Significance
Influenza virus is an important human pathogen. The circulating strains of influenza virus are constantly mutating and are acquiring resistance to all approved drugs. Therefore, development of influenza therapeutics against novel targets is urgently required. The hemagglutinin envelope glycoprotein (HA) is a promising target for small-molecule design. However, Arbidol is the only available antiviral drug that targets the HA. The absence of structural information on drug–HA complexes has hindered further therapeutic development efforts against this viral pathogen. Here, we report crystal structures of Arbidol in complex with influenza HAs. This structural information advances our understanding of how small molecules, such as Arbidol, can function as influenza fusion inhibitors and can be used for development of broad-spectrum, small-molecule therapeutics.
Keywords: influenza virus, hemagglutinin, X-ray crystallography, Arbidol, fusion inhibitor
Abstract
The broad-spectrum antiviral drug Arbidol shows efficacy against influenza viruses by targeting the hemagglutinin (HA) fusion machinery. However, the structural basis of the mechanism underlying fusion inhibition by Arbidol has remained obscure, thereby hindering its further development as a specific and optimized influenza therapeutic. We determined crystal structures of Arbidol in complex with influenza virus HA from pandemic 1968 H3N2 and recent 2013 H7N9 viruses. Arbidol binds in a hydrophobic cavity in the HA trimer stem at the interface between two protomers. This cavity is distal to the conserved epitope targeted by broadly neutralizing stem antibodies and is ∼16 Å from the fusion peptide. Arbidol primarily makes hydrophobic interactions with the binding site but also induces some conformational rearrangements to form a network of inter- and intraprotomer salt bridges. By functioning as molecular glue, Arbidol stabilizes the prefusion conformation of HA that inhibits the large conformational rearrangements associated with membrane fusion in the low pH of the endosome. This unique binding mode compared with the small-molecule inhibitors of other class I fusion proteins enhances our understanding of how small molecules can function as fusion inhibitors and guides the development of broad-spectrum therapeutics against influenza virus.
Influenza is a highly contagious viral infection of the respiratory tract that causes an enormous burden on the economy and public health worldwide. Influenza affects ∼5–20% of the US population with ∼3,000–49,000 deaths and ∼200,000 hospitalizations with influenza-related complications per annum (www.nfid.org/idinfo/influenza). In pandemic years, 1 million (1957–1958) to 50 million (1918–1919) deaths have occurred (1, 2). More recently, the emergence and global spread of H1N1 influenza from the 2009 pandemic and recent lethal cases of H5N1 and 2013 avian-origin H7N9 influenza demonstrate the limitations of currently available strategies to control influenza infection. Currently, the main flu interventions are the annual trivalent or quadrivalent vaccines (who.int/influenza/vaccines), but because of rapid antigenic drift and shift in influenza viruses, selection of appropriate vaccine strains is a formidable task (cdc.gov/flu/about/season/vaccine-selection.htm) (3–6). Furthermore, the small-molecule therapeutic space against influenza virus is currently limited to four licensed drugs: neuraminidase inhibitors oseltamivir (Tamiflu) and zanamivir (Relenza), which prevent release of nascent virions (7), and amantadine (Symmetrel) and rimantadine (Flumadine), which are M2 ion channel inhibitors (8). However, the emergence of drug-resistant influenza variants has led to a decline in the efficacy of these drugs (9–11). Therefore, new influenza therapeutics with novel mechanisms of action and against new targets are urgently required to combat the persistent threat of influenza viruses.
Therapeutic design strategies aimed at targeting the highly conserved functional regions of influenza proteins that are important for the early stages of viral infection may be highly effective and reduce the likelihood of generating escape mutants. Hemagglutinin (HA), the major glycoprotein on the surface of influenza virus, is involved in virus attachment to host cells and subsequent entry via fusion of the viral membrane with a host cell membrane. Structurally, HA is composed of head (HA1) and stem (HA2/HA1) domains (Fig. 1); the HA1 and HA2 chains are linked by a single disulfide bond (12). Fusion occurs after receptor binding on the host cell surface and subsequent virus entry by endocytosis into the endosome, where a drop in pH triggers an irreversible restructuring of the HA to expose the fusion peptide and initiate the fusion process. Interaction of the fusion peptide with the endosomal membrane initiates a cascade of events that enables the viral ribonucleoprotein particles to be released into the cytoplasm for viral replication (13). Thus, one therapeutic strategy is to target the highly conserved HA stem region involved in the fusion process and prevent the low pH-triggered conformational rearrangements that lead to fusion of the viral membrane with the endosome.
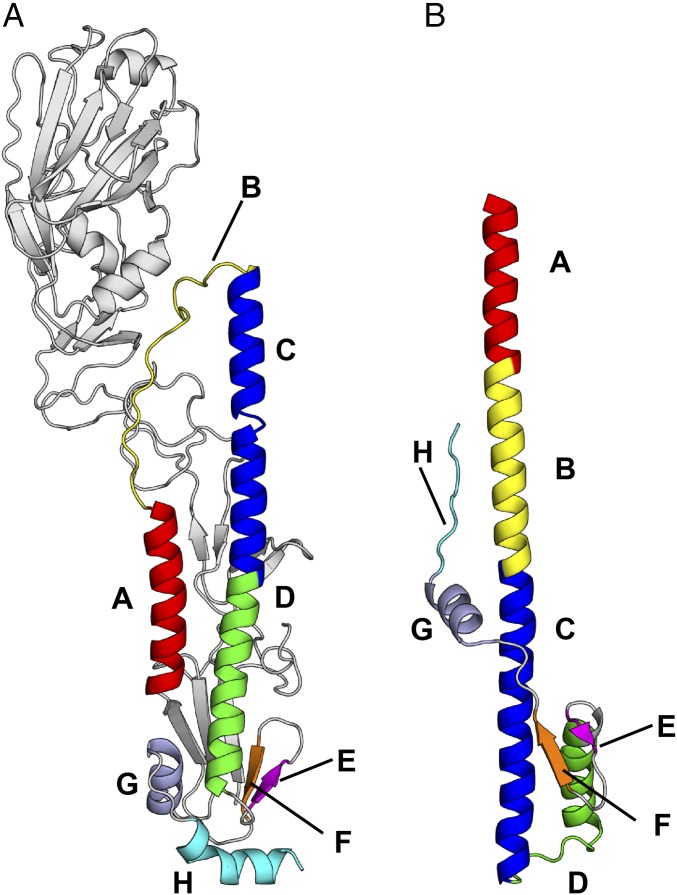
Fig. 1.
Prefusion and postfusion conformations of influenza HA. (A) Prefusion conformation of H7N9 A/Shanghai/2/2013 (H7/SH2; PDB 4LN6) HA at neutral pH. (B) Postfusion conformation of TBHA (PDB 1HTM) at fusion pH 5.0. The secondary structure elements that are involved in the large conformational rearrangements in going from the pre- to postfusion form of the HA mainly involve α-helices, but also loops and β-sheets. These elements are labeled from A to H in different colors.
Arbidol (umifenovir) is a broad-spectrum antiviral against a number of enveloped and nonenveloped viruses such as influenza, respiratory syncytial (RSV), adenovirus, Coxsackie B5, parainfluenza, Ebola (EBOV), and hepatitis B and C (14–20). Arbidol demonstrates broad activity against diverse strains and subtypes of influenza A and B viruses and is effective in mice infected with A/California/07/2009 (H1N1), A/Puerto Rico/8/1934 (H1N1), and A/Aichi/2/1969 (H3N2) influenza A viruses (14–18, 21). Arbidol is used primarily as an over-the-counter drug in Russia and China for prophylaxis and treatment of acute respiratory infections including influenza, but has not yet gained acceptance in other countries. Nevertheless, it is currently in phase IV clinical trials in the United States (https://clinicaltrials.gov/ct2/show/NCT01651663). One major drawback of Arbidol is that a large dose must be administered to achieve peak plasma concentration and therapeutic efficacy. To understand the molecule mechanism for its broad-spectrum inhibition of influenza viruses, various in vitro assays have been reported (14, 22–24), From the reported Kds with H1, H2, H3, H4, and H5 HA strains, Arbidol binds with higher affinity to group 2 HAs (Kds = 5.6–7.9 µM) than to group 1 HAs (Kds = 18.8–44.3 µM) (23). Mechanistically, Arbidol has been shown to increase influenza virus HA stability and prevent the low pH-induced HA transition to its fusogenic state, thereby blocking infection at one of the two key steps in cell entry, i.e., at viral fusion. However, its binding mode and molecular mechanism are largely unknown, thus hindering its further development as a specific and optimized influenza therapeutic.
