreated with RDE. The maximal amount of RDE that allowed full agglutination was measured. (G) Comparison of virus production in avian cells. Virus titers were determined from avian cells (LMH cells) infected with four variants of viruses as indicated at 48 hpi. (H) Western blot analysis of three variants of IAV after deglycosylation by the endoglycosidase mixture. The filter was probed with anti-HA antibody. (I) Following Fig. 1H, the mouse survival rate was measured after challenge with a lethal dose of WSN viruses. (B, E, F, and G) Mean ± SEM of three independent experiments. (I) Ten independent experiments are shown. *P < 0.001.
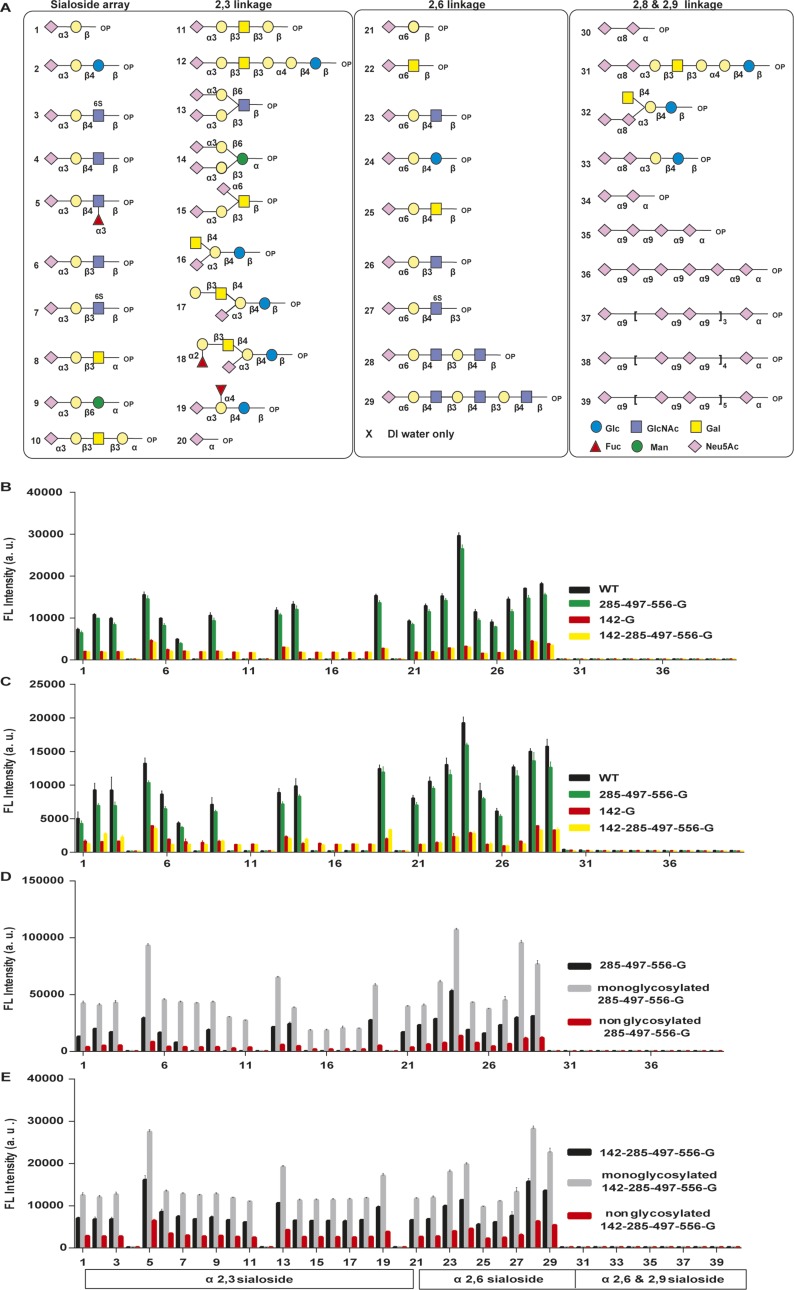
In the course of understanding the effect of glycosite 142 deletion on the life cycle of IAV, we found that glycosite 142 is very important for virus entry, because in the virus infectivity assay, very weak signals of HA and M1 were detected in the A549 cells infected with glycosite 142-deleted virus (142-G or 142-285-497-556-G virus; Fig. 1C and Fig. S1E). The glycan array analysis showed that the glycosite 142-deleted virus interacted with more sialosides than the WT virus (glycans 8, 10, 11, and 15–18), and the same results were also observed from the corresponding HA protein, suggesting that glycosylation at glycosite 142 affects the receptor binding specificity of HA (Fig. 1D and Fig. S2 A–C). In addition, glycosite 142 modulates the binding avidity of HA, because the glycosite 142-deleted virus showed lower fluorescence intensity in glycan array analysis, and had lower capability in hemagglutination and cell binding (Fig. 1 D and E and Fig. S1F). Although the glycosite 142-deleted virus can interact with more α2,3-sialosides in the glycan array analysis, it is not involved in the human and/or avian adoption of WSN HA, because the replication rate of 142-G or 142-285-497-556-G virus in LMH cell lines (from chicken hepatocellular carcinoma) was still two orders of magnitude lower than that of WT (Fig. S1G).
Fig. S2.
Glycan array analysis of HA with different glycosylations. (A) Overview of sialoside structures on the glycan array for IAV binding studies. This synthetic SA glycan array consisted of the following sialosides: 20 α2,3-glycans (1–20), 9 α2,6-glycans (21–29), and 10 α2,8 and α2,9 glycans (30–39), designed to study IAV binding. (B) Glycan array analysis of four variants of virus as indicated in the figure. (C) Glycan array analysis of four variants of HA protein. (D) Glycan array analysis of deglycosylated 285-497-556-G and (E) 142-285-497-556-G viruses. (B–E) Mean ± SEM of three independent experiments.
The molecular mechanism of avidity and specificity affected by the glycosylation at glycosite 142 was further studied and it was found that both avidity and specificity were modulated by the glycan composition. Treatment with a mixture of endoglycosidases (Endo-F1, F2, F3, and H) changed the interaction profile of the WT and 285-497-556-G viruses on the glycan array and the interaction patterns were similar to that of the 142-G and the 142-285-497-556-G viruses. Surprisingly, after treatment with the endoglcosidase mixture, the fluorescence intensity of the monoglycosylated virus on the glycan array was increased, but decreased in all types of nonglycosylated variants (PNGase F treatment; Fig. 1F and Figs. S1H and S2 D and E).
To study whether glycosite 142 is involved in the host immune response, we challenged mice with H5N1. The mice immunized with the inactivated 142-285-497-556-G virus survived for a shorter period and induced less HA antiserum compared with the mice immunized with inactivated WT virus, and the inactivated 285-497-556-G virus-immunized mice induced the same amount of HA antiserum but survived longer, and survived well in all cases after WSN challenge in the immunogenicity test (Fig. 1 G and H and Fig. S1I). This study suggests that glycosylation at glycosite 142 is important for the immunogenicity of IAV.
Glycosylation of NA and M2.