Here, we describe crystal structures of Arbidol in complex with influenza virus HA from the human H3N2 1968 pandemic strain and from the 2013 avian-origin H7N9. Arbidol binds in a hydrophobic cavity at the interface of the HA protomers in the upper region of the stem. This interaction creates an extensive network of noncovalent interactions that stabilize the prefusion conformation of HA to prevent its conformational rearrangement and ultimately inhibit membrane fusion. Our study also provides insights into the molecular mechanism of fusion inhibition by Arbidol in class I viral fusion proteins. The structural insights obtained here will facilitate the development of the next generation of influenza therapeutics.
Results
Crystal Structures of Arbidol in Complex with HAs.
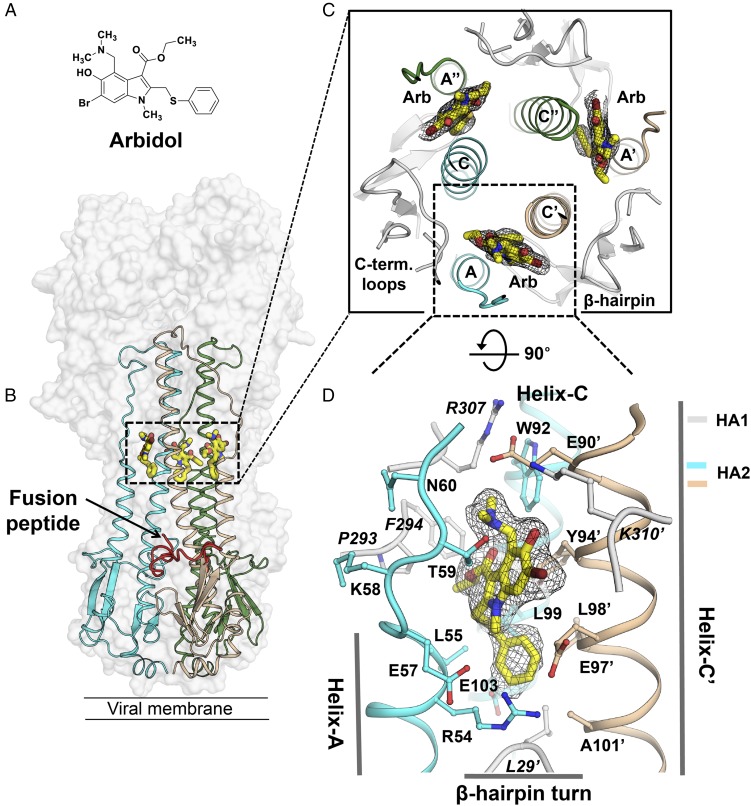
To understand the structural basis of fusion inhibition by Arbidol (Fig. 2A and Fig. S1), we determined the crystal structures of Arbidol in complex with HAs from pathogenic viruses H7N9 A/Shanghai/2/2013 (H7/SH2) and H3N2 A/Hong Kong/1/1968 (H3/HK68) at 2.37-Å and 2.54-Å resolution, respectively (Figs. 2–4, Table 1, and Fig. S2). The structures were obtained by soaking apo crystals of H7/SH2 and H3/HK68 HAs with ∼2.5 mM Arbidol. Earlier studies that used mass spectroscopy and in silico modeling predicted that Arbidol would bind in a cavity along the threefold symmetry axis of the HA trimer and interact with the fusion peptide (22, 25). However, the electron density maps clearly demonstrate that Arbidol binds further up the stem region, ∼16 Å from the fusion peptide, with three Arbidol sites per trimer (Fig. 2 B and C). The Arbidol-binding site is located at the interface between adjacent protomers of the HA trimer and is composed of A, C, and C′ α-helices from HA2 (“C′” refers to the adjacent protomer) and short C-terminal loops and an N-terminal β-hairpin from HA1 (Fig. 2 B–D). Protomer 1 contributes HA2 Arg54–Glu57 from helix-A, Lys58–Asn60 from loop-B, Trp92–Glu103 from helix-C, and HA1 Pro293, Phe294, and Arg307 from the C-terminal loops (Note: HA1 residues are written in italics throughout, and HA2 residues are written in regular font). Protomer 2 contributes Glu90′–Ala101′ from helix-C′, K310′ from a short loop close to the HA1 C terminus, and Leu29′ from a β-hairpin turn (Fig. 2D). The relative positions of helices A, C, and C′ on HA are shown in Figs. 1 and 2C. The binding site is disassembled in the postfusion form (Fig. 1B).
Fig. 2.
The Arbidol-binding site on influenza virus HA. (A) Molecular structure of Arbidol. (B) Crystal structure of Arbidol in complex with H7N9 A/Shanghai/2/2013 (H7/SH2) HA. The HA trimer is shown as a transparent gray surface, HA2 is shown in cyan, beige, and green secondary structure backbone traces, and Arbidol is shown as yellow sticks. One of the three N-terminal fusion peptides in HA2 in the trimer has been highlighted as a red ribbon. (C) Three identical Arbidol-binding sites are shown viewed along the threefold symmetry axis of the trimer. HA1 lateral short loops and the bottom β-hairpin turn are represented in the gray cartoons. (D) The Arbidol-binding site consists of helix-A and helix-C from protomer 1 (cyan) and helix-C′ from protomer 2 (beige). A 2Fo-Fc electron density map (black mesh) is contoured at 1σ around Arbidol.
Fig. S1.
Arbidol structure and its components. The structure of Arbidol and its component rings and side chains.
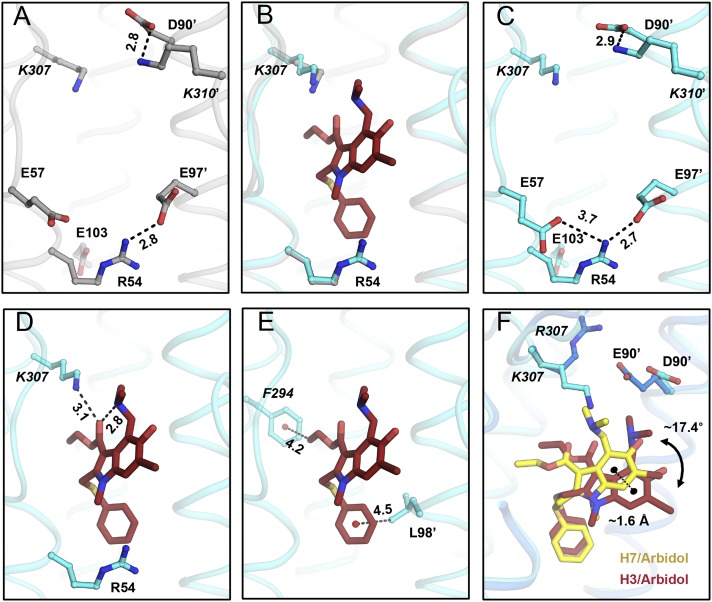
Fig. 4.
Molecular interactions in the Arbidol–H3/HK68 complex. (A) Charged residues in the Arbidol-binding site of the apo structure of H3N2 A/Hong Kong/1/1968 (H3/HK68; PDB ID code 4FNK) (gray sticks). (B) Superimposition of apo and Arbidol bound structures of H3/HK68. (C) Charged residues in the Arbidol-binding site of the Arbidol–H3/HK68 complex are shown as cyan sticks. (D and E) Noncovalent intramolecular interactions in the Arbidol–H3/HK68 complex. Arbidol is shown as reddish-brown sticks and HA residues are shown as cyan ball-and-sticks. Noncovalent intramolecular salt-bridge interactions in the Arbidol-binding site (A and C), inter- and intramolecular H-bonds made by Arbidol (D), and intramolecular CH–π bonds (E) are indicated by black dashed lines with distances in Ångstroms. (F) Superimposition of Arbidol-bound structures with H7/SH2 and H3/HK68 HAs. Arbidol bound to H7 is shown as yellow sticks, and Arbidol bound to H3 is shown as brown sticks. The distance between centroids of the two indole rings of Arbidol is ∼1.6 Å and is represented by a black dashed line; the torsional angle between the planes of the two different conformations of Arbidol is ∼17.4°.