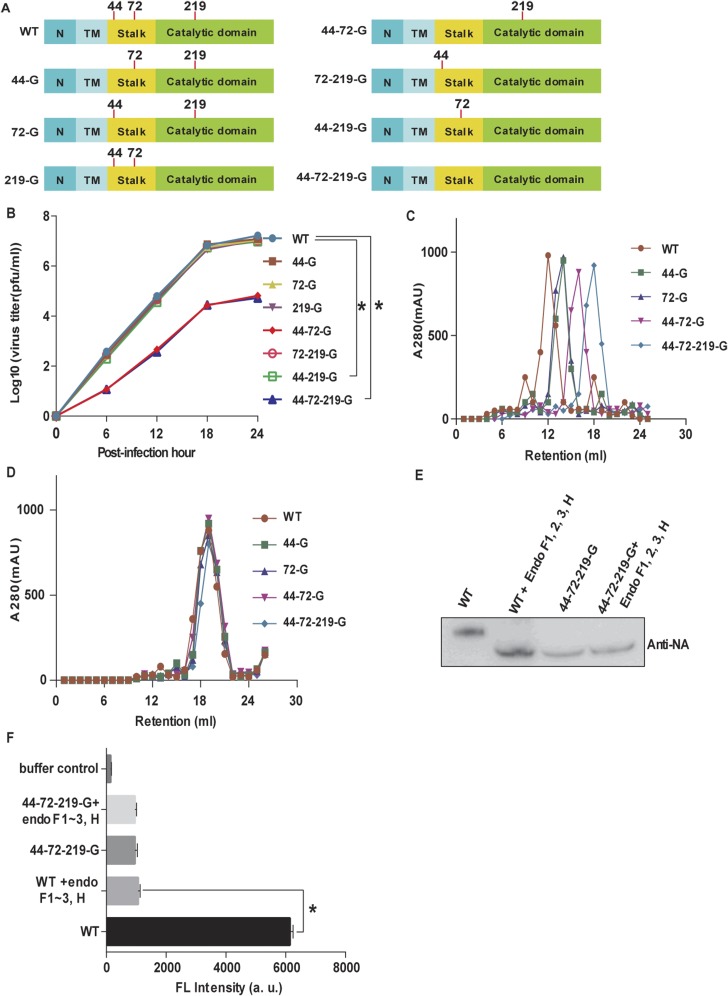
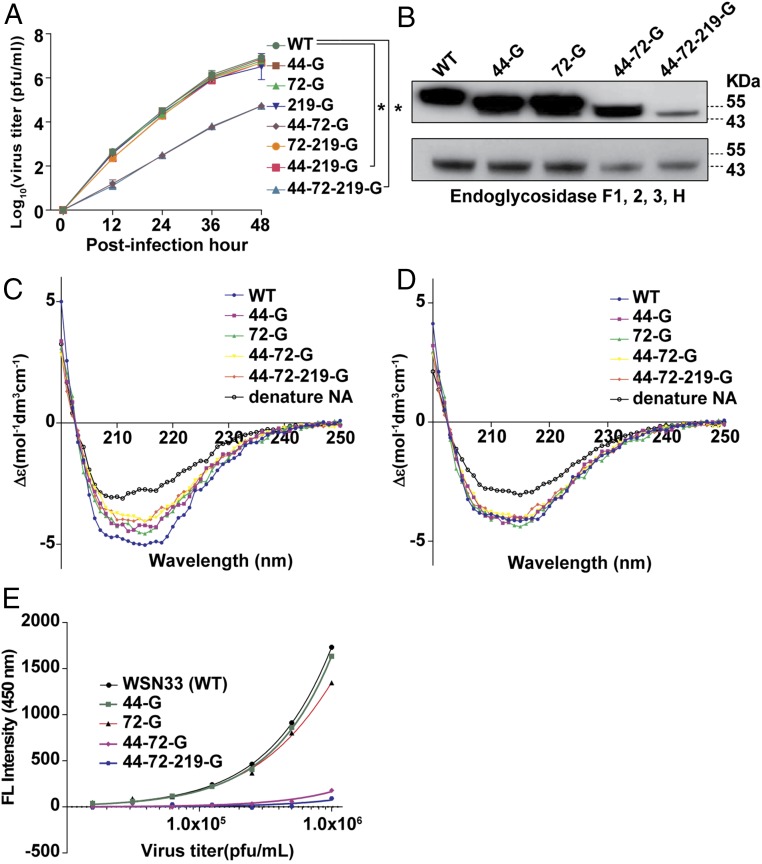
To understand whether NA glycosylation is involved in the life cycle of IAV, the same virus strain A/WSN/33 was used as a model for investigation of each glycosite by using reverse genetics (Fig. S3A). It was found that glycosites 44 and 72 (in the stalk domain) played an important role in virus replication, and the replication rate of the virus with deleted glycosites 44 and 72 (44-72-G or 44-72-219-G virus) was two orders of magnitude lower than the WT virus in both MDCK and A549 cells (Fig. 2A and Fig. S3B). Glycosites 44, 72, and 219 on NA were glycosylated to form variants of NA proteins with different molecular weights, but these variants showed similar molecular weights after treatment with the endoglycosidase mixture in Western blot analysis and gel filtration. (Fig. 2B and Fig. S3 C and D). Interestingly, the secondary structures of these variants were slightly different, but became the same after deglycosylation (Fig. 2 C and D). These results suggest that the glycans attached to glycosites 44, 72, and 219 are heterogeneous and affect the secondary structures of NA.
Fig. S3.
NA glycosylation and IAV. (A) Schematic overviews of eight IAVs with different glycosylation patterns on NA. Red lines, glycosite; N, N-terminal cytoplasmic domain; TM, transmembrane domain. All recombinant viruses were confirmed by genome sequencing. (B) Comparison of virus production in MDCK cells infected with eight variants of virus as indicated. Mean ± SEM for three independent experiments. (C) Gel filtration analysis of five types of NA proteins. (D) Gel filtration analysis of five types of deglycosylated NA proteins. (E) Western blot analysis of viruses after treatment with the endoglycosidase mixture. The filter was probed with anti-NA antibody. (F) Measurements of the NA activity of deglycosylated IAV by 4-MUNANA assay. Mean ± SEM for five independent experiments. *P < 0.001.
Fig. 2.
NA glycosylation and IAV. (A) Comparison of virus replication rates in A549 cells. (B) Western blot analysis of the molecular weights of glycosylated and deglycosylated variants as indicated. The filter was probed with anti-NA antibody. Mean ± SEM for three independent experiments. (C) Circular dichroism spectra of different types of NA as indicated. (D) Circular dichroism spectra of deglycosylated variants of NA. (E) NA activity of IAV measured by 4-MUNANA assay. Mean ± SEM for five independent experiments. *P < 0.001.
Glycans at N-44 and N-72 Affect NA Activity and Virulence.
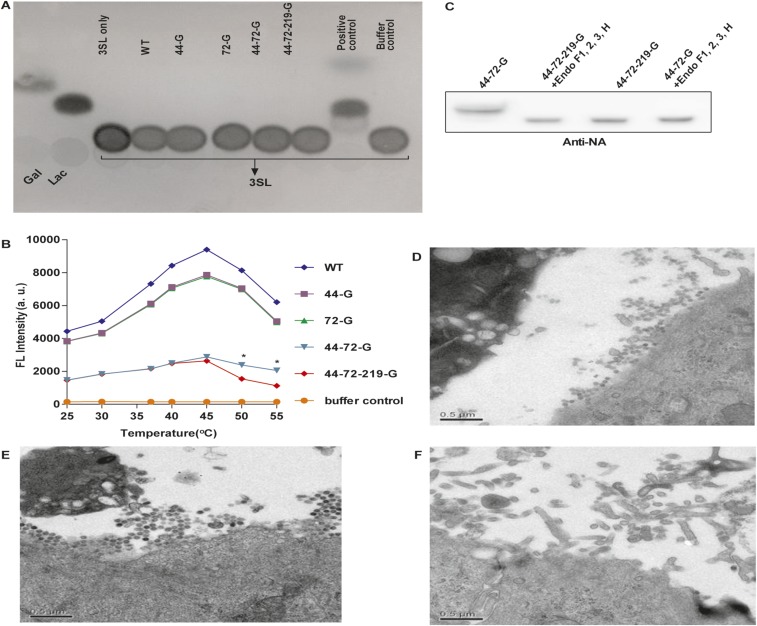
The glycans at glycosites 44 and 72 were also found to be important for the NA activity. The viruses without glycosites 44 and 72 (44-72-G and 44-72-219-G) showed significantly lower NA activity than the WT based on an assay using 2-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (4-MUNANA) as substrate (Fig. 2E). In addition, after the WT virus was treated with the endoglycosidase mixture, the molecular weight and NA activity were lower than that of the untreated virus (Fig. S3 E and F).