Table 1.
Data collection and refinement statistics for Arbidol complexes with H3 and H7 HAs
| X-ray data | Arbidol–H3/HK68 | Arbidol–H7/SH2 |
| Data collection | ||
| Beamline | APS-23-IDD | SSRL-11-1 |
| Wavelength, Å | 1.0332 | 0.9794 |
| Space group | C2221 | P21 |
| Unit cell, Å, degrees | a = 105.9, b = 151.8, c = 349.5 | a = 67.4, b = 231.4, c = 127.8 |
| β = 96.8 | ||
| Resolution range, Å* | 50–2.54 (2.58–2.54) | 50–2.37 (2.41–2.37) |
| Observations | 298,979 | 569,897 |
| Unique reflections | 91,320 (4,562) | 154,529 (7,188) |
| Completeness, % | 98.3(98.9) | 97.6 (90.8) |
| I/σ(I) | 8.7 (1.7) | 13.7 (1.4) |
| Rsym† | 0.14 (0.69) | 0.09 (0.45) |
| Rpim‡ | 0.07 (0.37) | 0.04(0.25) |
| CC1/2§ | 0.89 (0.61) | 0.96 (0.88) |
| Redundancy | 3.3 (3.1) | 3.7 (2.8) |
| Refinement | ||
| Resolution, Å | 48.22–2.54 | 48.50–2.37 |
| No. reflections¶ | 91,300 (4464) | 154,344 (7624) |
| Rcryst#/Rfree|| | 0.18/0.22 | 0.19/0.24 |
| No. atoms | ||
| Protein | 11,855 | 22,827 |
| Ligand/carbohydrate | 87/416 | 174/461 |
| Water /ions | 617/20 | 988/1 |
| Wilson B, Å2 | 34 | 36 |
| Average B value, Å2 | ||
| Protein | 33 | 45 |
| Ligand | 53 | 62 |
| Water/ions | 35/68 | 42/56 |
| Rmsd from ideal geometry | ||
| Bond length, Å | 0.014 | 0.007 |
| Bond angle, degrees | 1.2 | 0.9 |
| Ramachandran statistics, %** | ||
| Favored | 95.6 | 97.0 |
| Outliers | 0.4 | 0.0 |
Numbers in parentheses refer to outer shell statistics.
Rsym = Σhkl Σi | Ihkl,i − <Ihkl> |/Σhkl Σi Ihkl,i, where Ihkl,i is the scaled intensity of the ith measurement of reflection h, k, l, and < Ihkl> is the average intensity for that reflection.
Rpim = Σhkl [1/(n − 1)]1/2 Σi | Ihkl,i − <Ihkl> |/Σhkl Σi Ihkl,i, where n is the redundancy.
CC1/2 = Pearson correlation coefficient between two random half datasets
Value in parentheses refer to number of reflections in test set.
Rcryst = Σhkl | Fo - Fc |/Σhkl | Fo | × 100, where Fo and Fc are the observed and calculated structures factors.
Rfree was calculated as for Rcryst but on a test set of 5% of the data excluded from refinement.
Calculated using MolProbity.
Fig. S2.
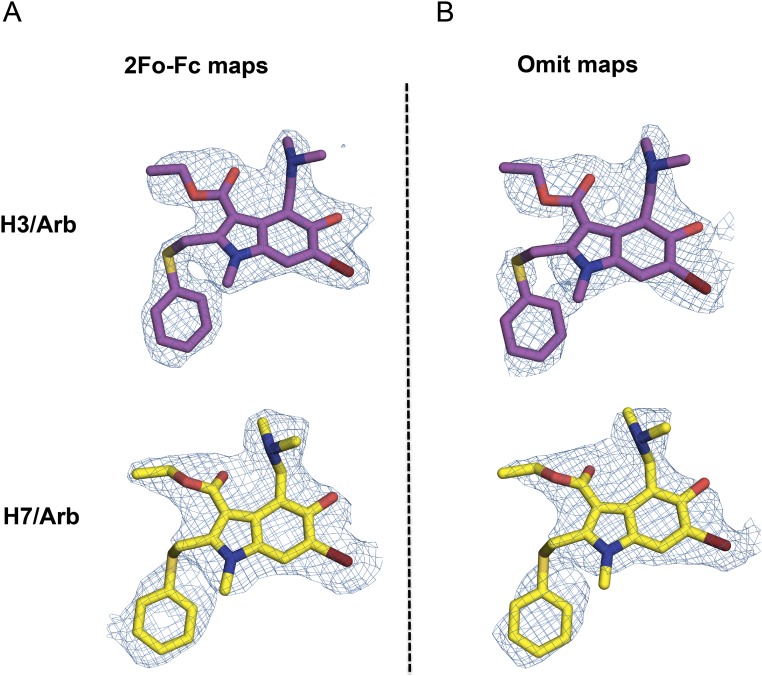
Electron density maps for the crystal structures of Arbidol in complex with H3 and H7 HAs. (A and B) 2Fo-Fc (A) and simulated annealing omit (B) maps, contoured at 1σ. For Arbidol in complex with H3/HK68, C, O, N, and Br are represented as magenta, red, blue, and brown sticks, respectively. For Arbidol in complex with H7/SH2, C, O, N, and Br are represented as yellow, red, blue, and brown sticks, respectively. Electron density maps are represented as a blue mesh.
Induced Fit in the Arbidol–H7/SH2 Complex.
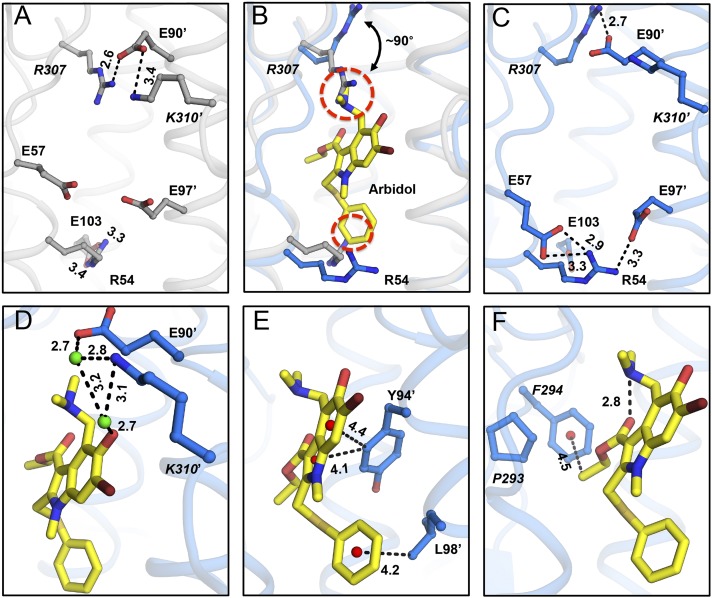
In the apo structure of H7/SH2 HA [Protein Data Bank (PDB) ID code 4LN6], the Arbidol-binding site contains four internal salt bridges formed by residues from adjacent protomers. The Glu90′ carboxyl from helix-C′ is involved in a salt-bridge network with Arg307 (protomer 1) and Lys310′ (protomer 2) from the short C-terminal loops of HA1. Furthermore, the Glu103 carboxyl from helix-C interacts with Arg54 from helix-A (Fig. 3A) within protomer 1. In the H7/SH2 HA apo conformation, Arbidol may experience some steric hindrance from Arg307 and Arg54 (Fig. 3B). However, upon binding of Arbidol, large conformational changes in these charged residues are observed in the binding site, likely because of induced fit. Some existing salt bridges are broken, and new ones are formed (Fig. 3 A–C). Arg307 rotates by ∼90°, and Glu90′ reorients to maintain the Glu90′–Arg307 interaction, whereas the Glu90′–Lys310′ interaction is broken. Similarly, the Glu103–Arg54 interaction is disrupted as Arg54 reorients to become more solvent-exposed, opening up the cavity so Arbidol can bind; Glu57 and Glu97′ reorient to form new salt bridges with Arg54 (Fig. 3 A–C).