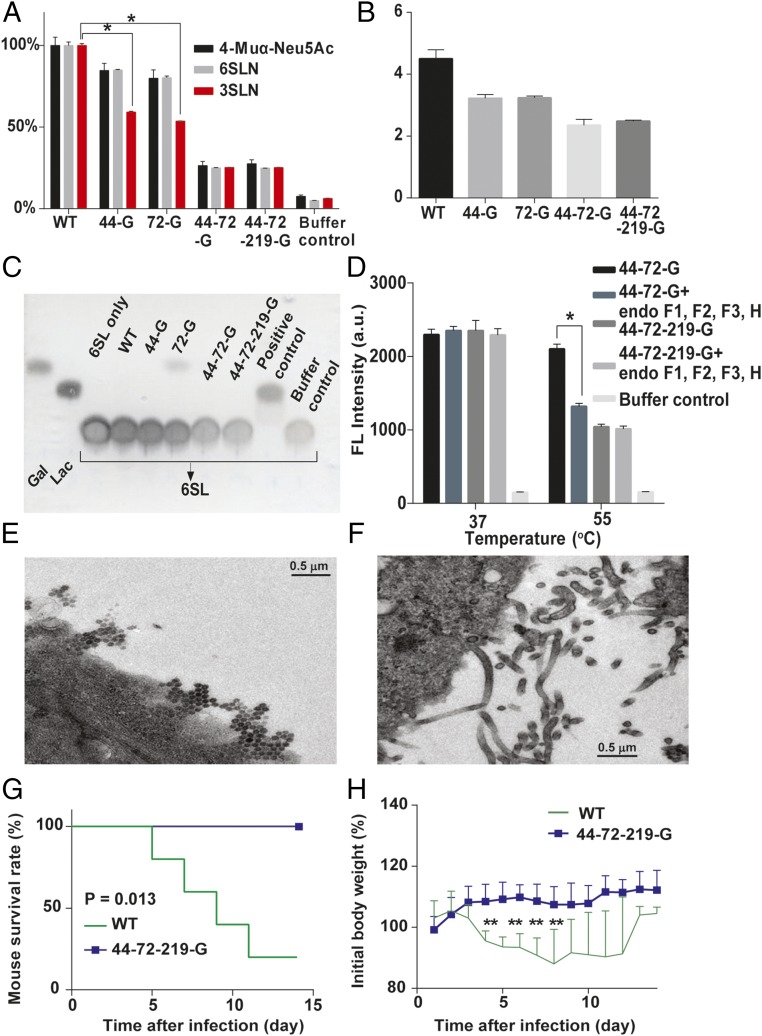
The glycans on NA affected the enzyme activity, affinity, and specificity, as the Vmax and the affinity value (Km) for different substrates were altered. When 2′(4-meyhylumbelliferyl)-α-d-N-acetylneuraminic acid (4-Muα-Neu5Ac), and 6′-sialyl-N-acetyllactosamine (6-SLN) were used as substrates, the NA without glycosites 44 and 72 had lower neurominidase activity but the Km values of all mutants were similar. Surprisingly, when 3-SLN was used as substrate, the activity of NA with glycosite 44 or 72 deleted decreased about 50%, but the Km value increased more than twofold (Fig. 3A and Table S1). Interestingly, the virus production rates of these variants in LMH cells were related to the NA activity on 3-SLN, which is an avian receptor (13) (Fig. 3B), suggesting that glycosylation on NA affected IAV replication in mammal and avian cells differently (Figs. 2A and 3B). In addition, whereas the NA without glycosite 72 (72-G) could interact with 6′-sialyllactose (6-SL) as substrate, none of the variants could interact with 3′-sialyllactose (3-SL; Fig. 3C and Fig. S4A). Interestingly, the activity of 44-72-219-G was similar to that of 44-72-G (with glycosite 219) from 25 to 40 °C, but lower at higher temperatures (45, 50, and 55 °C; Fig. S4B). After 44-72-G was treated with the endoglycosidase mixture, the activity of NA was lower than that without treatment at 55 °C, suggesting that the glycans on glycosite 219 (in the catalytic domain) affect the thermostability of NA (Fig. 3D and Fig. S4C).
Fig. 3.
Characterization of viruses with different glycosylation patterns on NA. (A) Measurements of NA activity using different glycoconjugates, 4-Muα-Neu5Ac, 6-SLN, and 3-SLN; the NA activity was relative to each WT substrate designated as 100%. (B) Comparison of virus production in chicken hepatocellular carcinoma cells (LMH cells). The virus titers were determined at 48 hpi. (C) TLC of variants of viruses that interact with 6-SL. (D) The 4-MUNANA assay was used to measure the NA activity of 44-72-G virus, deglycosylated 44-72-G virus, and 44-72-219-G virus at 37 and 55 °C. (E) Viral morphology as examined by transmission electron microscopy for WT virus. (F) 44-72-219-G virus. (G) After mice were infected with WT and glycosylation-defective NA virus (44-72-219-G), survival rate and (H) body weight were recorded for 14 d. (A, B, and D) Mean ± SEM for three independent experiments. (G and H) Mean ± SEM for five independent experiments. *P < 0.001. **P < 0.05.
Table S1.
Km and Vmax of NA proteins and viral variants
| Sample name | WT | 44-G | 72-G | 44-72-G | 44-72-219-G | Unit |
| Protein | ||||||
| For 4-Muα-Neu5Ac | ||||||
| Vmax | 5.025 | 4.95 | 4.45 | 1.15 | 1.2 | nM⋅s |
| Kcat | 514.3 | 468.5 | 426.8 | 100.3 | 105.8 | s |
| Km | 585.4 | 595.2 | 591 | 588.1 | 596.7 | µM |
| For 3SLN | ||||||
| Vmax | 7.04 | 4.09 | 3.89 | 1.46 | 1.48 | nM⋅s |
| Kcat | 688.2 | 389.8 | 364.4 | 136.4 | 139.1 | s |
| Km | 125.3 | 295.2 | 288.7 | 296.3 | 299.1 | µM |
| For 6SLN | ||||||
| Vmax | 1.07 | 0.92 | 0.85 | 0.24 | 0.21 | nM⋅s |
| Kcat | 104.2 | 90.6 | 83.6 | 19.8 | 20.7 | s |
| Km | 678.7 | 684 | 688 | 679 | 675 | µM |
| Virus | ||||||
| For 4-Muα-Neu5Ac | ||||||
| Specific activity | 9.7 | 7.78 | 7.34 | 2.3 | 2.42 | nmol⋅s⋅mg–1 |
| Km | 487 | 508 | 485 | 429 | 415 | µM |
| For 3SLN | ||||||
| Specific activity | 14.9 | 9 | 8.35 | 2.92 | 2.96 | nmol⋅s⋅mg–1 |
| Km | 115 | 274 | 268 | 285 | 281 | µM |
| For 6SLN | ||||||
| Specific activity | 2.25 | 1.88 | 1.78 | 0.48 | 0.42 | nmol⋅s⋅mg–1 |
| Km | 534 | 525 | 540 | 521 | 515 | µM |
Fig. S4.
Effect of NA glycosylation. (A) The TLC showed that all WSN NA mutated variants as shown could not cleave 3-SL. (B) Measurements of the NA activity of viruses as indicated at various temperatures by 4-MUNANA assay. Mean ± SEM for five independent experiments. *P < 0.001. (C) Western blot analysis of 44-72-G and 44-72-219-G viruses after treatment with the endoglycosidase mixture (F1, F2, F3, and H). The filter was probed with anti-NA antibody. Viral morphology as examined by transmission electron microscopy for (D) 44-G virus, (E) 72-G virus, and (F) 44-72-G virus.
Glycosites 44 and 72 also modulate the virus release and morphogenesis, because the cells infected with WT, 44-G, and 72-G viruses released many more spherical viral particles, but the cells infected with the 44-72-G or the 44-72-219-G virus produced mainly elongated and filamentous shaped particles, which were not observed in the WT virus-infected cells (Fig. 3 E and F and Fig. S4 D–F). In addition, mice infected with viruses without any glycans on NA (44-72-219-G virus) showed less-prominent changes in survival rate and body weight compared with the WT-infected mice, which had a 20% survival rate and considerable loss of body weight, suggesting that the glycosylation on NA affected virulence (Fig. 3 G and H).
Live Attenuated Vaccine Without the Stalk and Catalytic Domains of NA Showed Broad Protection With Strong CD8+ T-Cell Response.
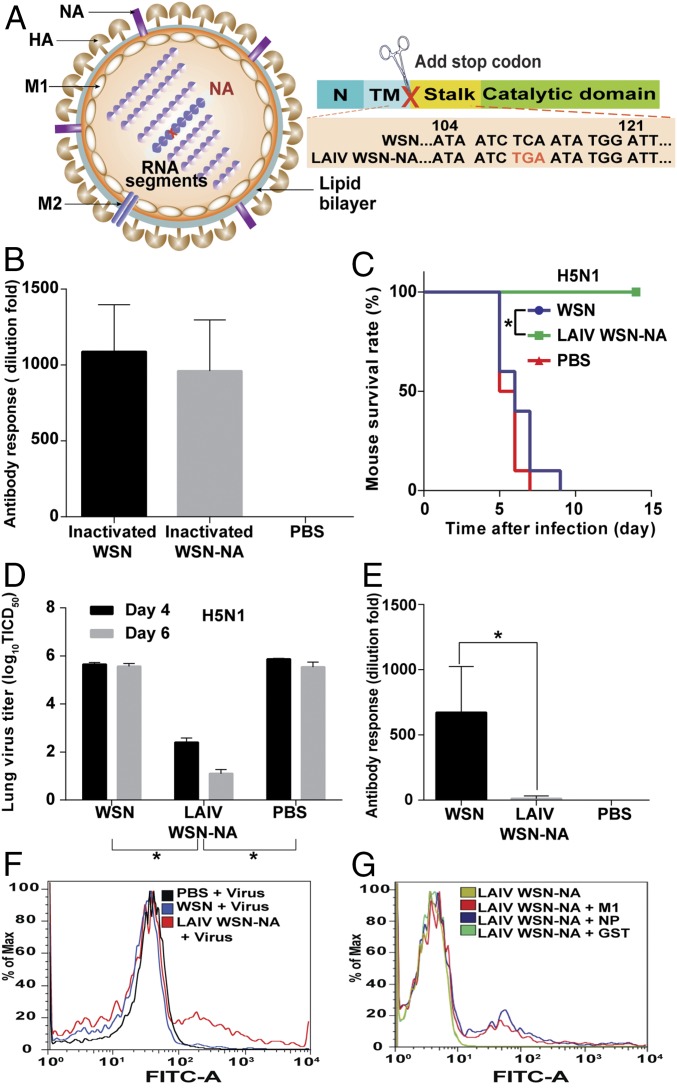
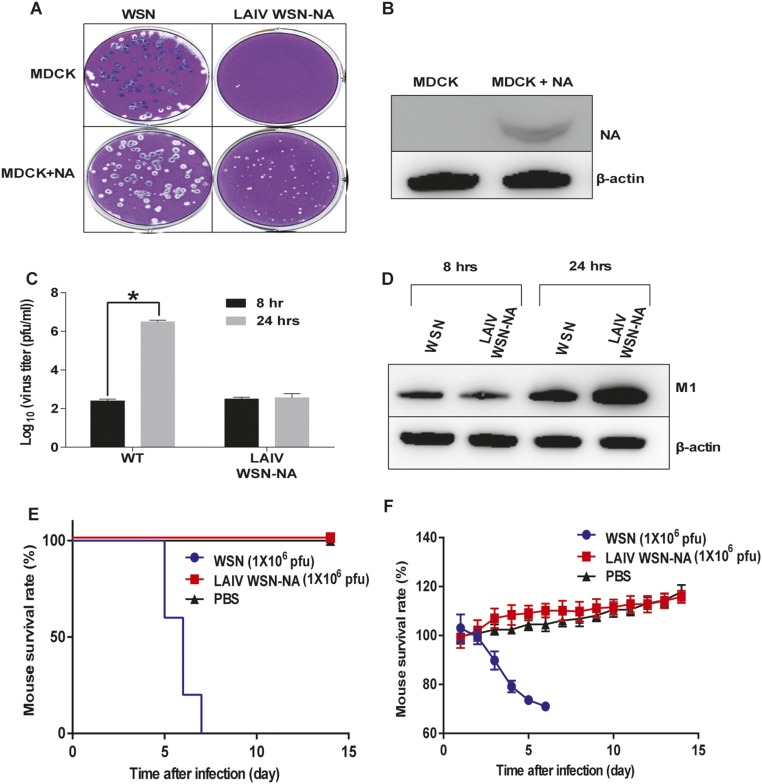
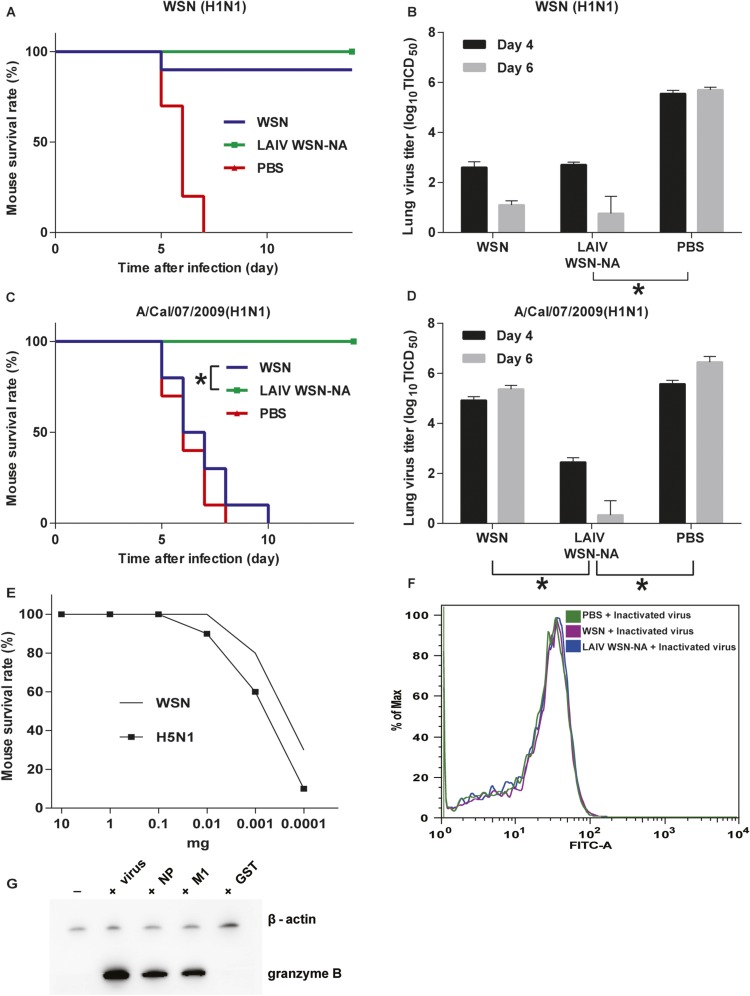
To study whether the NA activity would affect host immune response and its potential as live attenuated influenza vaccine (LAIV), we used the virus with nonglycosylated NA (44-72-219-G virus) as LAIV (LAIV WSN-44-72-219-G) because it had low NA activity and virulence. However, the nonglycosylated NA virus showed a similar immunogenicity as WT (Fig. S5). In addition, the NA activity had no effect on the efficiency of inactive vaccine, and the level of antibody produced was very low (14, 15). Therefore, in an attempt to enhance the immunogenicity of NA, we generated a virus without both the stalk and catalytic domains of NA (LAIV WSN-NA) by adding a stop codon to the RNA genome segment of NA (Fig. 4A). It was found that LAIV WSN-NA virus did not form the plaque in MDCK cells and could be rescued by expressing WT NA in the cell (Fig. S6 A and B). After 24 hours after postinfection (hpi), the A549 cells infected by LAIV WSN-NA virus showed fewer viruses released into the supernatant, and the intracellular viral protein M1 was accumulated, suggesting that LAIV WSN-NA virus had a defect in virus release (Fig. S6 C and D). The body weight of mice infected with 1 × 106 pfu of WSN viruses rapidly decreased, and these mice died at 6 d postinfection. However, the mice infected with 1 × 106 pfu of LAIV WSN-NA viruses via oral administration survived well and showed no body weight loss, indicating that LAIV WSN-NA virus is an effective LAIV with low pathogenicity, and attenuation of viruses in cells is essential for the efficacy of live vaccine (Fig. S6 E and F) (16). The immunogenicity of inactivated WSN-NA virus was similar to that of inactivated WSN virus in the immunogenicity test (Fig. 4B and Fig. S7). However, when LAIV WSN-NA virus was used as vaccine, it showed cross-strain and cross-subtype protection against WSN, A/Cal/07/2009, and H5N1 in the virus challenge study; the LAIV WSN-NA–treated mice survived well and cleared viruses from the lung, but did not induce any notable antibody response (Fig. 4 C–E and Fig. S8 A–D). In addition, the protective ability of LAIV WSN-NA also showed a dose-dependent response (Fig. S8E). After the virus infected the peripheral blood mononuclear cells (PBMC) from LAIV WSN-NA–treated mice, CD8+ T cells were specifically activated via IFN-γ and granzyme B expression, but the inactive virus did not show this activity, suggesting that LAIV WSN-NA can induce CD8+ T-cell activation upon virus infection (Fig. 4E and Fig. S8 F and G). Furthermore, the highly conserved viral epitopes NP and M1 could also stimulate CD8+ T-cell activation (17) (Fig. 4G and Fig. S8G). These results suggest that NA plays a key role in regulating the host immune response via CD8+ T-cell activation and LAIV WSN-NA is an effective vaccine.