Fig. 3.
Molecular interactions in the Arbidol–H7/SH2 complex. (A) Charged residues in the Arbidol-binding site of the apo structure of H7N9 A/Shanghai/2/2013 (H7/SH2; PDB ID code 4LN6) (gray sticks). (B) Superimposition of apo and Arbidol-bound structures of H7/SH2 HA. Arg54 and Arg307 in the apo conformation (gray sticks) would clash (red dashed circles) with Arbidol (yellow sticks), whereas in the Arbidol-bound conformation, Arg54 and Arg307 (blue sticks) move away from the binding pocket and form alternative intra- and interprotomer salt bridges. (C) Charged residues in the Arbidol-binding site of the Arbidol–H7/SH2 complex (blue sticks). (D–F) Noncovalent interactions in the Arbidol–H7/SH2 complex. Arbidol is represented by yellow sticks, HA residues as blue ball-and-stick models, centroids of rings as red spheres, and water molecules as green spheres. Noncovalent interaction mediated through salt-bridge networks (A and C), water molecules (D), and CH–π bonds (E and F) are highlighted using black dashed lines with distances in Ångstroms.
The interactions of Arbidol with HA in the Arbidol–H7/SH2 complex can be grouped into polar and nonpolar categories (Figs. 2D and 3). The central part of the binding pocket is hydrophobic with polar charged resides framing the top and bottom of the cavity. Because of the large number of nonpolar residues surrounding the binding cavity, Arbidol is positioned so that it does not form any direct H-bonds with binding-site residues, although water-mediated H-bonds with Glu90′ and Lys310′ are established (Fig. 3D). The conformation of Glu97′ is such that its aliphatic portion lines the binding site and packs across the face of the Arbidol thiophenyl ring (Fig. 2D), and the charged end is oriented away from the ring. The Arbidol thiophenol group occupies a cavity formed by hydrophobic residues Leu55, Leu99, Leu29′, Leu98′, and Ala101′ (Fig. 2D). The Cδ1 hydrogen (CH) from the aromatic ring of Tyr94′ of helix-C′ and the methyl of the ethylacetate moiety of Arbidol make CH–π interactions with the indole ring of Arbidol and Phe294, respectively (Fig. 3 E and F). Such unconventional H-bonds can contribute ∼1 Kcal/mol to the binding energy (26–28). Further, the carbonyl of the ethylacetate moiety forms an intramolecular H-bond with the Arbidol dimethylamine group that constrains the Arbidol conformation so that the ethylacetate moiety is directed toward the hydrophobic patch containing Pro293 and Phe294 and stabilizes the CH–π interactions (Fig. 3F).
Accommodation of Arbidol in the H3/HK68-Binding Pocket.
In the Arbidol-binding pocket in apo H3/HK68 HA, the carboxyl groups of Asp90′ and Glu97′ are involved in salt-bridge interactions with Lys310′ and Arg54, respectively (Fig. 4A). Unlike H7/SH2, these preexisting salt bridges do not pose a steric hindrance for binding Arbidol; therefore, they are maintained in the Arbidol–H3/HK68 complex (Fig. 4B). Additionally, upon Arbidol binding, a new salt bridge is formed between Glu57 and Arg54 (Fig. 4C). Overall, compared with H7/SH2, H3/HK68 accommodates Arbidol without undergoing any major conformational changes in the binding site.
Shape Complementarity in the Arbidol-Binding Site.
Arbidol has high shape complementarity with the binding sites in these two HAs. Although the Arbidol thiophenyl and indole rings are buried inside the cavity, the polar substituents, hydroxyl and bromine groups on the indole ring, are partially exposed to solvent. Overall Arbidol buries ∼565–580 Å2 of surface area on the HAs with high normalized shape complementarity of ∼0.73–0.80 (a value of 1 represents a perfect fit) (29).
Stabilization of the Prefusion Conformation of HA.
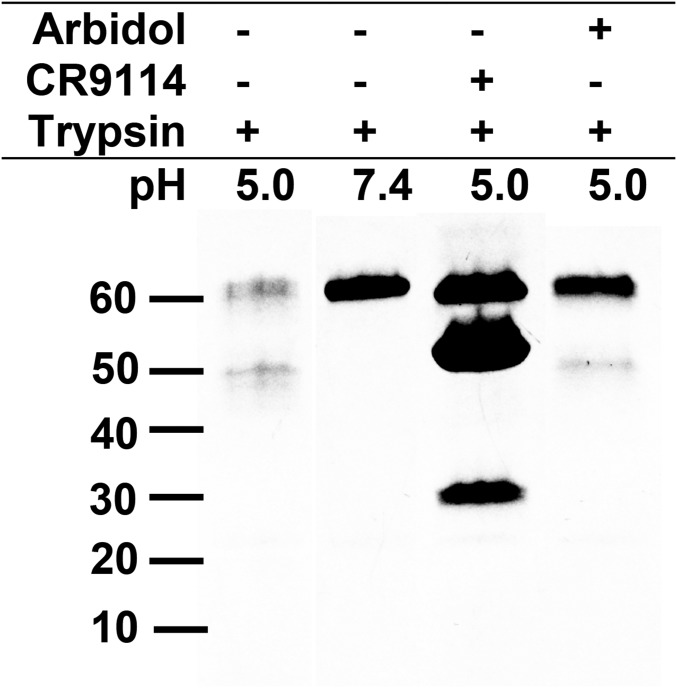
Acid-induced conformational changes in HA at low pH render HA susceptible to proteases and lead to the postfusion conformation (Fig. 1B). To assay the ability of Arbidol to prevent low pH-induced conformational changes in HA, we performed a trypsin-susceptibility assay (30). In the trypsin-susceptibility assay, group 1 H1 A/Puerto Rico/8/1934 (PR8) HA is readily converted to its protease-susceptible postfusion form at low pH (5.0) but not at neutral pH (7.4) (Fig. 5, lanes 1 and 2). Arbidol inhibits the conversion of PR8 HA to the postfusion conformation at low pH (5.0), thereby preventing its digestion by trypsin (Fig. 5, lane 4). This result is consistent with observations for the fusion-inhibiting stem-targeting antibody CR9114 Fab (Fig. 5, lane 3) (31).
Fig. 5.
Trypsin-susceptibility assay of Arbidol. SDS/PAGE analysis of the trypsin-susceptibility assay of Arbidol. Lane 1, acidified PR8 H1 HA treated with trypsin. Lane 2, PR8 HA treated with trypsin without prior acidification. Lane 3, acidified PR8-CR9114 Fab complex treated with trypsin (additional bands correspond to the Fab (∼50 kDa) and the heavy and light chains of the Fab (∼30 kDa). Lane 4, acidified PR8–Arbidol complex treated with trypsin.
Discussion
Mechanism of Arbidol-Binding and -Resistance Mutations.
Through structure-based characterization of Arbidol–HA complexes, we have shown that Arbidol binds in a hydrophobic cavity at the interface between two protomers within the HA trimer, with three identical binding sites being related by the threefold symmetry axis of the HA trimer. The binding site in group 2 HAs features an inner hydrophobic core and a solvent-exposed, highly charged periphery; residues in the binding site are contributed from α-helices A and C in one protomer and from helix-C′ from a second protomer. In H7/SH2, where steric clashes are likely to arise with the HA apo conformation, Arbidol appears to use an induced fit mechanism involving major and minor conformational changes in the binding pocket that results in the breaking of existing salt bridges and the formation of new ones. Because the apo conformation of the binding pocket of H3/HK68 HA does not pose a steric hindrance to Arbidol, significant conformational changes or salt-bridge alterations are not observed during the formation of the Arbidol–H3/HK68 complex. Because Arbidol interacts with two protomers in the trimer and establishes new interprotomer interactions (Figs. 3 and 4), it can serve to clamp the protomers together, thereby stabilizing the trimer and rendering it resistant to the low pH-mediated conformational changes that occur during membrane fusion.