Fig. S5.
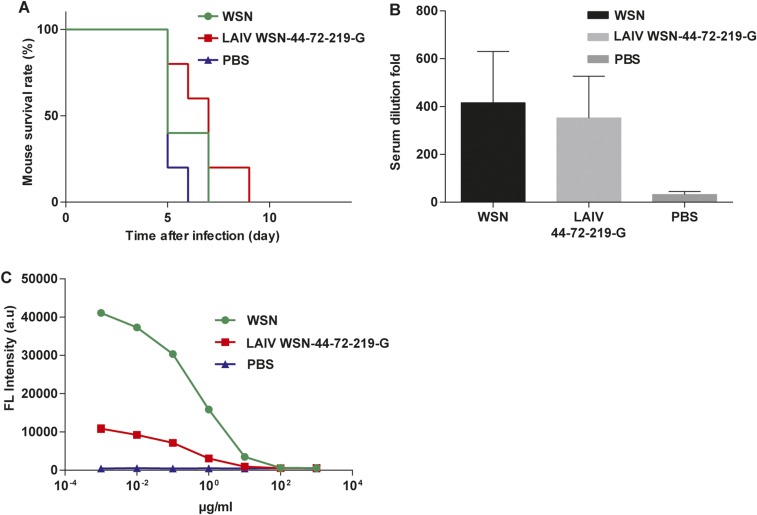
Immunogenicity of LAIV WSN-44-72-210-G. (A) Analysis of the survival rate of WSN, LAIV WSN-44-72-219-G, or PBS treatment mice after challenge with a lethal dose of H5N1. Five independent experiments shown. (B) The sera of the treated mice were analyzed using hemagglutination inhibition assay. (C) The titer of NA antibody was measured by neuraminidase inhibition assay. The IC50 of sera from WSN-treated mice was 4.3 μg/mL, and LAIV WSN-44-72-219-G was 3.2 μg/mL. (B and C) Mean ± SEM of five independent experiments.
Fig. 4.
Immunogenicity of LIAV WSN-NA. (A) Schematic overview of the LAIV WSN-NA design. The stalk and catalytic domains of NA were deleted by adding a stop code to the stalk domain of the RNA genome segment of NA, so the cells infected by the WSN without the stalk and catalytic domain cannot translate the stalk and catalytic domains of NA. Numbers refer to the nucleotide numbers from the 5′ end of the cRNA. Red TGA was the stop codon. N, N-terminal cytoplasmic domain; TM, transmembrane domain. (B) The sera of mice immunized with inactivated viruses as indicated were analyzed by using hemagglutination inhibition assay. (C) Analysis of the survival rate of WSN, LAIV WSN-NA, or PBS treatment mice after challenge with a lethal dose of H5N1. (D) H5N1 virus replication kinetics in the lungs of the treated mice as indicated on 4 and 6 postinfection days. (E) The sera of the treated mice were analyzed using hemagglutination inhibition assay to measure the antibody level. (F) After PBMC from WSN-, LAIV WSN-NA-, and PBS-treated mice were incubated with WSN virus (+virus), the analysis of INF-γ expression in CD8+ T cells was measured by flow cytometry. (F) Representative flow cytometry histograms of INF-γ expression in CD8+ T cells from LAIV WSN-NA–treated mice stimulated by M1 and NP epitopes. (B, D, and E) Mean ± SEM of 10 independent experiments. (C) Ten independent experiments are shown. (F and G) Data are representative of three similar experiments. *P < 0.001.
Fig. S6.
Characterization of LAIV WSN-NA virus. (A) The plaque formation of WSN and LAIV WSN-NA virus in WT MDCK and MDCK with NA expression (MDCK + NA). (B) Western blot analysis of NA expression in MDCK + NA cells. The filter was probed with anti-NA and anti–β-actin antibody. After A549 infection with virus at an MOI of 3, (C) culture fluids were collected at 8 and 24 h postinfection to determine the viral titers, and (D) total cell lysates were collected to analyze the intracellular viral protein M1 by Western blot. The filter was probed with anti-M1 and anti–β-actin antibody. After mice were infected with 1 × 106 pfu of WSN or LAIV WSN-NA viruses, survival rate (E) and body weight (F) were recorded for 14 days. (C) Mean ± SEM for three independent experiments. (E) Five independent experiments are shown. (F) Mean ± SEM for five independent experiments. *P < 0.001.
Fig. S7.
Comparison of host immune response to inactivated WSN and WSN-NA viruses. Mice were immunized with inactivated viruses as indicated and mouse survival rate was then measured after challenge with a lethal dose of (A) WSN and (B) H5N1 viruses. (C) The titer of NA antibody was measured by neuraminidase inhibition assay. IC50 of sera from WSN virus-treated mice was 5.2 μg/mL, and WSN-NA virus was 4.7 μg/mL. (A and B) Ten independent experiments are shown. (C) Mean ± SEM for 10 independent experiments.
Fig. S8.
Cross-strain protection from LAIV WSN-NA treatment. Mice were treated with WSN, LAIV WSN-NA, and PBS and mouse survival rate was then measured after challenge with a lethal dose of (A) WSN and (C) A/cal/07/2009 (H1N1). (B) WSN and (D) A/cal/07/2009 virus replication kinetics in the lung of the treated mice as indicated in 4 and 6 postinfection days. (E) The relationship between the dose of LAIV WSN-NA treatment and mouse survival rate after H5N1 and WSN challenge. (F) After PBMC from live WSN-, LAIV WSN-NA-, and PBS-treated mice were incubated with inactivated WSN virus, the analysis of INF-γ expression in CD8+ T cells were measured by flow cytometry. (G) Western blot analysis of granzyme B expression in CD8+ cells. CD8+ cells from LAIV WSN-NA treatment mice were incubated with live WSN virus (+virus), NP, and M1 epitopes (+NP and +M1). The filter was probed with anti-granzyme B and β-actin (for loading control) antibodies. (A, C, and E) Ten independent experiments are shown. (B and D) Mean ± SEM of three independent experiments. (F) Data are representative of three similar experiments. *P < 0.001.
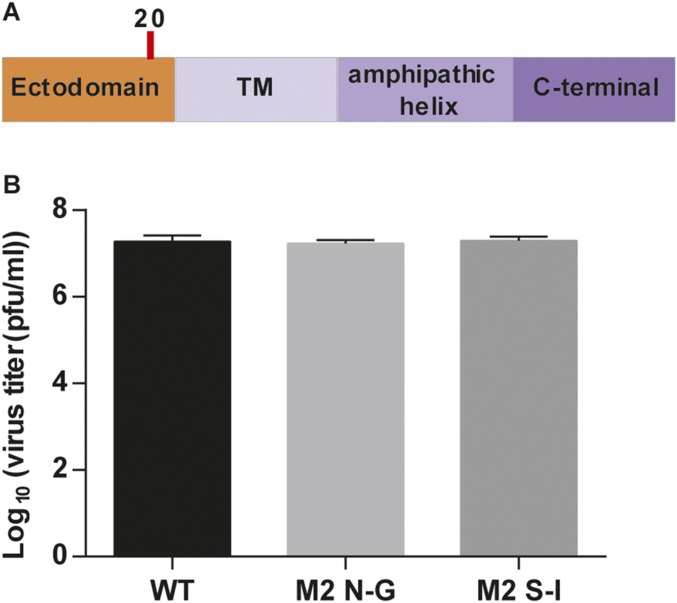
Finally, we found that there was one glycosite on M2, but it did not affect virus replication (Fig. S9).
Fig. S9.
Glycosylation on M2 did not affect flu replication. (A) Schematic overviews of M2 domain structure; the glycosites were highlighted with red line. The glycosite sequence on M2 was NDS. To study whether glycosylation on M2 affected flu replication, we changed N to G or S to I by reverse genetics. (B) Comparison of virus replication rates in MDCK cells. MDCK cells were infected with variants of flu at an MOI of 0.01. Culture fluids were collected at 24 h postinfection, and the virus titer was determined by plaque assay with MDCK cells. This result showed that these variants of flu had the same virus replication rate, suggesting that glycosylation did not affect the flu replication. Mean ± SEM of three independent experiments is shown.
Discussion
In the seasonal H1N1 strains, glycosite 142 is believed to play a significant role in evading the human immune response (8), and human H3N2 IAV also gains the glycosite in this region (glycosite 144 in H3 numbering) during evolution through positive selection (18). Surprisingly, we found that after IAV acquired glycosite 142, the efficiency of virus infectivity was promoted by the regulation of the HA–SA interaction, and the host immune response was altered. Therefore, we believe that glycosite 142 is an important factor that should be considered in the development of vaccines against human IAV.
Several studies have suggested that diversification in the stalk domain of NA is associated with the virulence and transmission of IAV, from ducks to land-based poultry, and its spread among humans via evolutionary processes, including sequence deletion and glycosite modification. However, the structural and functional roles of the stalk domain of NA were unknown thus far, and there was no report about the glycosylation of these canonical glycosites (19–23). In this study, we prove that the glycosites in the stalk domain of NA are glycosylated to regulate the activity, affinity, and specificity of NA to modulate IAV replication, suggesting that the glycans in the stalk domain of NA play an important role in the virulence and transmission of IAV. However, the relationship between NA activity and the diversity of the stalk domain, and whether the virus release is regulated by the glycosylation in the stalk domain is still unknown (24, 25). Our study here revealed that NA glycosylation is important for IAV virulence and transmission.
Recent development of universal influenza vaccines is focused on the use of conserved peptides or proteins as antigens with different adjuvants and administration methods to induce immune response (26–29). Expression of the conserved regions of IAV proteins by using viral vector systems can induce CD8+ T cells against lethal IAV challenge in animals, but it still only recognizes one target (30). Another type of vaccine based on the mixture of MVA-NP and M1 was also reported (31). In this study, the IAV without the stalk and catalytic domains of NA was found to significantly induce IAV-specific CD8+ T cells that recognize various strains and different subtypes of IAV in the absence of neutralizing antibodies. Although it is known that human IAV-specific CD8+ T cells provide cross-protection against influenza (32), it is important to further understand how the stalk and catalytic domains of NA affect host immune response. We believe that our findings provide an understanding of glycosylation on influenza cell surface and an innovative direction for the design of a universal influenza vaccine (33, 34).
Materials and Methods
Generation of Recombinant Viruses.
Eight fragments of A/WSN/33 viral genome were amplified by RT-PCR. We used the putative sequon N-X-S/T to create (from different amino acid to N) or delete (from N to A) the glycosites in the HA genome, and added a stop codon in the NA genome to remove the stalk and catalytic domain of NA by using site-directed mutagenesis. The viral cDNAs were inserted into pcDNA3.1 containing the pol I and CMV promoter similar to the generation of pHW2000. Recombinant viruses were generated by the eight-plasmid cotransfection method into MDCK/293T cells according to the published protocol (35). Supernatants were collected, titrated, and frozen at −80 °C until use. For the preparation of virus without NA stalk and catalytic domains (LAIV WSN-NA), we generated NA expression-stable MDCK/293T cell lines to rescue, maintain, and analyze the virus (16). Briefly, full-length NA (from WSN strain) was cloned to a cDNA expression lentivector (pLAS2w.Ppuro) to generate NA-expression lentivirus using the protocol provided by the National RNAi Core Facility, Academia Sinica, Taiwan (rnai.genmed.sinica.edu.tw). HEK293T and MDCK cells were infected with NA-expression lentivirus with triple multiplicity of infection in the presence of Polybrene (Sigma) at a final concentration of 8 μg/mL. Cells were incubated with virus for 24 h before replacing the medium with selective medium containing puromycin (3 μg/mL; Invitrogen). After a 3-d incubation, total cell lysate was collected to check the expression efficiency of NA by Western blot analysis.
Mice Treated with LAIV WSN-44-72-219-G and LAIV WSN-NA.
LAIV WSN-NA virus was cultured in MDCK cells with NA expression. A total of 25 µL of LAIV WSN-44-72-219-G or LAIV WSN-NA (nonlethal dose for WT WSN) were introduced into each nostril on days 0 and 21 while the mouse was conscious and the virus did not reach the lower respiratory tract. Then the immunogenicity test procedure was performed as described in SI Materials and Methods (14).
All animal experiments were evaluated and approved by the Institutional Animal Care and Use Committee of Academia Sinica. More details are provided in SI Materials and Methods.
SI Materials and Methods
Cell Lines and Virus.
MDCK and human embryonic kidney cells (HEK293T) were maintained in DMEM (Invitrogen). A549 human adenocarcinoma alveolar basal epithelial cells were kept in F-12K medium (Invitrogen), and LMH chicken hepatocellular carcinoma cells were cultured in Waymouth’s MB 752/1 medium (Invitrogen). All media were supplemented with 10% (vol/vol) heat-inactivated FBS (Thermo Scientific) and antibiotics (100 U/mL penicillin G and 100 gm/mL streptomycin). The influenza A virus A/WSN/33 strain was used in the studies.
Antibodies.
Mouse monoclonal anti-HA and anti-NA antibodies were obtained from Sino Biological. Mouse monoclonal anti-actin antibody was purchased from Millipore. Goat polyclonal anti-M1 and mouse monoclonal anti-6xHis antibodies were purchased from Santa Cruz Biotechnology. Rat monoclonal anti–INF-γ antibody was obtained from Abcam. Rabbit polyclonal anti-granzyme B antibody was purchased from Aviva Systems Biology. All commercial antibodies were validated for specificity by companies and us via Western blot.
Virus Replication Rate.
Monolayer cultures of MDCK, A549, and LMH cells in 12-well dishes were washed twice with 1× PBS. The cells were infected with variants of modified influenza virus at an MOI of 0.01, 0.1, and 0.1, respectively, in serum-free medium containing 0.1 (0.01 for LMH) g/mL l-(tosylamido-2-phenylethyl) chloromethyl ketone (TPCK)-trypsin (Pierce) and incubated at 37 °C for 1 h; cells were washed twice with 1× PBS and then incubated with the complete medium. At the time points shown in Fig. 1B, the supernatants were collected to determine the virus titer by performing plaque assay in MDCK cells.
Plaque Assay.
Monolayers of MDCK cells in six-well dishes were washed twice with 1× PBS. The cells were then inoculated with serial 10-fold dilutions of the virus in serum-free medium containing 0.5 μg/mL TPCK-trypsin and incubated at 37 °C for 1 h. Afterward, the cells were washed and overlaid with MEM containing 0.5% agarose (Lonza) and 0.5 gm/mL TPCK-trypsin. After 3 d, the cells were fixed with 10% (vol/vol) formaldehyde and stained with 0.1% crystal violet solution.
Protein Expression and Purification.
The plasmid that encodes the secreted HA and NA was transfected into the HEK293T cell lines using polyethyleneimine and cultured in Freestyle 293 expression medium (Invitrogen) supplemented with 0.5% bovine calf serum. The supernatant was collected 72 h after transfection and cleared by centrifugation. HA and NA proteins were purified with nickel-chelation chromatography as previously described (9, 37). The purified proteins were concentrated by a Millipore Amicon Ultra Filter and loaded onto a Superdex-200 gel filtration column (GE) preequilibrated in PBS buffer, and different fractions were collected.
Glycan Array.
Glycan microarrays were prepared by printing (AD3200; BioDot) the glycans with a pentylamine tail prepared in the laboratories to the NHS-activated glass slide (Nexterion H) by robotic pin (SMP2B; TeleChem International) at 25 °C with 60% humidity. Nexterion H slides were spotted with solutions of glycan 1–29 and 30–39 at 100 µM from bottom to top with three replicates horizontally in each grid and dried under vacuum (6). The resulting images were analyzed with GenePix Pro 6.0 (Molecular Devices) to locate and quantify the fluorescence intensity of all of the spots on the grid.
Virus-Binding Assay.