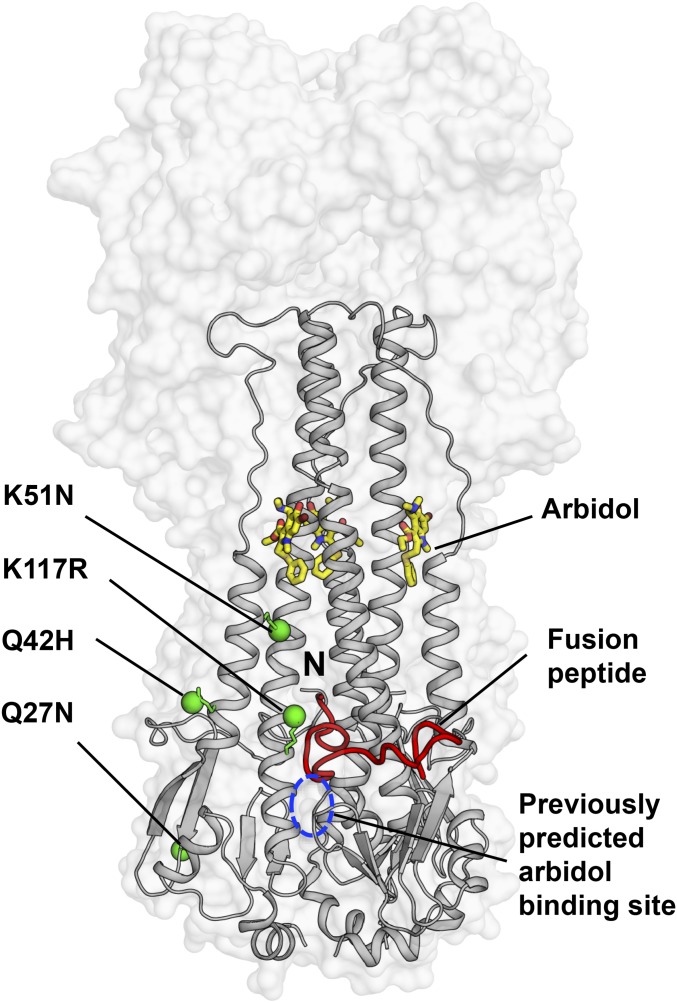
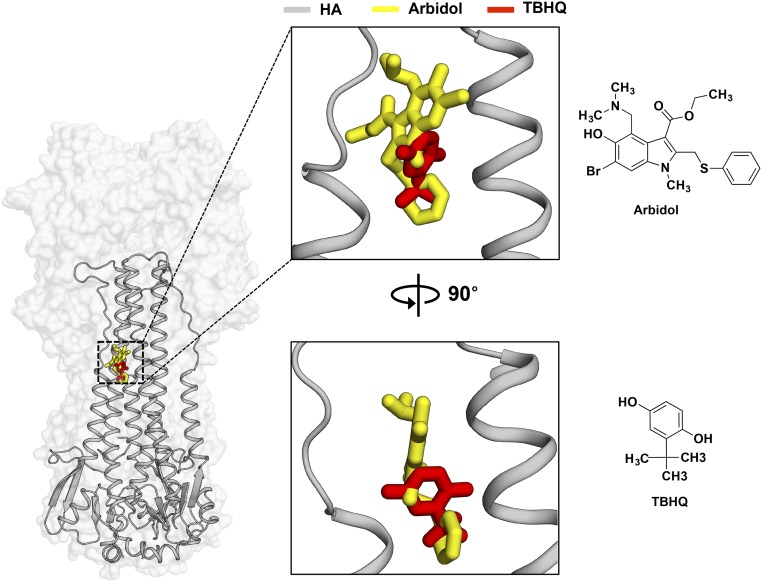
A prior study to investigate the mechanism of action of Arbidol using mass spectrometry and in silico docking had predicted a pocket close to the fusion peptide as the potential Arbidol-binding site (25). Similarly, characterization of resistance to Arbidol in influenza virus identified four single point mutations, K51N, K117R, Q27N, and Q42H, in the HA2 subunit of H7N7 A/chicken/Germany/27 HA (22). This study also proposed a model for Arbidol docking in a cavity close to the fusion peptide. However, because the resistance mutations did not map close to their predicted site, the authors refrained from deriving a mechanism of action for Arbidol. It is noteworthy that these resistance mutations also do not map on the actual Arbidol-binding site identified here in H3/HK68 and H7/SH2 HAs; only the K51N mutation is in the relative vicinity of the Arbidol-binding site (Fig. 6). Another study that used fluorescence quenching and thermal stability assays suggested the proximity of W92 and R54 residues to the Arbidol-binding site (23). Because these residues were earlier shown to be proximal to the binding site of another HA fusion inhibitor, tertiary butylhydroquinone (TBHQ) (32, 33), the authors predicted that Arbidol may bind to a similar site (23). Curiously, resistance mutations against TBHQ, which indeed binds to a portion of the Arbidol site (Fig. S3) and demonstrates limited interactions with group 2 HAs, do not correlate with any direct interactions in the smaller TBHQ-binding pocket (32, 33). This discrepancy may result from the complexity of the structural rearrangements induced in the HA by low pH. Therefore, additional studies on escape mutants, conducted in the light of the structures solved here, will be helpful in further understanding the mechanisms of Arbidol function and resistance.
Fig. 6.
Mapping of the Arbidol-resistance mutations on HA. The crystal structure of Arbidol in complex with H7N9 A/Shanghai/2/2013 (H7/SH2) HA is represented with the HA shown as a transparent gray surface. The HA2 is also shown in gray secondary structure backbone traces, and Arbidol is shown as yellow sticks. One of the three N-terminal fusion peptides in the trimer has been highlighted as a red ribbon, and its N terminus is labeled “N.” Previously reported Arbidol-resistance mutations K51N, K117R, Q42H, and Q27N in HA2 are shown as green spheres, and the previously predicted binding site for Arbidol from docking studies is marked by a blue dashed circle.
Fig. S3.
Binding modes of Arbidol and TBHQ on HA. (Left) Crystal structure of Arbidol in complex with H7N9 A/Shanghai/2/2013 (H7/SH2). The HA is shown as a transparent gray surface, HA2 is shown as gray secondary structure backbone traces, and Arbidol is shown as yellow sticks. One of the three binding sites of Arbidol is illustrated. TBHQ in complex with H14N5 A/mallard/Astrakhan/263/1982 (PDB ID code 3EYK) is superimposed on the H7/SH2 HA and is shown as red sticks. (Center) The same complex rotated by 90°. (Right) The chemical structures of Arbidol (Upper) and TBHQ (Lower).
Conformational Flexibility of Arbidol in Group 2 HAs.
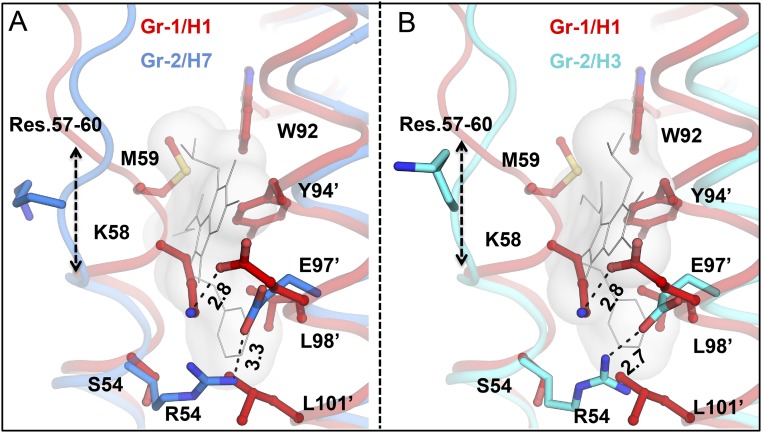
Although influenza H7/SH2 and H3/HK68 HAs belong to the same phylogenetic group, i.e., group 2 HAs, Arbidol adopts slightly different conformations in the binding sites of the two proteins. A comparison of the Arbidol-binding sites in the apo structures of H7/SH2 (PDB ID code 4LN6) and H3/HK68 (PDB ID code 4FNK) HAs reveals two differences: E90D′ and R307K in helix-C′ of protomer 2 and 1, respectively. These seemingly conservative substitutions have a significant effect on the Arbidol interaction with the HA. In apo H7/SH2 HA, Arg307 interacts with Glu90′ and provides potential steric hindrance to Arbidol binding, whereas in the Arbidol–H7/SH2 complex Arg307 flips by ∼90° so that its salt bridge with Glu90′ is retained but Arbidol binding is not hindered. If Arbidol were to adopt an identical binding mode in H3/HK68 HA, steric hindrance from Lys307 would be expected. However, Arbidol demonstrates conformational flexibility to accommodate the apo conformation of Lys307, whereas the thiophenol group binds identically to the two HAs. However, the indole ring exhibits a displacement of ∼1.6 Å and a dihedral angle shift of ∼17.4° between the two bound conformations (Fig. 4F). Notably, by displacing the indole ring, Arbidol engages Lys307 in an H-bond interaction (Fig. 4D), whereas Arg307 does not make any direct contacts with Arbidol (Fig. 3B).
Group-Specific Binding of Arbidol.
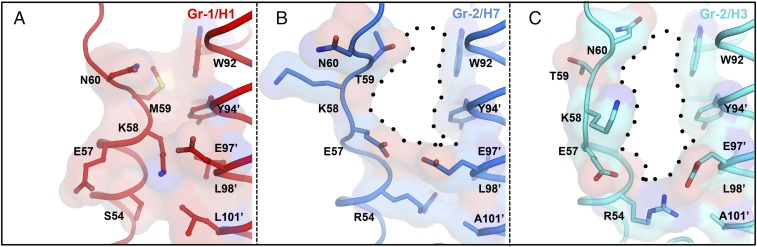
Previous in vitro and in vivo studies have shown that Arbidol has broad-spectrum activity against influenza viruses (16–18, 21). Comparison of the Arbidol-binding site in group 2 HAs against the corresponding region in group 1 HA [H1N1 A/Puerto Rico/8/1934 (H1/PR8; PDB ID code 1RU7)] reveals important structural differences between the two groups (Fig. 7 and Fig. S4). The volume of the Arbidol-binding pocket of group 2 HAs, H7/SH2 and H3/HK68, is ∼870–1,150 Å3, whereas the corresponding pocket in group 1 HAs is 10-fold smaller (∼90–150 Å3). In group 1, the Arbidol-binding site is primarily blocked by an extra turn of the short α-helix (HA2 residues 57–60). This helix is further stabilized by an interprotomer salt bridge between Lys58 and Glu97′ that causes the side chains of these two residues also to occupy the binding site. In group 2 HAs, Lys58 is disordered, and Glu97′ interacts with Arg54 and projects away from the Arbidol-binding pocket. Additionally, in group 1 HAs, the side chains of Met59 and Leu101′ also project inwards to block the Arbidol pocket. The corresponding Thr59 and Ala101′, respectively, in group 2 HAs have shorter side chains (Fig. 7). Other residues that, although conserved in the two groups, obstruct the Arbidol pocket in group 1 HAs are Trp92, Tyr94′, and Leu98′; these residues interact with Arbidol in group 2 HAs (Figs. 2D and 3). Therefore the structural analysis described here suggests either that large conformational rearrangements would have to occur in group 1 HAs in the site identified in group 2 HAs on Arbidol binding or that Arbidol occupies a distinct group-specific binding site in group 1 HAs. It similarly was noted that the binding site of TBHQ would be much more restricted in group 1 HAs than in group 2 HAs (33). Furthermore, in a previous study on Arbidol derivatives, one of the more potent inhibitors exhibited higher affinity to group 2 than to group 1 influenza A viruses, but the increases in affinity were not reflected in antiviral effects (23).
Fig. 7.
Group-specific binding mode of Arbidol. (A–C) Comparison of the location of the Arbidol-binding site in the apo structures of group 1 H1 A/Puerto Rico/8/1934 HA (PDB ID code 1RU7; red) (A) and in group 2 H7N9 A/Shanghai/2/2013 (H7/SH2; PDB ID code 4LN6; blue) (B) and H3N2 A/Hong Kong/1/1968 (H3/HK68; PDB ID code 4FNK; cyan) (C) HAs. The extra helical turn that blocks the Arbidol-binding pocket in group 1 H1 HAs is formed by residues 57–60, whereas the Arbidol-binding site (indicated by black dots) is accessible in group 2 HAs.
Fig. S4.
Group-specific binding of Arbidol. (A and B) Superimposition of group 1 H1 A/Puerto Rico/8/1934 (PR8) HA (PDB 1RU7; red) onto group 2 Arbidol-bound structures of H7N9 A/Shanghai/2/2013 (H7/SH2; blue) (A) and H3N2 A/Hong Kong/1/1968 (H3/HK68; cyan) (B) HAs. Arbidol is shown as a gray molecular surface. The extra helical turn that blocks the Arbidol-binding pocket in group 1 H1 is formed by HA2 residues 57–60. In H1 HA, Lys58 forms a salt bridge with Glu97′, further blocking the Arbidol-binding site, unlike the Glu97′–Arg54 salt bridge, which is more accommodating for Arbidol binding, in group 2 HAs. Hydrophobic residues from H1/PR8 HA that clash with the conformation of Arbidol in group 2 HAs are shown as red sticks.
Distinct Binding Modes of Fusion Inhibitors in Class I Fusion Proteins.
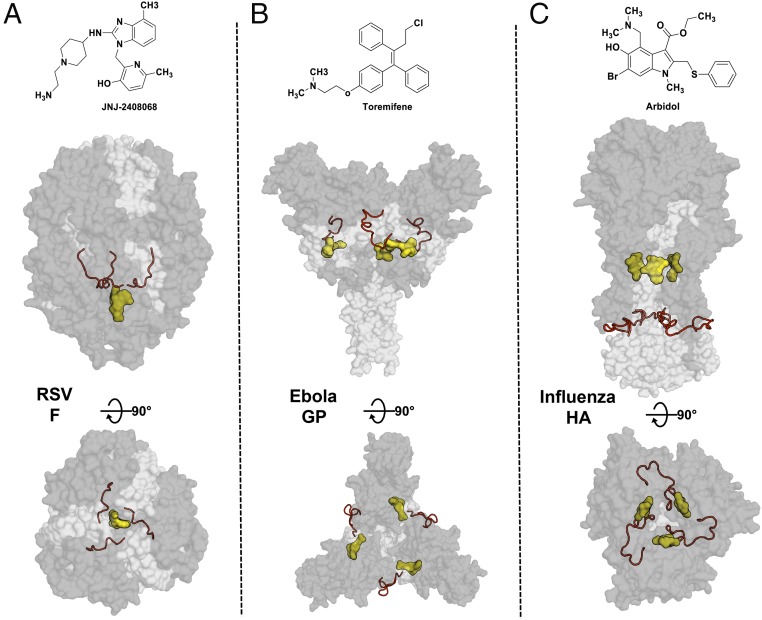
These Arbidol–HA structures can be compared with the structures of complexes of small-molecule fusion inhibitors against other class I fusion glycoproteins, such as the RSV fusion protein (RSV F) and EBOV glycoprotein (EBOV GP) (Fig. 8). Inhibitors of RSV F GP bind to a hydrophobic pocket around the threefold symmetry axis within the central cavity of the metastable prefusion structure. The pocket is adjacent to the fusion peptide and permits a stoichiometry of only one inhibitor per trimer of RSV F (Fig. 8A) (34). In EBOV GP, inhibitors bind with a stoichiometry of three ligands per trimer in a hydrophobic cavity between the attachment (GP1) and fusion (GP2) subunits at the base of the fusion peptide (Fig. 8B) (35). Unlike these inhibitors of other class I fusion proteins, Arbidol binds at a unique location, ∼16Å from the fusion peptide, with a stoichiometry of three ligands per HA trimer (Fig. 8C). Although these class I fusion glycoproteins are related to each other in their fusion mechanism, their inhibitor-binding sites are in completely different locations relative to the threefold symmetry axis and to the fusion peptide and with respect to the vertical location on the stem. However, they are all bound in hydrophobic pockets surrounded by surface-exposed charged residues.