The same amount of viruses was inactivated in a buffer containing the neuraminidase inhibitor Oseltamivir carboxylate (10 µM). Suspensions of the inactivated viruses with Oseltamivir carboxylate were overlaid onto the arrays and incubated at room temperature for 30 min. Slides were subsequently washed by successive rinses in PBS-0.05% Tween, PBS, and deionized water three times. Bound viruses were detected using the anti-H1 antibody. The slides were gently rocked at room temperature for 60 min. After repeating the washing steps, binding was detected by overlay with labeled secondary antibodies.
Infectivity Assay.
We used the PEG virus precipitation kit (BioVision) to concentrate viruses following the manufacturer’s protocol, and determined the amount of virus using Western blot. The same amounts of variant viruses were used to infect A549 cells in F-12K serum-free medium containing 0.5 μg/mL TPCK-trypsin and incubated at 37 °C for 30 min. Then, the cells were washed twice and overlaid with F-12K medium containing 10% (vol/vol) heat-inactivated FBS (Thermo Scientific) and antibiotics (100 U/mL penicillin G and 100 gm/mL streptomycin). After 10 hpi, the total cell lysate was collected and analyzed. MDCK cells were infected with variant viruses infected in serum-free medium containing 0.5 gm/mL TPCK-trypsin and incubated at 37 °C for 30 min. Then, the plaque assay procedure was followed.
Hemagglutination Assay.
The same amounts of IAV and HA proteins were serially diluted twofold in a total volume of 100 µL. Next, 25 μL of a 2% (vol/vol) turkey erythrocyte solution were added. The virus and erythrocytes were gently mixed and the hemagglutination was read after incubation for 60 min at room temperature.
Hemagglutination Inhibition Assay.
After serum samples were serially diluted twofold in a 96-well plate, four hemagglutination units of WT WSN were added to each well for 1 h at room temperature. After incubation, 25 µL of a 2% (vol/vol) turkey erythrocyte solution were added to give a total volume of 125 µL and incubated for 1 h at room temperature. The HAI titer of the individual serum sample was determined to be the inverse of the last dilution where cells were not agglutinated.
Cell-Binding Assay.
Turkey erythrocytes were pretreated with different amounts (0–60 µg/mL) of Vibrio cholerae neuraminidase [receptor destroying enzyme (RDE)] (Sigma) for 60 min at 37 °C. Then, the erythrocytes were washed once with PBS and made into 2% (vol/vol) erythrocyte solutions using PBS. Twenty-five microliters of each 2% solution were added to the same amount of IAV and HA proteins to have a total volume of 125 µL. The virus and RDE-treated erythrocytes were incubated for 1 h at room temperature, and agglutination was then measured. Data were expressed as the maximal concentration of RDE that still gave full agglutination (38).
Deglycosylation.
An aliquot of viruses or proteins was deglycosylated in a buffer solution purchased from Sigma-Aldrich. After ultracentrifugation, 200 μL infection medium containing virus (1 × 107 pfu) or purified proteins were mixed with protease inhibitor (Roche), 1 μg endoglycosidase F1, 1 μg endoglycosidase F2, 1 μg endoglycosidase F3, and 1 μg endoglycosidase H or 1 μg PNGase F at 37 °C for 24 h in the dark. The endoglycosidase mixture (endo F1, F2, F3, and H) or PNGase F can trim all of the glycan structure down to a single GlcNAc residue to produce monoglycosylated samples or nonglycosylated samples, individually (9). After deglycosylation, samples were checked by Western blot.
IAV Preparation for Immunogenicity Test.
WT, 285-497-556-G, and 142-285-497-556-G viruses were cultured in MDCK cells, and LAIV WSN-NA virus was cultured in MDCK cells with NA expression. These viruses were inactivated by using 0.1% BPL (Acros Organics) at room temperature for 24 h followed by dialysis for 24 h against HNE buffer (5 mM Hepes, 150 mM NaCl, 0.1 mM EDTA, pH 7.4) (39) and tested by performing serial passages on MDCK cells. Female C57BL/6 mice, aged 6–8 wk, were immunized intramuscularly with an equal amount of 6 µg HA per inactivated virus dose, and immunized twice s.c. on days 0 and 21. One week after the booster immunization, mice were divided into two groups, and every group contained 10 mice. One group was anesthetized and inoculated intranasally with 10 × LD50 of WSN/33 or H5N1 virus, and the serum sample of the other group was collected for analysis of the antiserum production. The mice were monitored daily for survival in a period of 14 d after challenge.
Transmission Electron Microscopy.
MDCK cells were grown on ACLAR embedding film with 7.8-mil thickness (EMS) for 1 d followed by infection with influenza virus at an MOI of 5. At 18 hpi, virus-infected cells were rinsed with 0.1 M cacodylate buffer and fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer at 4 °C for 30 min. Then cells were postfixed with 1% osmium tetroxide in 0.1 M cacodylate for 30 min, stained with 1% uranyl acetate and lead citrate for 1 h, dehydrated by using ethanol, and embedded with resin. After baking, the sample was cut into 80-nm-thin sections using ultramicrotome. Finally, samples were examined with Tecnai G2 Spirit TWIN (FEI).
Virulence Assay.
The virulence of recombinant viruses was measured using groups of five female 4- to 6-wk-old BALB/c mice that were intranasally inoculated with 50 μL of virus (5 × 105 PFU for WSN and LAIV 44-72-219-G virus in Fig. 1G, and 1 × 106 PFU for WSN and LAIV WSN-NA in Fig. S9E). Survival and body weight changes were recorded daily for 14 d after infection.
NA Activity.
The enzymatic activity of NA was measured by standardizing the virus samples with pfu and the protein samples as described (36). Aliquots of virus were prepared at a titer of 1 × 106 pfu and then incubated at 37 °C with 4-MUNANA (Sigma). After 30 min, the reaction was stopped with 0.14 M NaOH and 83% (vol/vol) ethanol, and the NA activity was measured with excitation at 360 nm and emission at 450 nm. The final concentration of the substrate ranged from 0 to 1,500 mM. Fluorescence was monitored every 5 min for 60 min (12 measures, 4-MUNANA assay). Km and Vmax of NA were calculated with Prism software (GraphPad) by fitting the data to the Michaelis–Menten equation using nonlinear regression. For measuring the kinetics of enzymatic cleavage of different glycoconjugates (4-Muα-Neu5Ac, 3-SLN, 6-SLN, 3-SL, and 6-SL; Sigma), the N-acetylneuraminic acid released from the glycoconjugates was determined by reaction with N-acetylmannosamine (ManNAc) dehydrogenase and sialic acid aldolase. ManNAc, the aldolase-cleaved product of N-acetylneuraminic acid, was interacted with ManNAc dehydrogenase and NAD+ to form the NADH by-product, and the fluorescence intensity of NADH at 340/450 nm was measured. In the reactions, 2 µg sialic acid aldolase (Pasteurella multocida, recombinant), 3 µg ManNAc dehydrogenase (Flavobacterium sp. 141-8, recombinant), 50 mM Mes buffer, pH 6.5, 0.2 mM NAD+, and an appropriate amount of NA were mixed, and the final concentration of the glycoconjugate was adjusted to a range from 0 to 1,500 mM. Fluorescence was monitored every 5 min for 30 min (six measures).
Neuraminidase Inhibition Assay.
Neuraminidase inhibition assay was used to analyze the production of NA antibody. Here, we use NA-Fluor Influenza Neuraminidase Assay Kit (Life Technologies) to perform this experiment. Briefly, the virus was incubated with the reaction buffer from the kit, then mixed and heat inactivated, followed by serial dilutions to different concentrations as indicated in Figs. S5C and S7C, and the fluorescence was monitored. Neuraminidase inhibition titers were calculated as the reciprocal of the highest dilution with at least 50% inhibition (15).
IAV-Specific CD8+ T-Cell Analysis.