Fig. 8.
Location of small-molecule inhibitors in class I fusion proteins. Prefusion structures of RSV F (PDB ID code 5EA3) (A), Ebola GP (PDB ID code 5JQ7) (B), and HA (C) are shown as transparent molecular surfaces with the small molecules JNJ2408068, toremifene, and Arbidol, respectively. The subunits in the glycoproteins RSV F (F1 and F2), Ebola GP (GP1 and GP2), and Influenza HA (HA1 and HA2) are represented by dark and light gray surfaces, respectively. Small molecules are shown as a yellow molecular surface, and fusion peptide is shown as a red ribbon. The top panels show the chemical structures of JNJ2408068, toremifene, and Arbidol, respectively. The middle and bottom panels represent the same complexes, rotated by 90° with a side view (middle panels) and a top view (bottom panels).
Implication of the Arbidol-Binding Site for Therapeutic Design.
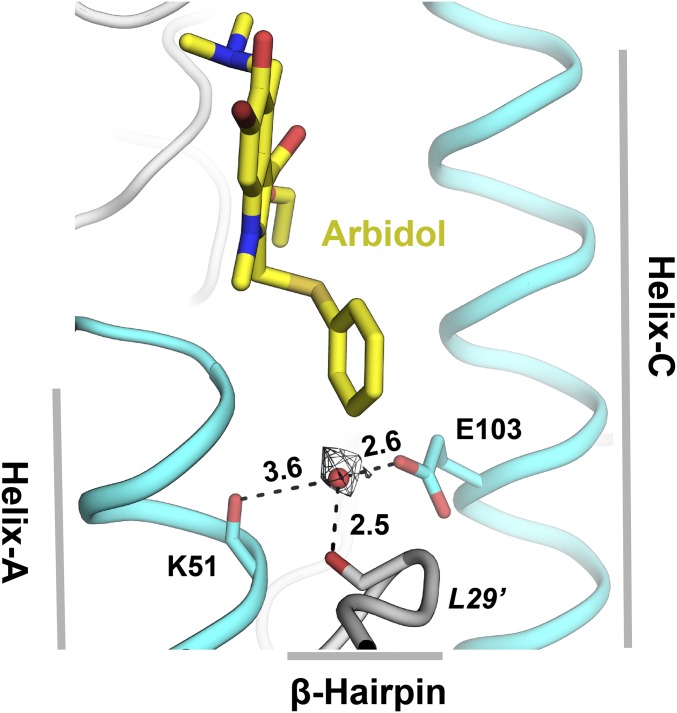
The Arbidol-binding sites in H7/SH2 and H3/HK68 HAs reveal many interesting features that can be further explored to develop optimized influenza therapeutics by a structure-based design. For example, indole ring substituents may be modified to improve hydrophobic interactions and introduce additional polar interactions. The thiophenol group is buried in a hydrophobic cavity containing an ordered water molecule, which mediates H-bonds between Glu103 and backbone carbonyls of Lys51 and Leu29′. Polar substituents at the meta position of the thiophenol ring could sustain the H-bond network by replacing the water molecule, thus allowing entropic gain (Fig. 2D and Fig. S5). Similarly, the indole ring substituents, the hydroxyl, dimethylamine, and ethylacetate groups, which are in the vicinity of Lys310′, Glu90′, Pro293, and Phe294, respectively (Fig. 2D), can be modified to improve Arbidol–HA interactions. The water-mediated H-bond between the hydroxyl and Lys310′ (Fig. 3D) may be improved further with a bulkier polar substituent that can displace the water molecule to gain binding entropy. Replacing the dimethylamine with a more charged moiety may result in the formation of a salt bridge with Glu90′. The hydrolysis-prone ethylacetate moiety may be replaced with a rigid hydrophobic group to improve molecular stability and gain nonpolar contacts with Pro293 and Phe294. Indeed, such a substitution might improve the bioavailability profile of Arbidol and result in a smaller therapeutic dosage. Overall Arbidol represents a remarkable scaffold for further optimization to improve its specificity, efficacy, and applicability as a specific influenza drug for therapeutic use.
Fig. S5.
Ordered water molecule in the Arbidol-binding site in H7/SH2 HA. The ordered water molecule (shown as a red sphere) mediates interactions (indicated by black dashed lines) between HA2 Lys51 of helix-A, Glu103 of helix-C (cyan), and the β-hairpin Leu29′ carbonyl (gray). Arbidol is shown as yellow sticks. Distances are in Ångstroms.
The structures of Arbidol–HA complexes solved here identify a mechanism of influenza virus fusion inhibition by small molecules that is distinct from the HA-targeting antibodies and from other class I fusion protein inhibitors. Arbidol occupies a discrete binding pocket similar to the pocket partially occupied by the much smaller, fragment-like molecule TBHQ and functions as a molecular glue to stabilize the HA trimer. By revealing the binding mode of Arbidol and its key interactions with HAs, these structures provide useful guidelines for structure–activity relationship studies to improve the efficacy of Arbidol. Overall, this work should accelerate the development of optimal inhibitors of the influenza virus fusion process.
Materials and Methods
Expression and Purification of HA.
The HA ectodomain was expressed using our baculovirus expression system as described previously (36, 37). Briefly, each HA was fused with a gp67 signal peptide at the N terminus and to a thrombin cleavage site, foldon trimerization domain, and His6-tag at the C terminus. Expressed HAs were purified using metal affinity chromatography using Ni-NTA resin. For crystallization studies, the HAs were digested with 5 mU trypsin (New England Biolabs) per milligram of HA for 16 h at 4 °C to produce uniformly cleaved HA1/HA2 subunits and to remove the trimerization domain and the His6-tag. The digested material was purified by gel filtration.
Crystallization and Structure Determination of Arbidol-H7/SH2 and H3/HK68 HA Complexes.
Gel filtration fractions containing the H7/SH2 and H3/HK68 HAs were concentrated to ∼10 mg/mL in 10 mM Tris (pH 8.0) and 100 mM NaCl. Crystallization screens were set up using sitting drop-vapor diffusion using previously reported crystallization conditions for both HAs: H7/SH2 [17–20% (wt/vol) PEG3350, 0.2 M ammonium acetate, pH 8.0, 20 °C] (38) and H3/HK68 (2.0 M ammonium sulfate, 100 mM sodium cacodylate, 200 mM sodium chloride, pH 6.5, 20 °C) (37). Within 3–7 d, diffraction-quality crystals were obtained. The resulting crystals were soaked with a final concentration of ∼2.5 mM Arbidol for 10 min, cryoprotected with 15% (vol/vol) glycerol, and then flash-cooled and stored in liquid nitrogen until data collection. Diffraction data were collected at 100 K on the General Medicine and Cancer Institutes Collaborative Access Team (GM/CA CAT) 23ID-D beamline at the Advanced Photon Source at the Argonne National Laboratory and at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 11-1. The diffraction data were processed with HKL-2000 (39). Initial phases were determined by molecular replacement using Phaser (40) with HA models from H7/SH2 (PDB ID code 4LN6) and H3/HK68 (PDB ID code 4FNK). Refinement was carried out in Phenix (41), alternating with manual rebuilding and adjustment in COOT (42). Detailed data collection and refinement statistics are summarized in Table 1.
Trypsin-Susceptibility Assay.
In the proteolysis experiment, ∼5 μM PR8 H1 HA was preincubated separately with ∼400 μM of Arbidol and ∼10 μM CR9114 Fab for 30 min at room temperature. Control reactions were incubated with 2% (vol/vol) DMSO. The pH of each reaction was lowered using 1-M sodium acetate buffer (pH 5.0). One reaction was retained at pH 7.4 to assess digestion at neutral pH. The reaction solutions then were mixed thoroughly and incubated for 20 min at 37 °C. After incubation, the reaction solutions were equilibrated at room temperature, and the pH was neutralized by the addition of 200 mM Tris buffer (pH 8.5). Trypsin-ultra (New England Biolabs, Inc.) was added to all samples at a final ratio of 1:50 by mass, and the samples were digested for 30 min at 37 °C. After incubation with trypsin, the reaction solutions were equilibrated at room temperature, quenched by the addition of nonreducing SDS buffer, and boiled for ∼2 min at 100 °C. All samples were analyzed by 4–20% SDS/PAGE gel and imaged using the Bio-Rad ChemiDoc imaging system.
Structural Analyses.
Surface areas buried on the HA upon binding of Arbidol were calculated with the Protein Interfaces, Surfaces and Assemblies (PISA) server at the European Bioinformatics Institute (43). MacPyMOL (DeLano Scientific) was used to render structure figures. The final coordinates were validated using MolProbity (44). The Arbidol-binding pocket volume was calculated using the CASTp program (45).
Acknowledgments
We thank J. P. Verenini for help in manuscript formatting and R. Stanfield, X. Zhu, N. Tzarum, and X. Dai for useful discussions on refinement. X-ray datasets were collected at the Advanced Photon Source, Argonne National Laboratory (beamline 23ID-D) and the Stanford Radiation Light Source (SSRL) (beamline 11-1). The GM/CA CAT is funded in whole or in part by federal funds from National Cancer Institute Grant Y1-CO-1020 and National Institute of General Medical Science (NIGMS) Grant Y1-GM-1104. Use of the Advanced Photon Source is supported by the US Department of Energy (DOE), Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. Use of the SSRL at the Stanford Linear Accelerator Center National Accelerator Laboratory is supported by the US DOE, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIGMS (including Grant P41GM103393). This work is supported by NIH Grant R56 AI117675 (to I.A.W.). R.U.K. is the recipient of an Early Mobility Postdoctoral Fellowship from the Swiss National Science Foundation. This is manuscript 29413 from The Scripps Research Institute. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or the NIH.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5T6N and 5T6S).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617020114/-/DCSupplemental.
References
- 1.Beveridge WI. The chronicle of influenza epidemics. Hist Philos Life Sci. 1991;13(2):223–234. [PubMed] [Google Scholar]
- 2.Nguyen-Van-Tam JS, Hampson AW. The epidemiology and clinical impact of pandemic influenza. Vaccine. 2003;21(16):1762–1768. doi: 10.1016/s0264-410x(03)00069-0. [DOI] [PubMed] [Google Scholar]
- 3.Salzberg S. The contents of the syringe. Nature. 2008;454(7201):160–161. doi: 10.1038/454160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davlin SL, et al. Influenza activity - United States, 2015-16 season and composition of the 2016-17 influenza vaccine. MMWR Morb Mortal Wkly Rep. 2016;65(22):567–575. doi: 10.15585/mmwr.mm6522a3. [DOI] [PubMed] [Google Scholar]
- 5.Hensley SE. Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr Opin Virol. 2014;8:85–89. doi: 10.1016/j.coviro.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houser K, Subbarao K. Influenza vaccines: Challenges and solutions. Cell Host Microbe. 2015;17(3):295–300. doi: 10.1016/j.chom.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Clercq E. Antiviral agents active against influenza A viruses. Nat Rev Drug Discov. 2006;5(12):1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beigel J, Bray M. Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res. 2008;78(1):91–102. doi: 10.1016/j.antiviral.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA. 2006;295(8):891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 10.Moscona A. Global transmission of oseltamivir-resistant influenza. N Engl J Med. 2009;360(10):953–956. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- 11.Sheu TG, et al. Dual resistance to adamantanes and oseltamivir among seasonal influenza A(H1N1) viruses: 2008-2010. J Infect Dis. 2011;203(1):13–17. doi: 10.1093/infdis/jiq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 13.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 14.Boriskin YS, Leneva IA, Pécheur EI, Polyak SJ. Arbidol: A broad-spectrum antiviral compound that blocks viral fusion. Curr Med Chem. 2008;15(10):997–1005. doi: 10.2174/092986708784049658. [DOI] [PubMed] [Google Scholar]
- 15.Leneva IA, Fediakina IT, Gus’kova TA, Glushkov RG. [Sensitivity of various influenza virus strains to Arbidol. Influence of Arbidol combination with different antiviral drugs on reproduction of influenza virus A] Ter Arkh. 2005;77(8):84–88. Russian. [PubMed] [Google Scholar]
- 16.Leneva IA, et al. Virus susceptibility and clinical effectiveness of anti-influenza drugs during the 2010-2011 influenza season in Russia. Int J Infect Dis. 2016;43:77–84. doi: 10.1016/j.ijid.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Brooks MJ, et al. Antiviral activity of Arbidol, a broad-spectrum drug for use against respiratory viruses, varies according to test conditions. J Med Virol. 2012;84(1):170–181. doi: 10.1002/jmv.22234. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, et al. Antiviral activity of Arbidol against influenza A virus, respiratory syncytial virus, rhinovirus, coxsackie virus and adenovirus in vitro and in vivo. Arch Virol. 2007;152(8):1447–1455. doi: 10.1007/s00705-007-0974-5. [DOI] [PubMed] [Google Scholar]
- 19.Pécheur EI, et al. The synthetic antiviral drug Arbidol inhibits globally prevalent pathogenic viruses. J Virol. 2016;90(6):3086–3092. doi: 10.1128/JVI.02077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaising J, Polyak SJ, Pécheur EI. Arbidol as a broad-spectrum antiviral: An update. Antiviral Res. 2014;107:84–94. doi: 10.1016/j.antiviral.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, et al. Antiviral and anti-inflammatory activity of Arbidol hydrochloride in influenza A (H1N1) virus infection. Acta Pharmacol Sin. 2013;34(8):1075–1083. doi: 10.1038/aps.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leneva IA, Russell RJ, Boriskin YS, Hay AJ. Characteristics of Arbidol-resistant mutants of influenza virus: Implications for the mechanism of anti-influenza action of Arbidol. Antiviral Res. 2009;81(2):132–140. doi: 10.1016/j.antiviral.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Brancato V, et al. Design of inhibitors of influenza virus membrane fusion: Synthesis, structure-activity relationship and in vitro antiviral activity of a novel indole series. Antiviral Res. 2013;99(2):125–135. doi: 10.1016/j.antiviral.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Fadeeva NI, et al. Inhibitors of the early stages of virus-cell interactions among derivatives of 3-ethoxycarbonyl-5-hydroxy-6-bromoindole. Chem Pharma J. 1992;26(9):676–680. [Google Scholar]
- 25.Nasser ZH, Swaminathan K, Müller P, Downard KM. Inhibition of influenza hemagglutinin with the antiviral inhibitor Arbidol using a proteomics based approach and mass spectrometry. Antiviral Res. 2013;100(2):399–406. doi: 10.1016/j.antiviral.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 26.Nishio M, Hirota M, Umezawa Y. The CH/π Interaction: Evidence, Nature, and Consequences. Wiley-VCH; Weinheim: 1998. pp. 1–232. [Google Scholar]
- 27.Levitt M, Perutz MF. Aromatic rings act as hydrogen bond acceptors. J Mol Biol. 1988;201(4):751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- 28.Kadam RU, et al. CH-π “T-shape” interaction with histidine explains binding of aromatic galactosides to Pseudomonas aeruginosa lectin LecA. ACS Chem Biol. 2013;8(9):1925–1930. doi: 10.1021/cb400303w. [DOI] [PubMed] [Google Scholar]
- 29.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15(4):327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 30.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337(6100):1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman LR, Kuntz ID, White JM. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: Irreversible inhibition of infectivity. J Virol. 1997;71(11):8808–8820. doi: 10.1128/jvi.71.11.8808-8820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell RJ, et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci USA. 2008;105(46):17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Battles MB, et al. Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat Chem Biol. 2016;12(2):87–93. doi: 10.1038/nchembio.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, et al. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535(7610):169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens J, et al. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303(5665):1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 37.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489(7417):526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu R, et al. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science. 2013;342(6163):1230–1235. doi: 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dundas J, et al. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(Web Server issue):W116-8. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]