The PBMC obtained from PBS-, WSN-, or LAIV WSN-NA–treated mice was incubated with live or inactivated WSN (UV treatment) at an MOI of 3 in RPMI medium 1640 containing 0.5 μg/mL TPCK-trypsin and incubated at 37 °C for 1 h. Then, the cells were washed twice and overlaid with RPMI medium 1640 containing 10% (vol/vol) heat-inactivated FBS and antibiotics. After incubation for 24 h, CD8+ T cells were isolated by Dynabeads Untouched Mouse CD8 Cells kit (Invitrogen) following the procedure from the company, and analyzed by flow cytometry and Western blot. For viral protein M1 and NP stimulation, the GM-CSF–cultured bone marrow-derived dendritic cells were incubated with or without 100 μM viral M1 and NP epitopes in complete RPMI medium 1640 for 24 h at 37 °C, then mixed with PBMCs from immunized mice at the ratio 1:1 (40). After incubation for 48 h, CD8+ T cells were isolated and analyzed by flow cytometry and Western blot. The M1 epitopes were GILGFVFTL, RLEDVFAGK, and ASCMGLIY, and NP epitopes were CTELKLSDY, SRYWAIRTR, and LELRSRYWA (17) (Mission Biotech). Peptides were dissolved in dimethyl sulfoxide at 5.0 mg/mL, diluted in RPMI 1640–100 μM, and stored at 20 °C.
Flow Cytometry.
Cells were harvested and suspended in FACS buffer [2% (vol/vol) FBS in PBS] at a density of 106/mL. The antibody used in this study was anti–INF-γ antibody. Cellular fluorescence intensity was analyzed by FACSCanto (BD Biosciences) and FCS Express 3.0 software.
Acknowledgments
We thank Dr. Jia-Tsrong Jan’s laboratory for mouse vaccination and challenge experiments, and Dr. Michael M. C. Lai for providing plasmids for the reverse genetics study. The pLAS2w.Ppuro vector was obtained from the National RNAi Core Facility at Academia Sinica. The Transmission Electron Microscope was from the imaging core in the Institute of Molecular Biology, Academia Sinica. Financial support was provided by Academia Sinica.
Footnotes
Conflict of interest statement: A provisional patent application on this work has been filed.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617174114/-/DCSupplemental.
References
- 1.Medina RA, García-Sastre A. Influenza A viruses: New research developments. Nat Rev Microbiol. 2011;9(8):590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Air GM. Influenza virus-glycan interactions. Curr Opin Virol. 2014;7:128–133. doi: 10.1016/j.coviro.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle TM, et al. The universal epitope of influenza A viral neuraminidase fundamentally contributes to enzyme activity and viral replication. J Biol Chem. 2013;288(25):18283–18289. doi: 10.1074/jbc.M113.468884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu CY, Jeng KS, Lai MM. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J Virol. 2011;85(13):6618–6628. doi: 10.1128/JVI.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao HY, et al. Differential receptor binding affinities of influenza hemagglutinins on glycan arrays. J Am Chem Soc. 2010;132(42):14849–14856. doi: 10.1021/ja104657b. [DOI] [PubMed] [Google Scholar]
- 7.Wagner R, Wolff T, Herwig A, Pleschka S, Klenk HD. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: A study by reverse genetics. J Virol. 2000;74(14):6316–6323. doi: 10.1128/jvi.74.14.6316-6323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei CJ, et al. Cross-neutralization of 1918 and 2009 influenza viruses: Role of glycans in viral evolution and vaccine design. Sci Transl Med. 2010;2(24):24ra21. doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang CC, et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc Natl Acad Sci USA. 2009;106(43):18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JR, et al. Vaccination of monoglycosylated hemagglutinin induces cross-strain protection against influenza virus infections. Proc Natl Acad Sci USA. 2014;111(7):2476–2481. doi: 10.1073/pnas.1323954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P, Wang Z, Zhang L, Li D, Lin J. The mechanism by which 146-N-glycan affects the active site of neuraminidase. PLoS One. 2015;10(8):e0135487. doi: 10.1371/journal.pone.0135487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310(5745):77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. Structure and receptor binding preferences of recombinant hemagglutinins from avian and human H6 and H10 influenza A virus subtypes. J Virol. 2015;89(8):4612–4623. doi: 10.1128/JVI.03456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MA, Ganesan AP, Luckashenak N, Mendonca M, Eisenlohr LC. Endogenous antigen processing drives the primary CD4+ T cell response to influenza. Nat Med. 2015;21(10):1216–1222. doi: 10.1038/nm.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monto AS, et al. Antibody to influenza virus neuraminidase: An independent correlate of protection. J Infect Dis. 2015;212(8):1191–1199. doi: 10.1093/infdis/jiv195. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe S, Watanabe T, Kawaoka Y. Influenza A virus lacking M2 protein as a live attenuated vaccine. J Virol. 2009;83(11):5947–5950. doi: 10.1128/JVI.00450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boon AC, et al. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J Virol. 2002;76(2):582–590. doi: 10.1128/JVI.76.2.582-590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki Y. Positive selection for gains of N-linked glycosylation sites in hemagglutinin during evolution of H3N2 human influenza A virus. Genes Genet Syst. 2011;86(5):287–294. doi: 10.1266/ggs.86.287. [DOI] [PubMed] [Google Scholar]
- 19.Munier S, et al. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J Virol. 2010;84(2):940–952. doi: 10.1128/JVI.01581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Air GM. Influenza neuraminidase. Influenza Other Respi Viruses. 2012;6(4):245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, Zhong Y, Qin Y, Sun S, Li Z. The evolutionary pattern of glycosylation sites in influenza virus (H5N1) hemagglutinin and neuraminidase. PLoS One. 2012;7(11):e49224. doi: 10.1371/journal.pone.0049224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutvisuttinunt W, et al. Viral subpopulation diversity in influenza virus isolates compared to clinical specimens. J Clin Virol. 2015;68:16–23. doi: 10.1016/j.jcv.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka Y, et al. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J Virol. 2009;83(9):4704–4708. doi: 10.1128/JVI.01987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yondola MA, et al. Budding capability of the influenza virus neuraminidase can be modulated by tetherin. J Virol. 2011;85(6):2480–2491. doi: 10.1128/JVI.02188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leyva-Grado VH, et al. Modulation of an ectodomain motif in the influenza A virus neuraminidase alters tetherin sensitivity and results in virus attenuation in vivo. J Mol Biol. 2014;426(6):1308–1321. doi: 10.1016/j.jmb.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YN, Lee YT, Kim MC, Gewirtz AT, Kang SM. A novel vaccination strategy mediating the induction of lung-resident memory CD8 T cells confers heterosubtypic immunity against future pandemic influenza virus. J Immunol. 2016;196(6):2637–2645. doi: 10.4049/jimmunol.1501637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nachbagauer R, et al. Age dependence and isotype specificity of influenza virus hemagglutinin stalk-reactive antibodies in humans. MBio. 2016;7(1):e01996–e15. doi: 10.1128/mBio.01996-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He B, et al. Adenovirus-based vaccines against avian-origin H5N1 influenza viruses. Microbes Infect. 2015;17(2):135–141. doi: 10.1016/j.micinf.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Sandt CE, et al. Differential recognition of influenza A viruses by M158-66 epitope-specific CD8+ T cells is determined by extraepitopic amino acid residues. J Virol. 2015;90(2):1009–1022. doi: 10.1128/JVI.02439-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He F, Leyrer S, Kwang J. Strategies towards universal pandemic influenza vaccines. Expert Rev Vaccines. 2016;15(2):215–225. doi: 10.1586/14760584.2016.1115352. [DOI] [PubMed] [Google Scholar]
- 31.Berthoud TK, et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin Infect Dis. 2011;52(1):1–7. doi: 10.1093/cid/ciq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sridhar S, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. 2013;19(10):1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 33.Pica N, Palese P. Toward a universal influenza virus vaccine: Prospects and challenges. Annu Rev Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 34.Impagliazzo A, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349(6254):1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000;97(11):6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benton DJ, Martin SR, Wharton SA, McCauley JW. Biophysical measurement of the balance of influenza a hemagglutinin and neuraminidase activities. J Biol Chem. 2015;290(10):6516–6521. doi: 10.1074/jbc.M114.622308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu ZL, Ethen C, Hickey GE, Jiang W. Active 1918 pandemic flu viral neuraminidase has distinct N-glycan profile and is resistant to trypsin digestion. Biochem Biophys Res Commun. 2009;379(3):749–753. doi: 10.1016/j.bbrc.2008.12.139. [DOI] [PubMed] [Google Scholar]
- 38.Hensley SE, et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326(5953):734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budimir N, et al. Induction of heterosubtypic cross-protection against influenza by a whole inactivated virus vaccine: The role of viral membrane fusion activity. PLoS One. 2012;7(1):e30898. doi: 10.1371/journal.pone.0030898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lutz MB, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223(1):77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]